Abstract
The ability to generate well-defined gradients in cell density on engineered scaffolds is critical to the regeneration of many types of tissues. Herein we describe a simple and versatile method for generating gradients in cell density on glass slides and nonwoven mats of electrospun nanofibers. By inserting a glass slide (without or with a mat of nanofibers on the surface) into a suspension of cells at a tilt angle, a gradient in cell density along the direction of insertion can be established on the top surface of the glass slide due to the variation in the number of cells available for sedimentation. Different gradients in cell density can be readily obtained by simply varying the tilt angle. This method can also be applied to generate reverse gradients on the same substrate with two different cell types, which may find potential use in interface tissue engineering.
Keywords: cell culture, cell density gradient, interface tissue engineering
Gradients in cell types, cell quantities, and extracellular matrix (ECM) molecules represent a common feature of many biological systems. At the junction of unmineralized and mineralized tissues in the tendon-to-bone insertion, for example, the cell phenotype gradually changes from fibroblasts in the tendon to fibrochondrocytes at the interface and osteoblasts in the bone.[1] Similar gradations also exist at the myotendinous junction[2] and between various tissue layers that make up the skin.[3] The gradients in cell density regulate both quantity and quality of cell-cell interactions and thus the functions of relevant tissues.[4] In a sense, the gradients determine the number of physical connections that a single cell is able to establish with surrounding cells, which in turn regulates cell behavior by influencing the intracellular signaling pathways.[5] Perturbation of cell-cell interactions, as typically occurs during an injury, triggers a cascade of events that either re-establishes tissue homeostasis or more often leads to scar formation.[6] In addition to physical connections, spatial variations in cell density may lead to the establishment of gradations in biochemical and ECM cues, as a result of which cell migration, proliferation, and differentiation can all be affected.[7] As a result, the ability to generate controllable gradients in cell density is critical to the recreation of functionality for many types of tissues.
Various methods have been pursued to generate gradients in cell density. In most cases, the gradient was established as a result of variation in mechanical,[8] chemical,[9] or electrical[10] cues. Despite their different sources of stimuli, these methods all relied on directed cell migration for the development of a gradient in cell density. In general, directed cell migration is plagued by problems such as slow migration rate,[11] low controllability,[12] and limited reproducibility.[13] The unpredictable differences among batches would make the clinical applications of these products challenging. An alternative to these approaches, live cells can be printed with an inkjet printer to deliver cells in a pre-determined pattern.[14] Although this technique overcomes the low reproducibility of directed cell migration and enables the accurate fabrication of gradients in cell density,[15] it tends to suffer from other drawbacks such as frequent blockage of the printer head, limitation of substrates that are both “printable” and biocompatible, and low cell viability due to the harsh printing process.[16] These and other issues associated with the existing methods for generating gradients in cell density motivated the current study.
In the present work, we developed a simple and versatile method for generating gradients in cell density. It involved the insertion of a substrate into a homogeneous suspension of cells at a tilt angle (Fig. 1). We hypothesized that, due to the varying volumes of cell suspension above the substrate, the number of cells available for sedimentation onto the substrate would vary along the direction of insertion, naturally leading to the establishment of a gradient in cell density on the substrate. Different gradients in cell density could be obtained by altering the tilt angle of the substrate. The capability of this method could also be extended to generate opposing gradients of two different types of cells on the same substrate, which may find use in the regeneration of the aforementioned biological interfaces.
Figure 1.

A schematic of the experimental setup. A glass slide is placed at a tilt angle in a beaker filled with a homogeneous suspension of cells. Due to the varying volumes of cells available for sedimentation above the slide, the cells are deposited on the slide with a gradient in density.
