Abstract
We assessed several methods of 13C metabolic flux analysis (MFA) and found that isotopically nonstationary MFA achieved maximum flux resolution in cultured P493-6 B-cells, which have been engineered to provide tunable expression of the Myc oncoprotein. Comparison of metabolic flux maps obtained under oncogenic (High) and endogenous (Low) Myc expression levels revealed network-wide reprogramming in response to ectopic Myc expression. High Myc cells relied more heavily on mitochondrial oxidative metabolism than Low Myc cells and globally upregulated their consumption of amino acids relative to glucose. TCA cycle and amphibolic mitochondrial pathways exhibited 2- to 4-fold flux increases in High Myc cells, in contrast to modest increases in glucose uptake and lactate excretion. Because our MFA approach relied exclusively upon isotopic measurements of protein-bound amino acids and RNA-bound ribose, it is readily applicable to more complex tumor models that are not amenable to direct extraction and isotopic analysis of free intracellular metabolites.
Keywords: metabolic flux analysis, isotopomer analysis, cancer metabolism, Warburg effect, mass spectrometry, lymphoma
1. INTRODUCTION
The reprogramming of energy metabolism is emerging as an important molecular hallmark of cancer cells (Hanahan and Weinberg, 2011). In particular, understanding the so-called Warburg effect, described as the avid conversion of glucose to lactate by tumor cells under aerobic conditions, has become a high priority in cancer research (Hsu and Sabatini, 2008; Koppenol et al., 2011; Vander Heiden et al., 2009). Recent discoveries linking specific metabolic alterations to cancer development have strengthened the idea that deregulated metabolism is more than a side effect of malignant transformation, but may in fact be a functional driver of tumor growth and progression in some cancers (Dang et al., 2009; DeBerardinis et al., 2008; Vander Heiden et al., 2010). Furthermore, prior studies have demonstrated that the Warburg effect can be reversed by either inhibiting lactate production (Fantin et al., 2006; Le et al., 2010) or altering the expression of specific glycolytic enzymes (Christofk et al., 2008), which correlates with a reduction in the ability of isogenic cancer cells to form tumors in nude mouse xenografts. Because of these and other discoveries, deregulated metabolic pathways have become attractive targets for cancer therapeutics (Evans et al., 2005; Kroemer and Pouyssegur, 2008; Michelakis et al., 2008).
To guide the search for new therapeutic targets and to better understand the mechanisms of metabolic reprogramming in tumor cells, integrative approaches are needed to fully characterize the metabolic phenotypes of cancer cells and to determine how they are influenced by specific molecular alterations. In particular, the ability to map intracellular carbon flows using 13C metabolic flux analysis (MFA) provides an attractive platform to elucidate the functional behavior of entire biochemical networks, rather than individual reactions or nodes in isolation (Sauer, 2006). By feeding cells a 13C-labeled substrate and subsequently measuring the patterns of isotope incorporation in downstream metabolic products, extensive information about the intracellular distribution of carbon flux can be obtained. This enables system-wide quantification of reversible, parallel, and cyclic metabolic pathways that would be otherwise unidentifiable based solely upon measurements of extracellular nutrient uptake and product excretion (Zamboni et al., 2009).
While 13C MFA provides a rich source of phenotypic information, the application of this technique to mammalian systems presents unique challenges. In particular, the presence of subcellular compartmentation, complex media formulations, and slow labeling dynamics can lead to significant difficulties in experimental design and data interpretation (Zamboni, 2011). As a result, most mammalian MFA studies have relied on direct extraction and isotopomer analysis of free intracellular metabolites, rather than more slowly labeled—but highly abundant and stable—macromolecular species (Zamboni et al., 2009). Isotopically non-stationary MFA (INST-MFA) provides one approach to circumvent these limitations through computational analysis of metabolite labeling patterns obtained during the transient labeling period prior to isotopic steady state (Wiechert and Noh, 2005). This approach offers several advantages over steady-state MFA, including shorter experimental times and the ability to determine fluxes with increased precision (Nöh and Wiechert, 2011; Young and Walther, 2008).
In this contribution, we investigated the metabolic alterations caused by differential expression of the MYC oncogene in a human B-cell line. MYC encodes the transcription factor c-Myc (herein termed Myc), which is a global regulator of cell growth, metabolism, and apoptosis (Dang, 1999). Myc exhibits deregulated expression in approximately 30% of human cancers (Dang et al., 2008) and is one of four transcription factors that collectively can reprogram differentiated adult cells back to a pluripotent stem cell state (Takahashi and Yamanaka, 2006). Although a few prior studies have applied isotopomer analysis to investigate the metabolic fates of 13C-labeled glucose and glutamine tracers in Myc-expressing cells, these approaches were not capable of integrating numerous isotopic measurements into a comprehensive flux map that encompasses all major pathways of central carbon metabolism (Le et al., 2012; Morrish et al., 2008; Wise et al., 2008). Furthermore, these studies were focused on discovering metabolic differences between Myc-expressing and non-expressing cells, rather than between cells with oncogenic (High) and endogenous (Low) Myc expression levels. Our study, on the other hand, applied rigorous 13C flux analysis to quantify metabolic phenotypes of P493-6 B-cells, which have been engineered to provide three distinct levels of Myc expression (No, Low, or High) depending on culture conditions.
We compared several steady-state and isotopically nonstationary MFA approaches to identify the best approach for analysis of P493-6 cells based on isotopomer measurements of protein-bound amino acids and ribose-bound RNA. We concluded that 13C INST-MFA was the most effective strategy for flux determination in these cells, and that ribose isotopomer measurements were important for maximizing flux identifiability. We then applied this approach to quantify fluxes in both High and Low Myc P493-6 cells (Fig. 1) and found significant reprogramming of central metabolism in response to ectopic Myc expression. High Myc cells relied more heavily on mitochondrial metabolism than Low Myc cells and globally upregulated their consumption of amino acids relative to glucose. The oxidative pentose phosphate (PP) pathway exhibited minimal activity under both High and Low Myc conditions, with negligible flux through the non-oxidative PP branch. Based on these results, we expect that 13C INST-MFA will become a powerful tool for analysis of tumor cell physiology and for identification of critical metabolic nodes that can be targeted to inhibit cancer growth.
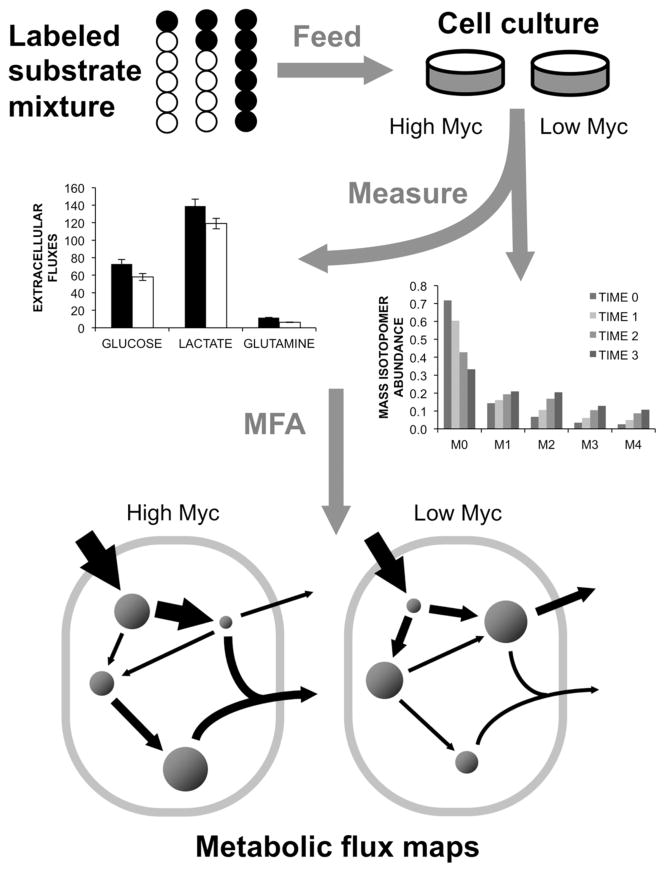
Figure 1. Overview of MFA study design.
A mixture of 13C-labeled glucose tracers were fed to P493-6 growing under High or Low Myc conditions. Extracellular medium concentrations and intracellular metabolite labeling were measured at various time points throughout exponential phase. A computational model was applied to map fluxes by minimizing the lack of fit between simulated and measured labeling data and extracellular flux measurements.
2. MATERIALS AND METHODS
2.1 Cell Culture
The human P493-6 B-cell line expresses an EBNA2-estrogen receptor fusion protein and contains a tetracycline (Tet)-repressible human MYC construct (Schuhmacher et al., 1999). Addition of 1 μg/mL Tet completely represses MYC expression, while the co-addition of 1 μM beta-estradiol (BES, MP Biomedicals, Solon, OH) induces a low level of endogenous MYC expression driven by the EBNA2 viral protein (Yustein et al., 2010). This allows for three distinct levels of Myc expression: High (no addition), Low (Tet + BES), and None (Tet alone). Only the High and Low expression conditions were examined in this study. Cells were cultured in RPMI 1640 medium (2 g/L glucose and 2 mM glutamine) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin (PS) at 37°C and 5% CO2. All cell culture supplies were purchased from Invitrogen (Carlsbad, CA). For tracer experiments, glucose-free medium was supplemented with the following mixture of 13C-labeled substrates: 28% [U-13C6]glucose, 20% [1-13C]glucose, and 52% [1,2-13C2]glucose. All tracers were purchased from Cambridge Isotope Laboratories (Andover, MA).
2.2 Oxygen Uptake Rates
High-resolution O2 consumption measurements were conducted at 37°C in RPMI 1640 medium using the OROBOROS O2K Oxygraph (Oroboros Instruments, Innsbruck, Austria). Cells were adjusted to a density of one million cells/mL and allowed to equilibrate in the instrument for a minimum of ten minutes. Cells were stirred at 750 rpm in atmospheric conditions without CO2 control. To confirm that oxygen uptake was dependent on cellular respiration, we treated cells with the Complex I inhibitor rotenone at a concentration of 100 nM (Kim et al., 2006). Higher rotenone concentrations produced erratic measurements and did not result in further reductions in the O2 uptake rate.
2.3 Specific Rate Determination
Extracellular uptake and excretion rates were determined in triplicate growth experiments. Three separate T-75 tissue culture flasks were seeded at a density of 150,000 cells/mL. Every 12–16 hours, 300 μL of cell suspension was removed from each flask after gentle mixing using a pipettor. 50 μL were used for counting on a hemacytometer while the remainder was centrifuged to remove cells, and the conditioned cell-free medium was frozen at −80°C. Concentrations of medium glucose and lactate were determined using a YSI 2300 Stat Plus Glucose and Lactate Analyzer (YSI, Yellow Springs, OH). Medium amino acid concentrations were determined using high-performance liquid chromatography (HPLC, Agilent 1200 series) with a gradient elution method on a reverse-phase column (Greene et al., 2009). Briefly, samples were derivatized immediately prior to injection with orthophthaldildehyde (OPA) and injected onto a ZORBAX Eclipse PLUS C18 column (Agilent Technologies, 4.6 × 150 mm, 3.5 μm). Mobile phase A was composed of 10 mM Na2HPO4, 10 mM Na2B4O7, and 8 ppm NaN3. Mobile phase B was a 9:9:2 mixture of methanol:acetonitrile:water. The gradient profile was as follows: 2% B for 0.5 minutes, ramp linearly to 47% for 15.5 minutes, ramp linearly to 100% B in 0.1 minutes, hold at 100% B for 3.4 minutes, ramp linearly to 2% B in 0.1 minutes, and hold for 1.4 minutes for a total time of 21 minutes. The flow rate was 1.5 mL/min, and the column was held at 40°C for the duration of the run.
The specific growth rate (μ) and specific production rate (qi) for each measured metabolite were defined as follows:
| (1) |
| (2) |
where X is the cell density, ki is the first-order degradation rate of the ith metabolite, and Ci is the concentration of the ith metabolite. The method of Glacken et al. (1988) was applied to estimate specific production rates by regression analysis of extracellular time-course measurements. The spontaneous degradation of glutamine to ammonia and pyrrolidonecarboxylic acid was included in the calculations (Ozturk and Palsson, 1990). The degradation rate was determined to be 0.0031 h−1 by measuring glutamine disappearance in control experiments performed in the absence of cells. Evaporation rates determined in control T-75 flasks without cells were found to be negligible in comparison to cell specific metabolic rates.
2.4 Steady-State Labeling Experiment
In order to achieve steady-state labeling, it was necessary to culture cells continuously in the presence of tracers throughout multiple platings. Cells were seeded at an initial density of 150,000 cells/mL in a T-25 flask. Every two days, cells were counted and replated at the same initial density in fresh tracer-containing medium. After 4 platings (approximately 8 cell doublings), the cells were harvested by centrifugation at 1500 RPM, washed with PBS, and extracted to isolate total cellular protein and RNA. Each experiment was performed in triplicate.
2.5 Isotopically Non-stationary Labeling Experiment
INST-MFA requires the measurement of isotopic enrichment at multiple time points during the transient labeling period. For High Myc cells, samples were taken at 6, 12, 24, 36, 48, and 72 hours. For Low Myc cells, which exhibited a slower growth rate, samples were taken at 12, 24, 36, 48, 72, and 96 hours. For each time point, three separate T-75 flasks were seeded at the appropriate density to achieve a final cell number of approximately ten million cells per flask at the time of sampling. Prior to seeding, the growth medium was removed by centrifugation and the cells were resuspended in glucose labeled medium. At the sample times indicated previously, cells were harvested by centrifugation at 1500 RPM, washed with phosphate buffered saline (PBS), and extracted to isolate total cellular protein and RNA.
2.6 Extraction, Hydrolysis and Derivatization of Total Cellular Protein and RNA
Extraction of protein and RNA was achieved using the TRIzol method, with the exception that proteins were precipitated using acetone instead of isopropanol (Simms et al., 1996). Protein samples were hydrolyzed to their constituent amino acids by incubating in 6N HCl for 20 hours at 110°C in a vacuum hydrolysis tube (Pierce). Similarly, RNA samples were hydrolyzed to ribose by incubating in 2N HCl at 100°C for 2 hours under vacuum. Protein hydrolysate samples were evaporated to dryness at 60°C under air flow. These dried samples were redissolved in 200 μL of ddH2O, filtered through a 0.2 μm filter, and re-dried prior to derivatization. RNA hydrolysate samples were evaporated to dryness at 60°C under air flow prior to derivatization.
To enable GC-MS analysis, amino acids were converted to their tert-butyl dimethylsilyl (TBDMS) derivatives by dissolving in 50 μL pyridine and 70 μL MTBSTFA + 1% TBDMCS (Pierce), followed by incubation at 60°C for 30 minutes. Ribose was converted to its aldonitrile pentapropionate derivative to enable GC-MS analysis (Antoniewicz et al., 2011; Lee et al., 1998). Briefly, dried samples were dissolved in 50 μL of 2 wt. % hydroxylamine hydrochloride in pyridine (Sigma) and incubated for 60 minutes at 90°C. Next, 100 μL of propionic anhydride (Sigma) was added, and the samples were incubated at 60°C for an additional 30 minutes. Samples were then centrifuged and evaporated at 60°C. The dried samples were dissolved in 100 μL of ethyl acetate prior to GC-MS analysis.
2.7 Medium Glucose Derivatization
Medium glucose labeling was assessed by GC-MS analysis. 100 μL of medium was washed with three volumes of cold acetone and centrifuged to remove protein. Samples were then evaporated to dryness at 60°C under air flow. Glucose was then converted to its aldonitrile pentapropionate derivative using the same procedure described in the previous section for ribose analysis.
2.8 Gas Chromatography Mass Spectrometry (GC-MS) Analysis
Derivatized sugar and amino acid samples were analyzed by GC-MS using a HP5-MS capillary column (30 m × 0.25 mm i.d. × 0.25 μm; Agilent J&W Scientific) installed in an Agilent 7890A gas chromatograph (GC). The injection volume was 1 μL, and all samples were run in split mode with an inlet temperature of 270°C and a split ratio of 10:1. Helium flow was controlled electronically at 0.73 mL/min for amino acid analysis and 1.65 ml/min for ribose and glucose analysis. The GC was interfaced to an Agilent 5975C mass spectrometer (MS) operated in electron impact mode with temperatures of 230°C for the ion source and 150°C for the quadrupole. The GC temperature program for amino acid analysis was: 150°C for 2 minutes, ramp at 5°C/min to 280°C, hold for 2 minutes. Mass spectra were obtained in scan mode over the range 100–500 m/z. The GC temperature program for glucose and ribose analysis was: 80°C for 1 minute, ramp at 20°C/min to 280°C, hold for 4 minutes. Mass spectra were obtained in scan mode over the range 100–700 m/z. Raw ion chromatograms were integrated using a custom MATLAB M-file that applies consistent integration bounds and baseline correction to each ion (Antoniewicz et al., 2007a).
2.9 Isotopomer Network Model
A detailed isotopomer model for mammalian B-cell metabolism was constructed. The metabolic network contains reactions for glycolysis, pentose phosphate pathway, TCA cycle, amphibolic pathways, amino acid catabolism, and biomass synthesis (Supplementary Table II). The network comprises 54 reactions with carbon atom transitions specified for all reactions. The network includes 8 extracellular substrates (glucose, arginine, asparagine, cysteine, glutamine, isoleucine, leucine, and serine), 6 metabolic products (biomass, lactate, alanine, glutamate, glycine, and lipids) and 35 balanced intracellular metabolites. Constraints from cofactor metabolites such as ATP and NAD(P)H were excluded because these balances have been shown to produce unreliable results in mammalian systems (Bonarius et al., 1998). Refer to Supplementary Materials for a detailed description of the model formulation and assumptions.
2.10 Flux Determination and Statistical Analysis
The elementary metabolite unit (EMU) framework was applied to efficiently simulate the labeling state of measurable metabolites represented in the isotopomer model (Antoniewicz et al., 2007b; Young et al., 2008). Both steady-state MFA and INST-MFA approaches involve solving an inverse problem whereby metabolic fluxes are determined by least-squares regression of measured extracellular fluxes and metabolite labeling patterns. The flux parameters of the isotopomer model were iteratively adjusted using a Levenberg-Marquardt algorithm until optimal agreement with experimental data was obtained. Flux estimation was repeated a minimum of 50 times from random initial values to ensure a global minimum was achieved. All results were subjected to a chi-square statistical test to assess goodness-of-fit, and accurate 95% confidence intervals were computed for all estimated parameters by evaluating the sensitivity of the sum-of-squared residuals (SSRES) to parameter variations (Antoniewicz et al., 2006).
2.11 Isotopomer Spectral Analysis (ISA)
Isotopomer spectral analysis (ISA) is an alternative method to determine fluxes from nonstationary isotopomer measurements. It assumes that any deviations from steady state are due to the presence of unlabeled material that has not yet washed out of the system, and adjustable parameters are introduced that represent the fraction of unlabeled material that persists in both precursor and product pools; these parameters are denoted as D and G, respectively. The ISA framework was originally developed by Kelleher and Masterson (1992) and has been applied by Antoniewicz et al. (2007c) to determine fluxes in a nonstationary model of E. coli metabolism.
As an alternative to INST-MFA, we developed an ISA-based nonstationary model by including two G dilution parameters, G1 and G2, into our steady-state isotopomer model. G1 represents the fraction of labeled protein in biomass and G2 represents the fraction of labeled RNA. Hence, (1-G1) and (1-G2) represent the amount of unlabeled protein and RNA, respectively, that remain at a given time point. Theoretical values for G1 and G2 were determined based on the doubling time of the cells as described in Antoniewicz et al. (2007c). Fluxes were determined at each sample time point of the nonstationary labeling experiment using the ISA-based model. Further details of the ISA procedure and its underlying assumptions are provided in the Supplementary Materials.
2.12 Biomass Equation
The dry weight of each cell was determined to be approximately 150 pg. This value was estimated by drying 70–150×106 cells in triplicate at 80°C overnight. The composition of the dry cell mass was estimated from the literature (Quek et al., 2010; Sheikh et al., 2005). Non-protein components included in the equation were nucleotides, lipids, and glycogen. The composition used for each purine was assumed to be: one R5P, one GLY, one CO2, and two MEETHF. The pyrimidine composition was assumed to be: one R5P, one CO2, and 1 ASP, except for thymine which has one additional MEETHF. One G6P was assumed to make one glycogen monomer. The phospholipid fraction was assumed to be composed of the following lipids: phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, phosphatidylinositol, phosphatidylglycerol, and cardiolipin. In each case, these molecules were modeled as having 17.43 AcCoA molecules and one DHAP with the following additional requirements: phosphatidylserine requires one SER, phosphatidylinositol requires one G6P, phosphatidylglycerol requires an additional DHAP, and cardiolipin requires an additional 17.43 AcCoA and 2 DHAP. (17.43 is the AcCoA requirement for each pair of fatty acid side-chains based on the average lipid composition reported by Sheikh et al. (2005)). Glycolipids were assumed to be represented wholly by sphingomyelin which was modeled as requiring 17.43 AcCoA and 1 SER. Sterols were modeled as cholesterol, which was assumed to require 18 AcCoA molecules for biosynthesis. Stoichiometric coefficients were determined by multiplying the estimated fraction of each biomass component by the cell dry weight and converting to units of nmol/million cells (Zamorano et al., 2010).
3. RESULTS
3.1 Cell Metabolic Phenotypes
The calculated growth rates and extracellular fluxes for both High and Low Myc cells are shown in Table I. Specific uptake and excretion rates were determined for 18 of 20 amino acids, but only 10 of those were consumed (or produced) in stoichiometric excess of the amounts required for biomass synthesis. Growth of Low Myc cells was 40% slower than High Myc cells, while glucose uptake was reduced by only 21%. Both High and Low Myc cells exhibited a highly glycolytic phenotype, with the majority of incoming carbon excreted as lactate. This was most clearly indicated by the lactate-to-glucose (L/G) ratios of 1.9 ± 0.2 and 2.1 ± 0.3 for High and Low Myc cells, respectively. Besides glucose, glutamine is the other major carbon substrate for mammalian cell cultures (Vander Heiden et al., 2009). Glutamine uptake supplied 9% of total carbon to High Myc cells and 7% to Low Myc cells. The uptake of most other amino acids was similarly elevated in High Myc cells. The total amount of carbon contributed by amino acids decreased from 28% in High Myc cells to 20% in Low Myc cells, with uptake fluxes of arginine, glutamine, histidine, isoleucine, leucine, lysine, methionine, serine, tyrosine, and valine all significantly elevated in the High Myc cells. Excretion fluxes of alanine, aspartate, and glycine were also significantly increased in High Myc cells.
TABLE I. Extracellular fluxes for High and Low Myc conditions.
Fluxes have units of nmol/106 cells/h except for biomass, which has units of h−1. Fluxes that were included in the MFA flux estimations are marked with a ✓. The uptake rates of other amino acids (marked with a ×) were stoichiometrically matched to the growth rate, indicating that they were solely used for biomass synthesis. These amino acids were not included in the MFA flux estimations because their catabolism was assumed to be negligible. Significance is indicated for the comparison between Low and High Myc conditions based on a two-tailed Student’s t-test with p < 0.05.
| High Myc | Low Myc | |||
|---|---|---|---|---|
|
| ||||
| Metabolite | Flux (nmol/106 cells/h) | Flux (nmol/106 cells/h) | Included in MFA | p < 0.05 |
| Biomass (h−1) | 0.0293 ± 0.0008 | 0.0176 ± 0.0006 | ||
|
| ||||
| Uptake Fluxes | ||||
| Glucose | 73 ± 5 | 58 ± 4 | ||
| Arginine | 4.7 ± 0.4 | 2.03 ± 0.15 | ||
| Asparagine | 0.9 ± 0.2 | 1 ± 0.2 | ||
| Cystine | 0.73 ± 0.11 | 0.94 ± 0.15 | ||
| Glutamine | 11.5 ± 0.3 | 6.3 ± 0.2 | ||
| Histidine | 0.78 ± 0.08 | 0.19 ± 0.03 | ||
| Isoleucine | 2.5 ± 0.2 | 1.4 ± 0.2 | ||
| Leucine | 3.3 ± 0.2 | 1.1 ± 0.1 | ||
| Lysine | 2.09 ± 0.15 | 0.61 ± 0.05 | ||
| Methionine | 0.67 ± 0.07 | 0.13 ± 0.03 | ||
| Phenylalanine | 0.13 ± 0.17 | 0.32 ± 0.07 | ||
| Serine | 4.1 ± 0.2 | 1.44 ± 0.01 | ||
| Threonine | 0.27 ± 0.04 | 0.61 ± 0.06 | ||
| Tyrosine | 0.58 ± 0.06 | 0.15 ± 0.03 | ||
| Valine | 1.42 ± 0.09 | 0.58 ± 0.05 | ||
|
| ||||
| Excretion Fluxes | ||||
| Lactate | 139 ± 8 | 119 ± 6 | ||
| Alanine | 1.65 ± 0.11 | 0.76 ± 0.12 | ||
| Aspartate | 0.36 ± 0.08 | −0.57 ± 0.12 | ||
| Glutamate | 2.8 ± 0.3 | 3.11 ± 0.03 | ||
| Glycine | 0.84 ± 0.11 | 0.02 ± 0.06 | ||
3.2 Isotopic Steady-State MFA – High Myc Cells
Because the extracellular flux measurements do not provide sufficient constraints to estimate intracellular fluxes involved in cyclic or parallel pathways, we sought to apply isotope labeling experiments and 13C MFA to generate a comprehensive flux map of High Myc P493-6 cells. At least 6 doublings of the cell population are required for the protein fraction to approach steady-state labeling. We estimated fluxes based on the steady-state labeling experiment both with and without ribose labeling data. After adding the ribose measurements to the model, the SSRES increased slightly above the acceptable 95% confidence threshold of the associated chi-square distribution (from SSRES = 11.7 with 8 degrees of freedom (DOF) to SSRES = 36.1 with 20 DOF). Despite this marginal lack of fit, we hypothesized that the addition of ribose labeling measurements would improve the precision of the flux estimates, especially within the PP pathway where ribose precursors are generated. This hypothesis was tested by computing the root-mean-square (RMS) error of net flux estimates within glycolysis, PP pathway, and TCA cycle (Table II). We found that the addition of ribose labeling data decreased the RMS error of net PP pathway reactions from 57% to 34% and resulted in an overall improvement in RMS error from 27% to 21%. These findings confirm our hypothesis that the addition of ribose labeling measurements has a substantial impact on the precision of flux estimates within the PP pathway.
TABLE II. Root-mean-square (RMS) percentage errors for selected net flux estimations.
RMS errors were calculated by first computing the percentage relative standard error of each net flux (i.e., si max(vi, 1)× 100%, where si is standard error and vi is the net flux value). The resulting relative errors were combined by taking the square root of the sum of squared errors divided by the number of fluxes in each pathway. Reactions included in glycolysis, PP pathway, and TCA cycle are listed in Supplementary Table II. (SS w/w/o Ribose = steady-state with/without ribose measurements included.)
| Method | Glycolysis | Pathway PPP | TCA Cycle | Overall |
|---|---|---|---|---|
| SS w/o Ribose | 5.7 | 57 | 31 | 27 |
| SS w/ Ribose | 5.5 | 34 | 32 | 21 |
| ISA 24 Hour | 5.5 | 54 | 42 | 29 |
| ISA 36 Hour | 5.5 | 54 | 30 | 26 |
| ISA 48 Hour | 5.0 | 62 | 27 | 27 |
| ISA 72 Hour | 4.9 | 60 | 22 | 26 |
| INST-MFA | 4.8 | 32 | 18 | 19 |
3.3 Isotopically Nonstationary MFA – High Myc Cells
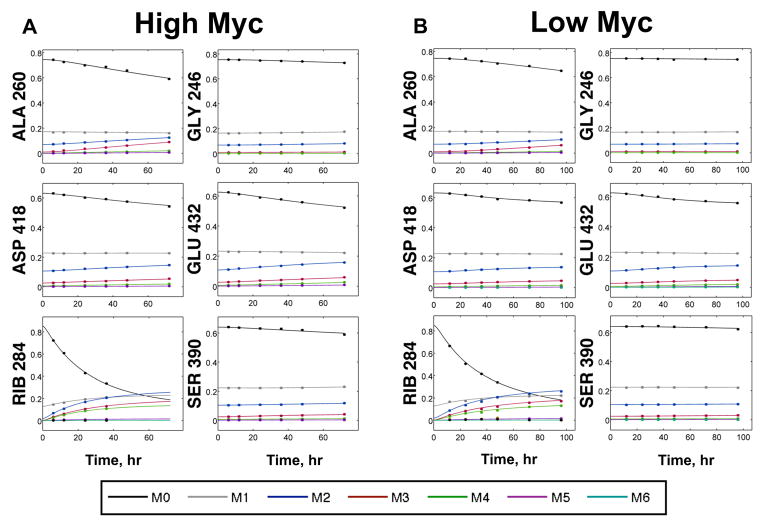
Because of the long experimental times required to reach isotopic steady state, we performed a transient labeling experiment on High Myc cells and analyzed the data using INST-MFA. Metabolic fluxes were estimated using isotopomer data collected at six time points: 6, 12, 24, 36, 48, and 72 hours after tracer introduction. This resulted in 376 independent mass isotopomer measurements, which were combined with the 13 extracellular fluxes indicated in Table I to estimate metabolic fluxes and their 95% confidence intervals. Each time point included proteinogenic amino acid and RNA-ribose isotopomer measurements, except for 48 and 72 hours, which only included amino acid measurements. Fig. 2A shows the dynamic labeling trajectories of several selected GC-MS fragment ions along with the INST-MFA model fits. (Fits for the remaining isotopomer measurements are shown in Supplementary Fig. 2A.) The model was overdetermined by 240 measurements, and the fit was accepted based on a chi-square test with SSRES = 91.7.
Figure 2. Labeling dynamics of selected GC-MS fragment ions.
GC-MS ions shown are for alanine (ALA), glycine (GLY), serine (SER), aspartate (ASP), glutamate (GLU), and ribose (RIB). Each panel shows the experimentally measured mass isotopomer abundances (data points) and INST-MFA model fits (solid lines) for a single fragment ion measured under the (A) High or (B) Low Myc condition. Raw mass isotopomer data are shown without correction for natural isotope abundance.
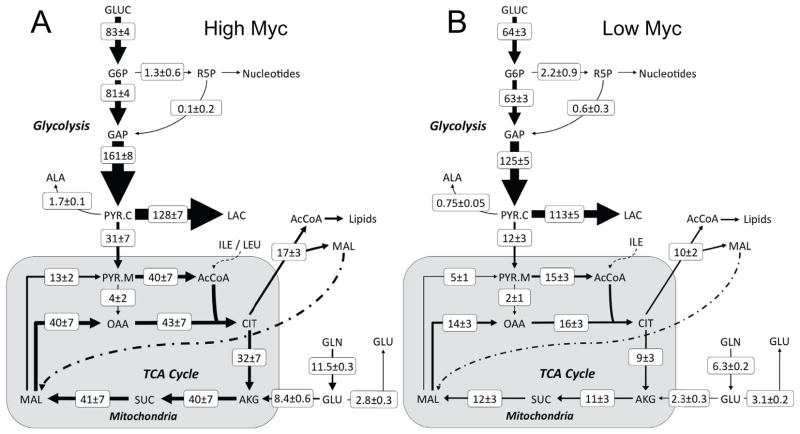
The High Myc flux map determined by INST-MFA is shown in Fig. 3A. (Refer to Supplementary Tables III and IV for a full listing of flux values and 95% confidence intervals. Pool sizes were completely unidentifiable or exhibited large 95% confidence intervals for most metabolites except for those where labeling was directly measured. Identifiable pool sizes and their 95% confidence intervals are shown in Supplementary Table V.) The oxidative PP pathway exhibited negligible activity, with only 2% of the incoming glucose diverted into this branch. Approximately 19% of the pyruvate synthesized from glucose entered the TCA cycle, 79% was excreted as lactate, and the remainder was converted to alanine. The majority (~73%) of glutamine consumed by the cell was metabolized to α-ketoglutarate, with the remainder excreted as glutamate. Both ATP-citrate lyase and mitochondrial malic enzyme were highly active, consuming more than 35% of the citrate and 25% of the malate produced by the cell, respectively. These enzymes are hypothesized to play an important role in supplying carbon for lipid biosynthesis in tumor cells (Moreadith and Lehninger, 1984).
Figure 3. P493-6 B-cell flux maps determined under (A) High and (B) Low Myc conditions.
Net fluxes are shown in units of nmol/106 cells/hour. Fluxes are represented as M ± SE, where M is the median of the 95% flux confidence interval and SE is the estimated standard error of M calculated as (UB95 – LB95)/3.92. (UB95 and LB95 are the upper and lower bounds of the 95% confidence interval, respectively.) Arrow thickness is scaled proportional to net flux. Some fluxes included in the isotopomer model are not shown in the figure to enhance clarity.
3.4 Isotopomer Spectral Analysis (ISA)-based Flux Estimation – High Myc Cells
As an alternative to INST-MFA, we applied an ISA-based approach to estimate fluxes from nonstationary labeling data collected at 24, 36, 48, and 72 hours after tracer introduction. The 24- and 36-hour time points were overdetermined by 19 measurements while those at 48 and 72 hours were overdetermined by 6 measurements due to the lack of ribose labeling information. All estimates returned SSRES values that were within the expected 95% confidence range of the appropriate chi-square distribution. As shown in Table III, the G1 parameter, which represents the fraction of newly synthesized protein estimated from isotope labeling measurements, was unidentifiable at time points prior to 48h. This is due to the low 13C enrichment of amino acids at these time points and the inability of the ISA model to distinguish between internal and external sources of isotope dilution. At later times, the estimated G1 values differed significantly from theoretical values derived from a simple dilution calculation (Antoniewicz et al., 2007c). Because Antoniewicz et al. (2007c) provided a simple bacterial growth medium that contained glucose as the sole carbon substrate, they observed close agreement between experimentally determined and theoretically predicted G parameter values. However, our mammalian culture medium contained high concentrations of amino acids and other unlabeled carbon sources. This additional source of isotope dilution resulted in a large mismatch between the experimental and theoretical G1 parameters. In contrast, the ribose portion of RNA was apparently derived exclusively from glucose carbon, and therefore the trajectory of G2 values matched closely with the theoretically predicted values in Table III.
TABLE III. Experimentally determined and theoretically predicted G1 and G2 dilution parameters.
Theoretical values are the same for both the G1 and G2 parameters and were determined based on the High Myc condition growth rate of 0.0293 h−1. RNA samples were not available for the 48 and 72 hour time points, which prevents the calculation of the G2 parameter at these times. Optimized parameter values and their standard errors are shown.
| Time Point (Hours) | Dilution Parameters | Theory | |
|---|---|---|---|
| G1 | G2 | ||
| 6 | 3.3 ± 25.3 | 16.3 ± 0.7 | 16.1 |
| 12 | 22.8 ± 24.0 | 30.7 ± 1.1 | 29.6 |
| 24 | 34.4 ± 21.3 | 54.5 ± 0.9 | 50.5 |
| 36 | 35.3 ± 19.7 | 66.9 ± 1.1 | 65.1 |
| 48 | 40.7 ± 8.7 | N/A | 75.4 |
| 72 | 51.1 ± 5.5 | N/A | 87.9 |
3.5 Comparison of MFA Approaches
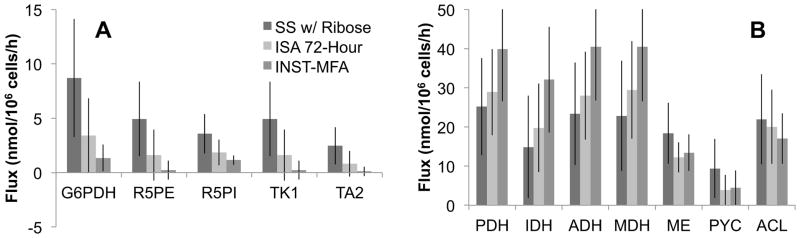
In order to select the most appropriate method for further MFA studies, we compared the precision and accuracy of each flux estimation approach. We compared steady-state MFA, 24-, 36-, 48-, and 72-hour ISA, and INST-MFA based on the 95% confidence intervals of net flux estimates. INST-MFA provided noticeably tighter confidence intervals for PP pathway and TCA cycle fluxes (Fig. 4 and Table II). We attribute this increase in precision primarily to the transient labeling measurements of alanine and ribose ion fragments, which are particularly sensitive to changes in G6PDH flux (Supplementary Fig. 3). Furthermore, the overall RMS errors of INST-MFA flux estimates were lower than those obtained from any other method (Table II). These comparisons reveal that the INST-MFA method is the most robust approach to determine fluxes in our system because of its enhanced flux precision as well as its ability to integrate labeling data obtained at multiple time points during the isotopically nonstationary period. Despite these noticeable differences in precision, the methods produced overlapping 95% confidence intervals for the majority of flux estimates. All but 8 of the 56 net fluxes exhibited overlap of their 95% confidence intervals when compared across all methods. The only disagreements were between the steady-state and INST-MFA experiments, which exhibited nonoverlapping confidence intervals in the PP pathway (Fig. 4). This could be attributable to biological variation between the experiments or disturbances introduced by periodic replating of cells in the steady-state labeling experiment. Furthermore, the SSRES of the steady-state experiment was slightly outside the acceptable range, which suggests that isotope labeling may not have been fully equilibrated at the time of sampling. All other pair-wise comparisons (steady-state vs. ISA or ISA vs. INST-MFA) exhibited overlapping confidence intervals across all fluxes, indicating statistical agreement. Overall, the methods were remarkably consistent in light of the different labeling strategies (single vs. multiple platings), seed densities, and modeling assumptions applied in each case. In addition to providing superior flux resolution, INST-MFA also imposes the least restrictive modeling assumptions (e.g., no isotopic steady-state assumption) and is therefore expected to be free from potential biases introduced by the other methods.
Figure 4. Intracellular fluxes in the High Myc condition as determined by alternative MFA methods.
A comparison of three flux estimation methods is shown based on 12 fluxes of central metabolism obtained under the High Myc condition (SS w/ Ribose = steady-state with ribose measurements included). Error bars represent 95% confidence intervals on the flux estimates, and the plotted values represent the medians of the confidence intervals. The fluxes shown were chosen because their confidence intervals exhibited the most variability across the three methods. (A) PP pathway net fluxes: glucose-6-phosphate dehydrogenase (G6PDH), ribulose-5-phosphate epimerase (R5PE), ribulose-5-phosphate isomerase (R5PI), transketolase 1 (TK1), and transaldolase 2 (TA2). (B) TCA cycle and amphibolic net fluxes: pyruvate dehydrogenase (PDH), isocitrate dehydrogenase (IDH), α-ketoglutarate dehydrogenase (ADH), malate dehydrogenase (MDH), malic enzyme (ME), pyruvate carboxylase (PYC), and ATP citrate lyase (ACL).
We also sought to determine whether the increased precision we observed in the INST-MFA flux estimates was due to the increased total number of labeling measurements or to an inherent increase in sensitivity associated with the transient labeling measurements. We copied the steady-state labeling data five times to simulate six replicate measurement sets. This produced the same total number of isotopomer measurements as in the INST-MFA data set. We re-estimated fluxes based on the replicated steady-state measurements and found that there was significant improvement in net flux precision over the original steady-state dataset, but that INST-MFA still provided superior identifiability of net fluxes in glycolysis and TCA cycle (Supplementary Table IX). INST-MFA was also able to resolve a greater number of exchange fluxes in comparison to steady-state MFA, and the number of identifiable exchange fluxes was not impacted by replicating the steady-state isotopomer measurements. Taken together, these results indicate that the increase in total number of labeling measurements can partially explain the improved precision of INST-MFA, but that the transient isotopomer measurements contain some inherent flux information that is not obtainable from the steady-state isotopomer measurements.
3.6 Isotopically Nonstationary MFA – Low Myc cells
We next applied INST-MFA to estimate fluxes in Low Myc cells because of its superior precision in determining fluxes of High Myc cells. Furthermore, the short labeling time required for INST-MFA is especially important in experiments with the more slowly growing Low Myc cells, because it would require nearly two weeks to achieve isotopic steady state in the protein fraction of these cells. Fluxes were estimated from labeling data obtained at six time points: 12, 24, 36, 48, 72, and 96 hours after tracer introduction. We fit the isotopomer model to 402 independent mass isotopomer abundances and 12 external flux measurements. Figure 2B shows the dynamic labeling measurements of several selected GC-MS fragment ions along with model simulations based on the best-fit parameters. (Fits for the remaining isotopomer measurements are shown in Supplementary Fig. 2B.) The system was overdetermined by 263 measurements, and the fit was accepted based on a chi-square test with SSRES = 89.0. (Refer to Supplementary Tables VI, VII, and VIII for a full listing of estimated flux and pool size values and 95% confidence intervals.) The data clearly indicate slower labeling in the Low Myc cells when compared to the High Myc cells, which agrees with the lower rates of growth and substrate uptake shown in Table I.
A comparison of the flux maps of the High and Low Myc cells in Figure 3 shows several noticeable differences in nutrient utilization, as well as some unexpected similarities. Overall, the distribution of glycolytic and PP pathway fluxes was quite similar in High and Low Myc cells, which was surprising in light of the 42% reduction in growth rate exhibited by the Low Myc cells. Despite a reduced rate of glucose uptake, the Low Myc cells exhibited a slightly higher L/G ratio and a non-significant reduction in lactate excretion rate. Small oxidative PP pathway fluxes were observed in both conditions, which was just sufficient to meet the biosynthetic demand for ribose-5-phosphate (R5P). On the other hand, the most striking differences were found in mitochondrial metabolic pathways, where the Low Myc cells exhibited 2- to 4-fold reductions in all TCA cycle and amphibolic fluxes. Furthermore, the Low Myc cells channeled a significantly lower percentage of pyruvate into the TCA cycle (10% versus 19%). Glutamine uptake was halved in Low Myc cells, but glutamate secretion remained the same, resulting in a near 4-fold reduction in anaplerotic flux from glutamine to alpha-ketoglutarate. This directly correlated with a near 3-fold reduction in mitochondrial malic enzyme flux, which functions to balance the flow of carbon leading to citrate synthesis. The extrusion of citrate into the cytosol and its subsequent degradation to AcCoA, a process that supplies carbon for fatty acid synthesis and protein acetylation, did not change as drastically as other mitochondrial fluxes and was closely matched to growth rate. This fate accounted for 53% of the citrate produced in Low Myc cells, but only 35% of the citrate produced in High Myc cells, indicating a clear shift toward increased oxidative metabolism in High Myc cells.
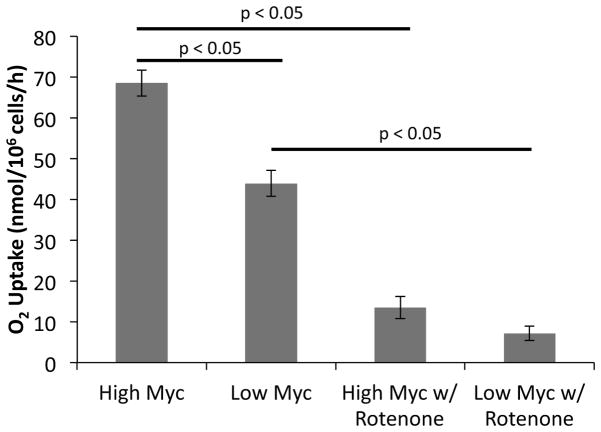
3.7 Oxygen Uptake Rates
The flux maps in Fig. 3 indicate a decrease in overall mitochondrial metabolism as a result of reduced Myc expression. We hypothesized that this change would correlate with a decrease in oxygen uptake rate (OUR) for respiratory processes. This hypothesis was tested by direct measurement of OUR, which confirmed that the Low Myc cells consumed oxygen at a rate of approximately 60% that of High Myc cells (Fig. 5). Oxygen uptake was strongly dependent on mitochondrial Complex I, as it was almost completely abolished in the presence of the Complex I inhibitor rotenone in both High and Low Myc cells.
Figure 5. Oxygen uptake rate of P493-6 cells under High and Low Myc conditions.
Oxygen uptake rates were measured in units of nmol/106 cells/hour. Rates were determined either in untreated cells (n=11) or cells treated with 100 nM rotenone (n=2). Error bars indicate standard error of the mean values. Significance was determined using a two-tailed Student’s t-test with p<0.05.
4. DISCUSSION
The ability to quantitatively map intracellular carbon fluxes using isotope tracers and metabolic flux analysis (MFA) provides a powerful approach to identify functional network states and regulatory mechanisms that characterize cell metabolism. Although a handful of prior studies have used 13C MFA to examine the role of specific oncoproteins (Gaglio et al., 2011; Kim and Forbes, 2007) or cell signaling pathways (Forbes et al., 2006; Grassian et al., 2011) in promoting global metabolic adaptations of tumor cells, we postulated that applying 13C MFA to map fluxes in P493-6 B-cells would allow us to quantify the direct metabolic consequences of oncogenic Myc expression while minimizing the confounding effects of clonal variability. The P493-6 B-cell line is an EBV-immortalized line with a tetracycline-repressible Myc expression construct (Pajic et al., 2001; Pajic et al., 2000). This cell system therefore provides a unique platform to investigate the effects of varying Myc expression within an isogenic background, and it has been used successfully by other groups to assess the role of Myc in regulating cell growth, metabolism, and apoptosis (Gao et al., 2009; Hatzivassiliou et al., 2005; Kim et al., 2007; Kim et al., 2004; Le et al., 2012; Liu et al., 2008; Schlosser et al., 2005). Furthermore, because Low Myc cells are nontumorigenic while High Myc cells resemble human Burkitt lymphoma cells, comparison of metabolic phenotypes between Low and High Myc cells is expected to reveal specific differences between normal proliferating cells and cancerous cells (Yustein et al., 2010).
Using this B-cell model system, we proceeded to quantify metabolic phenotypes under both Low and High Myc conditions. We measured time courses of cell density and extracellular metabolite concentrations throughout exponential phase, followed by regression analysis to estimate specific rates of growth, substrate consumption, and product excretion. Based on these specific rates, we observed that High Myc cells significantly increased their growth rate and the magnitude of most nutrient uptake and excretion fluxes in comparison to Low Myc cells. This is consistent with previous reports of the general stimulating effect of Myc on cell growth and metabolism (Fan et al., 2010; Morrish et al., 2009; Morrish et al., 2008). In particular, Myc has been shown to enhance flux through the glycolytic pathway by direct transactivation of GLUT1, HK2, PFKM, TPI1, GAPD, ENO1 and LDHA genes in studies of Rat1a fibroblasts and P493-6 cells (Kim et al., 2004; Osthus et al., 2000; Shim et al., 1997). Although our results show a modest increase in glucose consumption and lactate production as a result of ectopic Myc expression, the relative changes in these glycolytic fluxes were sub-proportional to the change in specific growth rate we observed. On the other hand, most amino acid uptake fluxes were increased 2- to 3-fold in High Myc cells relative to Low Myc cells, which exceeded the 1.7-fold change in specific growth rate. Therefore, ectopic Myc expression impacted amino acid fluxes more strongly than glycolytic fluxes in P493-6 cells.
Prior work has shown that glutamine metabolism is under direct control of Myc (Gao et al., 2009; Wise et al., 2008) and that Myc-overexpressing cells are particularly sensitive to glutamine withdrawal or inhibition of anaplerotic glutamine flux entering the TCA cycle (Fan et al., 2010; Wise et al., 2008; Yuneva et al., 2007). Our results show that, in addition to strongly upregulating their glutamine consumption, High Myc cells exhibited significant increases in most other incoming and outgoing amino acid fluxes. Furthermore, nearly half of the incoming amino acids were consumed in excess of their biosynthetic requirements, which indicates that they were partially catabolized to meet the energetic or redox demands of P493-6 cells. Aside from one study that identified serine hydroxymethyltransferase (SHMT2) as a direct Myc target gene (Nikiforov et al., 2002), little is known about how Myc stimulates metabolism of other amino acids besides glutamine. Based on our results, elucidating the mechanisms by which Myc influences global patterns of amino acid utilization in tumor cells represents a promising area of further investigation.
Despite much prior work to investigate the altered nutrient requirements of Myc-overexpressing cells, quantitative information about flux through intracellular metabolic pathways is not obtainable based on analysis of extracellular measurements alone. Therefore, we applied isotope labeling and 13C MFA to further elucidate the intracellular flux distributions of P493-6 cells under Low and High Myc conditions. After considering several alternative approaches, we concluded that a transient isotope labeling experiment followed by INST-MFA would provide the best overall accuracy and precision for flux determination in our system. A comparison of flux maps generated under High and Low Myc conditions not only confirmed the previously described global increases in glucose and amino acid metabolism exhibited by High Myc cells, but also revealed dramatic alterations in mitochondrial metabolism. In contrast to the small relative shift in glycolytic flux, we observed near 4-fold enhancements in most TCA cycle and amphibolic fluxes in response to ectopic Myc expression. We also observed a significantly higher rate of oxygen consumption by High Myc cells in comparison to Low Myc cells, although the relative increase in oxygen consumption was less dramatic than the increase in TCA cycle flux determined by MFA. This could be attributable to increased utilization of NAD(P)H for anabolic metabolism in High Myc cells or to unavoidable differences in culture conditions that were required to determine oxygen uptake rate (OUR). Despite these differences, however, both the MFA and OUR measurements are qualitatively consistent with an overall upregulation of mitochondrial metabolism in High Myc cells. This increase in mitochondrial activity is likely necessary to meet the increased ATP demands of High Myc cells, since the observed increase in glycolytic flux is insufficient to support the change in growth rate. Assuming a biosynthetic ATP requirement of 43 mmol/gDW (Sheikh et al., 2005) and our measured cell dry weight of 150 pg/cell, the difference in ATP demand for growth between High and Low Myc cells is approximately 75 nmol/106 cells/h. However, the difference in glycolytic flux accounts for only half of this ATP production, the remainder of which must be supplied by oxidative phosphorylation. The increase in TCA cycle flux and oxygen uptake exhibited by High Myc cells would more than compensate for the ATP deficit attributable to glycolysis.
In addition to providing ATP to support enhanced growth of High Myc cells, increased mitochondrial metabolism likely also plays a role in promoting availability of biosynthetic precursors, such as AcCoA that is needed for lipid biosynthesis and post-translational modification of proteins (Morrish et al., 2009; Morrish et al., 2010). Several recent studies have reported that tumor cells growing under hypoxic conditions or with mitochondrial defects shift to a reductive carboxylation pathway to supply carbon for lipid biosynthesis (Metallo et al., 2012; Mullen et al., 2012; Wise et al., 2011). This involves conversion of glutamine to citrate by reversal of the isocitrate dehydrogenase (IDH) reaction, which normally functions to oxidize citrate in the TCA cycle. Although we could not precisely assess the reversibility of IDH based on our measurements, the net TCA cycle flux was determined to operate strictly in the forward direction in both Low and High Myc cells. This does not preclude the possibility of glutamine carbons becoming incorporated into citrate or fatty acids through reversible or cyclic action of IDH1/2 isoforms, even in the presence of a net forward TCA cycle flux. However, Le et al. (2012) have recently shown that when P493-6 cells were grown in the presence of [U-13C5]glutamine under either aerobic or hypoxic conditions, the labeling patterns of citrate reflect a predominantly oxidative mode of glutamine metabolism rather than reductive carboxylation or its conversion to lactate.
One surprising finding of our study was the overall low level of oxidative PP pathway flux in both High and Low Myc cells. This could be due to the previously reported effects of Myc to enhance mitochondrial capacity and thereby decrease production of reactive oxygen species (ROS) in Myc-overexpressing cells (Morrish et al., 2008). Oxidative stress is a key regulator of oxidative PP pathway flux, which has been shown to decrease dose-dependently in response to treatments that reduce ROS levels (Tuttle et al., 2007). It is also possible that NADP-dependent isoforms of isocitrate dehydrogenase (IDH1 or IDH2) or malic enzyme (ME1 or ME3) provide the dominant source of NADPH required for biosynthesis and redox homeostasis in P493-6 cells, and thereby diminish the cellular demand for oxidative PP pathway activity.
To assess isotope labeling in our system, we relied exclusively on GC-MS measurements of RNA-bound ribose and protein-bound amino acids, following extraction and hydrolysis of total cellular protein and RNA. The labeling patterns of these macromolecular building blocks serve as proxies of the precursor metabolites from which they were biosynthetically derived. This “retrobiosynthetic” approach is commonly used in microbial and plant systems; however, the vast majority of recent 13C MFA studies performed on mammalian cells have relied exclusively upon direct extraction and isotopomer analysis of free intracellular metabolites (Niklas and Heinzle, 2011). Lee et al. (1998) previously applied mass isotopomer measurements of RNA-derived ribose and lipid-derived palmitate to investigate PP pathway fluxes in HepG2 cells but did not attempt to integrate these labeling data within a comprehensive flux model of central metabolism. Furthermore, we could identify only two previous examples where 13C MFA has been applied to a mammalian system based on isotopomer measurements of protein-bound amino acids rather than free metabolites (Goudar et al., 2010; Sriram et al., 2008). Although there are advantages and disadvantages to both approaches, and the preferred method will clearly depend upon the biological system and questions to be addressed, the most important benefits of the retrobiosynthetic approach are (i) the high signal-to-noise ratio that is obtained due to the abundance of protein and RNA within the cell and (ii) the long-term stability of the macromolecule pools, which obviates the need for sophisticated sample quenching and extraction methods that are required to preserve the in vivo labeling state of labile intracellular metabolites. The latter consideration is particularly germane to the possible future extension of 13C MFA to 3D culture systems or other cellular environments where rapid sample collection is not practical. For example, harvesting cells from semi-solid substrates such as collagen or Matrigel typically requires incubation at altered temperatures, possibly in the presence of proteolytic enzymes, in order to depolymerize the matrix. These procedures are not compatible with current metabolite extraction methods that have been developed for 2D adherent cultures or suspension cultures where cells can be readily recovered after quenching. One potential drawback of the retrobiosynthetic approach is that it is only applicable to actively proliferating cells; however, this would still encompass the vast majority of cancer biology studies that are aimed at identifying potential drug targets or biomarkers that are specifically upregulated in growing tumors. We anticipate that further development of MFA approaches based on isotopomer analysis of protein, nucleotide, and lipid building blocks will open the door to novel investigations of tumor cell metabolism in non-traditional culture systems, and perhaps eventually to in vivo tumors.
Prior to initiating MFA studies under both Low and High Myc conditions, we compared several different approaches for collecting and analyzing isotope labeling data as a function of time. To date, most MFA studies have relied upon steady-state isotope labeling measurements as inputs for flux estimation, rather than transient isotopomer measurements (Niklas and Heinzle, 2011). This simplifies the sampling procedure and furthermore reduces the isotopomer model to a system of algebraic equations, which can be solved more efficiently in comparison to the ODE-based models that are required for INST-MFA (Quek et al., 2010). A major drawback of the retrobiosynthetic approach, however, is the slow labeling that occurs in protein and RNA fractions. Because the turnover of these pools is linked to cell growth, at least 6 cell doublings must be achieved under balanced growth conditions and in the presence of tracer to achieve isotopic steady state. Therefore, we examined the possibility of using a transient isotope labeling approach to circumvent the practical difficulties associated with prolonged tracer experiments.
We analyzed isotope labeling data collected at multiple time points during exponential growth of P493-6 cells in simple flask cultures using steady-state MFA, INST-MFA, and an ISA-based pseudo-steady-state method. Comparison of all approaches revealed that their net flux estimates were largely in agreement, at least within the errors of the respective methods. The precision of INST-MFA flux estimates was best overall and was dramatically superior to other methods in resolving PP pathway fluxes. We attribute this to enhanced sensitivity of the SSRES to transient measurements of ribose and alanine labeling, particularly the Ala232 and Rib284 fragment ions, which constrained the nonoxidative branch of the PP pathway and indicated low net flux through the pathway as a whole. INST-MFA has the further advantage that it integrates all of the transient labeling data into a single flux map, and it does not depend on isotopic steady-state or pseudo-steady-state assumptions. Although INST-MFA has been previously applied to other mammalian systems (Ahn and Antoniewicz, 2011; Maier et al., 2009; Maier et al., 2008; Munger et al., 2008; Young et al., 2008) and bacterial systems (Nöh et al., 2007; Schaub et al., 2008; Young et al., 2011), this is the first time that it has been used to analyze transient labeling of macromolecular components such as proteinogenic amino acids or RNA-ribose. Furthermore, only one prior study has presented a systematic comparison between steady-state and INST-MFA methods in the same system, and this work was restricted to bacterial cultures (Noack et al., 2011).
5. CONCLUSION
Overall, 13C INST-MFA was the most effective strategy for flux determination in P493-6 cells based on isotopic measurements of protein-bound amino acids and RNA-bound ribose. This approach has the advantage of relying exclusively on isotopomer measurements derived from highly stable and abundant macromolecular pools, while avoiding the long experimental times that would be required to achieve isotopic steady state. We were able to precisely quantify the rates of all major glycolytic, PP pathway, TCA cycle, and amphibolic fluxes in P493-6 cells under both High and Low Myc expression levels. High Myc cells relied more heavily on mitochondrial metabolism than Low Myc cells and globally upregulated their consumption of amino acids relative to glucose. Most TCA cycle and amphibolic mitochondrial pathways exhibited near 4-fold flux increases in High Myc cells, in contrast to modest increases in glucose uptake and lactate excretion. The oxidative pentose phosphate pathway exhibited minimal activity under both High and Low Myc conditions. This approach can be readily extended to investigate the metabolic adaptations of tumor cells and other proliferating mammalian cells, as well as their response to specific pharmacologic or genetic interventions.
Supplementary Material
Isotopically nonstationary MFA provided maximum flux resolution in cultured B-cells (83)
Flux maps were compared under oncogenic (High) and endogenous (Low) Myc expression (82)
High Myc cells relied more heavily on oxidative phosphorylation than Low Myc cells (82)
High Myc cells also upregulated their consumption of amino acids relative to glucose (84)
MFA approach relied on labeling patterns of hydrolyzed protein and RNA constituents (83)
Acknowledgments
This work was supported by NIH R21 CA155964 and a Vanderbilt Discovery Award. The authors thank Matt Lang and Mike Betenbaugh for critical readings of the manuscript prior to submission. We also thank Rob Egnatchik for his assistance with the oxygen uptake measurements and Irina Trenary for her assistance with cell culture.
ABBREVIATIONS
- AcCoA
Acetyl-Coenzyme A
- AcCoA.C
cytosolic Acetyl-Coenzyme A
- ACL
ATP-citrate lyase
- ADH
α-ketoglutarate dehydrogenase
- AKG
α-ketoglutarate
- ALA
alanine
- ARG
arginine
- ASP
asparate
- CIT
citratre
- FUM
fumarate
- G6P
glucose-6-phosphate
- G6PDH
glucose-6-phosphate dehydrogenase
- GAP
glyceraldehyde-3-phosphate
- GLN
glutamine
- GLU
glutamate
- GLY
glycine
- IDH
isocitrate dehydrogenase
- LAC
lactate
- MAL
malate
- MCT
monocarboxylate transporter
- MDH
malate dehydrogenase
- ME
malic enzyme
- OAA
oxaloacetate
- PDH
pyruvate dehydrogenase
- PYC
pyruvate carboxylase
- PYR.C
cytosolic pyruvate
- PYR.M
mitochondrial pyruvate
- R5P
ribose-5-phosphate
- R5PE
ribulose-5-phosphate epimerase
- R5PI
ribulose-5-phosphate isomerase
- SER
serine
- TA2
transaldolase 2
- TK1
transketolase 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn WS, Antoniewicz MR. Metabolic flux analysis of CHO cells at growth and non-growth phases using isotopic tracers and mass spectrometry. Metab Eng. 2011;13:598–609. doi: 10.1016/j.ymben.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006;8:324–37. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. Anal Chem. 2007a;79:7554–7559. doi: 10.1021/ac0708893. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab Eng. 2007b;9:68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Measuring deuterium enrichment of glucose hydrogen atoms by gas chromatography/mass spectrometry. Anal Chem. 2011;83:3211–6. doi: 10.1021/ac200012p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR, Kraynie DF, Laffend LA, Gonzalez-Lergier J, Kelleher JK, Stephanopoulos G. Metabolic flux analysis in a nonstationary system: fed-batch fermentation of a high yielding strain of E. coli producing 1,3-propanediol. Metab Eng. 2007c;9:277–92. doi: 10.1016/j.ymben.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonarius HP, Timmerarends B, de Gooijer CD, Tramper J. Metabolite-balancing techniques vs. 13C tracer experiments to determine metabolic fluxes in hybridoma cells. Biotechnol Bioeng. 1998;58:258–262. doi: 10.1002/(sici)1097-0290(19980420)58:2/3<258::aid-bit20>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Heiden MGV, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–3. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–6. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Evans JM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD. Metformin and reduced risk of cancer in diabetic patients. BMJ. 2005;330:1304–5. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Dickman K, Zong W. Akt and c-Myc differentially activate cellular metabolic programs and prime cells to bioenergetic inhibition. J Biol Chem. 2010;285:7324. doi: 10.1074/jbc.M109.035584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–34. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- Forbes NS, Meadows AL, Clark DS, Blanch HW. Estradiol stimulates the biosynthetic pathways of breast cancer cells: Detection by metabolic flux analysis. Metab Eng. 2006;8:639–52. doi: 10.1016/j.ymben.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Gaglio D, Metallo CM, Gameiro PA, Hiller K, Danna LS, Balestrieri C, Chiaradonna F, Alberghina L, Stephanopoulos G. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol Sys Biol. 2011;7:1–15. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–5. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glacken MW, Adema E, Sinskey AJ. Mathematical descriptions of hybridoma culture kinetics: I. Initial metabolic rates. Biotechnol Bioeng. 1988;32:491–506. doi: 10.1002/bit.260320412. [DOI] [PubMed] [Google Scholar]
- Goudar C, Biener R, Boisart C, Heidemann R, Piret J, de Graaf A, Konstantinov K. Metabolic flux analysis of CHO cells in perfusion culture by metabolite balancing and 2D [13C, 1H] COSY NMR spectroscopy. Metab Eng. 2010;12:138–49. doi: 10.1016/j.ymben.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Grassian AR, Metallo CM, Coloff JL, Stephanopoulos G, Brugge JS. Erk regulation of pyruvate dehydrogenase flux through PDK4 modulates cell proliferation. Genes Dev. 2011;25:1716–33. doi: 10.1101/gad.16771811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene J, Henderson JW, Jr, Wikswo JP. Rapid and Precise Determination of Cellular Amino Acid Flux Rates Using HPLC with Automated Derivatization with Absorbance Detection. Agilent Technologies; 2009. pp. 1–8. [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G, Zhao F, Bauer D, Andreadis C, Shaw A, Dhanak D, Hingorani S, Tuveson D, Thompson C. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Kelleher JK, Masterson TM. Model equations for condensation biosynthesis using stable isotopes and radioisotopes. Am J Physiol. 1992;262:E118–25. doi: 10.1152/ajpendo.1992.262.1.E118. [DOI] [PubMed] [Google Scholar]
- Kim BJ, Forbes NS. Flux analysis shows that hypoxia-inducible-factor-1-alpha minimally affects intracellular metabolism in tumor spheroids. Biotechnol Bioeng. 2007;96:1167–82. doi: 10.1002/bit.21205. [DOI] [PubMed] [Google Scholar]
- Kim J, Tchernyshyov I, Semenza G, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metabolism. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–93. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Zeller KI, Wang Y, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol Cell Biol. 2004;24:5923–36. doi: 10.1128/MCB.24.13.5923-5936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–82. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037–42. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, Zimmerman LJ, Liebler DC, Slebos RJ, Lorkiewicz PK, Higashi RM, Fan TW, Dang CV. Glucose-Independent Glutamine Metabolism via TCA Cycling for Proliferation and Survival in B Cells. Cell Metab. 2012;15:110–21. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WN, Boros LG, Puigjaner J, Bassilian S, Lim S, Cascante M. Mass isotopomer study of the nonoxidative pathways of the pentose cycle with [1,2–13C2]glucose. Am J Physiol. 1998;274:E843–51. doi: 10.1152/ajpendo.1998.274.5.E843. [DOI] [PubMed] [Google Scholar]
- Liu YC, Li F, Handler J, Huang CR, Xiang Y, Neretti N, Sedivy JM, Zeller KI, Dang CV. Global regulation of nucleotide biosynthetic genes by c-Myc. PLoS One. 2008;3:e2722. doi: 10.1371/journal.pone.0002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier K, Hofmann U, Bauer A, Niebel A, Vacun G, Reuss M, Mauch K. Quantification of statin effects on hepatic cholesterol synthesis by transient (13)C-flux analysis. Metab Eng. 2009;11:292–309. doi: 10.1016/j.ymben.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Maier K, Hofmann U, Reuss M, Mauch K. Identification of metabolic fluxes in hepatic cells from transient 13C-labeling experiments: Part II. Flux estimation. Biotechnol Bioeng. 2008;100:355–70. doi: 10.1002/bit.21746. [DOI] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, Kelleher JK, vander Heiden MG, Iliopoulos O, Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nat Letters. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelakis ED, Webster L, Mackey JR. Dichloroacetate (DCA) as a potential metabolic-targeting therapy for cancer. Br J Cancer. 2008;99:989–994. doi: 10.1038/sj.bjc.6604554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreadith RW, Lehninger AL. The pathways of glutamate and glutamine oxidation by tumor cell mitochondria. J Biol Chem. 1984;259:6215–21. [PubMed] [Google Scholar]
- Morrish F, Isern N, Sadilek M, Jeffrey M, Hockenbery DM. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene. 2009;28:2485–91. doi: 10.1038/onc.2009.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish F, Neretti N, Sedivy JM, Hockenbery DM. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle. 2008;7:1054–66. doi: 10.4161/cc.7.8.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish F, Noonan J, Perez-Olsen C, Gafken PR, Fitzgibbon M, Kelleher J, VanGilst M, Hockenbery D. Myc-dependent mitochondrial generation of acetyl-CoA contributes to fatty acid biosynthesis and histone acetylation during cell cycle entry. J Biol Chem. 2010;285:36267–74. doi: 10.1074/jbc.M110.141606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen P-H, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nature Biotechnology. 2008;26:1179–86. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov MA, Chandriani S, O’Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy JM, Cole MD. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol. 2002;22:5793–800. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklas J, Heinzle E. Metabolic Flux Analysis in Systems Biology of Mammalian Cells. Adv Biochem Eng Biotechnol. 2011 doi: 10.1007/10_2011_99. [DOI] [PubMed] [Google Scholar]
- Noack S, Nöh K, Moch M, Oldiges M, Wiechert W. Stationary versus non-stationary (13)C-MFA: a comparison using a consistent dataset. J Biotechnol. 2011;154:179–190. doi: 10.1016/j.jbiotec.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Nöh K, Grönke K, Luo B, Takors R, Oldiges M, Wiechert W. Metabolic flux analysis at ultra short time scale: Isotopically non-stationary 13C labeling experiments. J Biotechnol. 2007;129:249–267. doi: 10.1016/j.jbiotec.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Nöh K, Wiechert W. The benefits of being transient: isotope-based metabolic flux analysis at the short time scale. Appl Microbiol Biotechnol. 2011;91:1247–1265. doi: 10.1007/s00253-011-3390-4. [DOI] [PubMed] [Google Scholar]
- Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- Ozturk SS, Palsson BO. Chemical decomposition of glutamine in cell culture media: effect of media type, pH, and serum concentration. Biotechnol Prog. 1990;6:121–128. doi: 10.1021/bp00002a005. [DOI] [PubMed] [Google Scholar]
- Pajic A, Polack A, Staege MS, Spitkovsky D, Baier B, Bornkamm GW, Laux G. Elevated expression of c-myc in lymphoblastoid cells does not support an Epstein-Barr virus latency III-to-I switch. J Gen Virol. 2001;82:3051–5. doi: 10.1099/0022-1317-82-12-3051. [DOI] [PubMed] [Google Scholar]
- Pajic A, Spitkovsky D, Christoph B, Kempkes B, Schuhmacher M, Staege MS, Brielmeier M, Ellwart J, Kohlhuber F, Bornkamm GW. Cell cycle activation by c-myc in a Burkitt lymphoma model cell line. International journal of cancer Journal international du cancer. 2000;87:787–793. doi: 10.1002/1097-0215(20000915)87:6<787::aid-ijc4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Quek L-E, Dietmair S, Krömer JO, Nielsen LK. Metabolic flux analysis in mammalian cell culture. Metab Eng. 2010;12:161–171. doi: 10.1016/j.ymben.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Sauer U. Metabolic networks in motion: 13C-based flux analysis. Mol Sys Biol. 2006;2:62. doi: 10.1038/msb4100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub J, Mauch K, Reuss M. Metabolic flux analysis in Escherichia coli by integrating isotopic dynamic and isotopic stationary 13C labeling data. Biotechnol Bioeng. 2008;99:1170–85. doi: 10.1002/bit.21675. [DOI] [PubMed] [Google Scholar]
- Schlosser I, Holzel M, Hoffmann R, Burtscher H, Kohlhuber F, Schuhmacher M, Chapman R, Weidle UH, Eick D. Dissection of transcriptional programmes in response to serum and c-Myc in a human B-cell line. Oncogene. 2005;24:520–4. doi: 10.1038/sj.onc.1208198. [DOI] [PubMed] [Google Scholar]
- Schuhmacher M, Staege MS, Pajic A, Polack A, Weidle UH, Bornkamm GW, Eick D, Kohlhuber F. Control of cell growth by c-Myc in the absence of cell division. Curr Biol. 1999;9:1255–8. doi: 10.1016/s0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- Sheikh K, Förster J, Nielsen LK. Modeling hybridoma cell metabolism using a generic genome-scale metabolic model of Mus musculus. Biotechnol Prog. 2005;21:112–121. doi: 10.1021/bp0498138. [DOI] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94:6658–63. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms D, Cizdziel PE, Chomczynski P. TRIzol A New Reagent for Optimal Single-Step Isolation of RNA. Focus. 1996;15:99–102. [Google Scholar]
- Sriram G, Rahib L, He J, Campos A, Parr L, Liao J, Dipple K. Global metabolic effects of glycerol kinase overexpression in rat hepatoma cells. Mol Genet Metab. 2008;93:145–159. doi: 10.1016/j.ymgme.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tuttle SW, Maity A, Oprysko PR, Kachur AV, Ayene IS, Biaglow JE, Koch CJ. Detection of reactive oxygen species via endogenous oxidative pentose phosphate cycle activity in response to oxygen concentration: implications for the mechanism of HIF-1alpha stabilization under moderate hypoxia. J Biol Chem. 2007;282:36790–6. doi: 10.1074/jbc.M700327200. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, Christofk HR, Wagner G, Rabinowitz JD, Asara JM, Cantley LC. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiechert W, Noh K. From stationary to instationary metabolic flux analysis. Adv Biochem Eng Biotechnol. 2005;92:145–172. doi: 10.1007/b98921. [DOI] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–7. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, Dematteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–6. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J, Walther J. An elementary metabolite unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnol Bioeng. 2008 doi: 10.1002/bit.21632. [DOI] [PubMed] [Google Scholar]
- Young JD, Shastri AA, Stephanopoulos G, Morgan JA. Mapping photoautotrophic metabolism with isotopically nonstationary (13)C flux analysis. Metab Eng. 2011;13:656–65. doi: 10.1016/j.ymben.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD, Walther JL, Antoniewicz MR, Yoo H, Stephanopoulos G. An elementary metabolite unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnol Bioeng. 2008;99:686–699. doi: 10.1002/bit.21632. [DOI] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yustein JT, Liu Y-C, Gao P, Jie C, Le A, Vuica-Ross M, Chng WJ, Eberhart CG, Bergsagel PL, Dang CV. Induction of ectopic Myc target gene JAG2 augments hypoxic growth and tumorigenesis in a human B-cell model. Proc Natl Acad Sci U S A. 2010;107:3534–3539. doi: 10.1073/pnas.0901230107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni N. 13C metabolic flux analysis in complex systems. Curr Opin Biotechnol. 2011;22:103–108. doi: 10.1016/j.copbio.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Zamboni N, Fendt SM, Rühl M, Sauer U. 13C-based metabolic flux analysis. Nature protocols. 2009;4:878–892. doi: 10.1038/nprot.2009.58. [DOI] [PubMed] [Google Scholar]
- Zamorano F, Wouwer AV, Bastin G. A Detailed Metabolic Flux Analysis of an Underdetermined Network of CHO Cells. J Biotechnol. 2010;150:497–508. doi: 10.1016/j.jbiotec.2010.09.944. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.