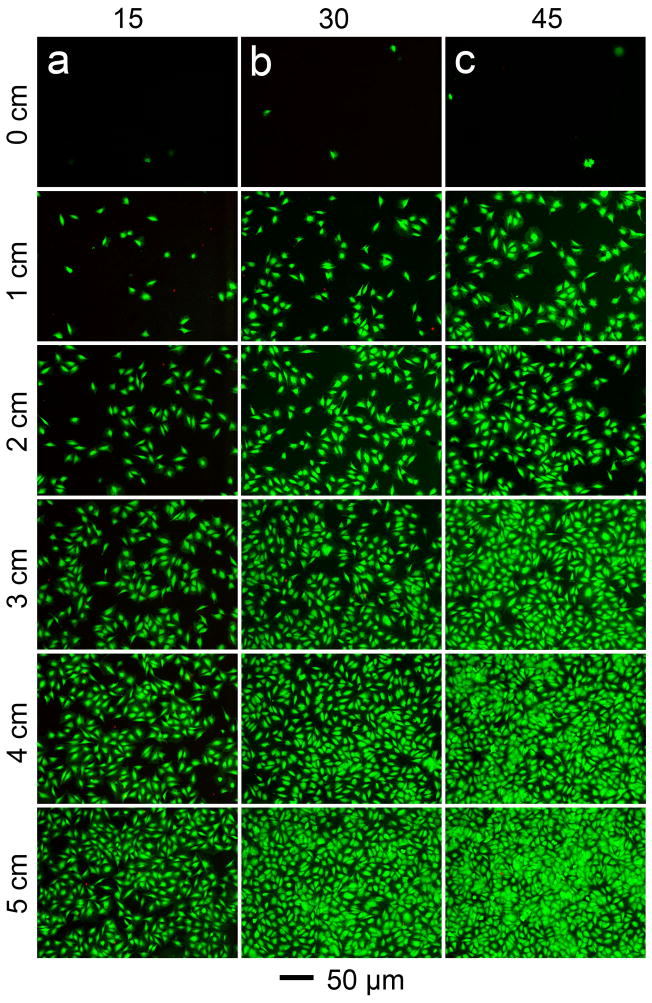
We first used glass slides and MC3T3 preosteoblasts to demonstrate that this technique could generate different gradients in cell density by varying the tilt angle. The experiment was carried out with the glass slides being tilted at 15, 30, and 45 degrees, respectively. In order to improve cell attachment, the glass slides were pre-coated with fibronectin and then inserted into homogeneous suspensions of MC3T3 preosteoblasts at a concentration of 2×105/mL (one slide per cell suspension). This concentration of cells was chosen to allow for a clear observation of cell density by fluorescence staining. In principle, the concentration of cells can also be varied to alter the gradient pattern of cells. The cells were allowed to sediment for 2 h so that they could attach to the glass slide. The slides were then extracted and rinsed briefly with phosphate buffered saline (PBS) to wash off any unattached or loosely-bound cells. All the glass slides were 5 cm in length. The highest point on a slide during the sedimentation process was denoted as the starting point and the lowest point was denoted as the position of 5 cm. The schematic in Figure S1 of the Supporting Information shows a detailed description of the experimental design. The volume of the cell suspension was set in such a way that the starting point of each slide was barely immersed for each tilt angle. Therefore, the depth of the cell suspension in the beaker was 5 × sin(15°) = 1.3 cm, 5 × sin(30°) = 2.5 cm, and 5 × sin(45°) = 3.53 cm for tilt angles of 15, 30, and 45 degrees, respectively. Similarly, the relationship between the height of any given point on the slide, D, its location along the length of the slide, L, and the tilt angle θ is: D = L·sin(θ) (Fig. S1). In this way, all of the gradient patterns started from a density of zero at the starting point (i.e., there were no cells at this position available for sedimentation). In an alternative protocol, one could fix the volume of cell suspension for all samples so all of the slides would have the same cell density at the position of 5 cm. However, this scenario would result in variable cell densities at the starting point, and the pattern of the gradient would not stand out as clearly. Therefore, the former protocol was adopted for the entire study.
Live/dead staining was performed immediately after sedimentation to help visualize the cells, where live cells were stained green and dead cells red (Fig. 2). As expected, all the samples had cell viabilities close to 100% after sedimentation. While all the three groups showed gradient patterns, the cell density increased faster at a larger tilt angle. There were almost no cells present at the starting point in any of the three groups. Different cell densities were observed at the 5-cm position depending on the tilt angle, with the lowest cell density at 15 degrees (Fig. 2a), intermediate cell density at 30 degrees (Fig. 2b), and the highest cell density at 45 degrees (Fig. 2c). These results suggest that the slope of the gradient can be tuned by simply changing the tilt angle of the glass slide during cell seeding. The relationship between the fluorescence intensity of the cells and their location on the glass slides is plotted in Figure S2. In all cases, a linear regression provided a close fit to the data, indicating that local cell density increased linearly with position on the glass slide. The slopes of the regression lines were calculated to be 4.0, 7.5, and 10.5 for tilt angles of 15, 30, and 45 degrees, respectively. The linear relationship between tilt angle and cell density supported our hypothesis that the density of cells on the slide depends on the depth in the cell suspension.
Figure 2.
Fluorescence micrographs showing gradients in cell density generated on glass slides at tilt angles of (a) 15, (b) 30, and (c) 45 degrees, respectively. The cells were stained with a live/dead kit immediately after seeding to give live and dead cells green and red colors, respectively.
As expected, the slope of the regression also adopted a linear relationship with sinθ (Fig. S3). To test the predictive power of this relationship, we calculated the expected cell density for a tilt angle of 75 degrees using the regression equation and seeded cells onto a slide placed in a cell suspension at a tilt angle of 75 degrees. The slope of regression for the cell density gradient at a tilt angle of 75 degrees was predicted to be 14.5. However, seeding cells at 75 degrees for 2 h did not support this estimation, with very few cells adhering to the entire glass slide (Fig. 3a). The regression curve from the experimental results had a slope of 1.8, even smaller than the slope for the gradient created at 15 degrees (Fig. S4). The likely reason for the observed low cell density is that, at large tilt angles, most of the cells rolled down to the bottom of the container before they had a chance to adhere to the substrate as they sedimented. Since 2·sin29° was roughly the same as sin75°, we performed two sequential seeding procedures at 29 degrees to obtain the gradient in cell density estimated for a tile angle of 75 degrees. Figure 3b shows the gradient after the first seeding process at 29 degrees. Figure 3c shows the cumulative gradient after two sequential seeding procedures at 29 degrees. The fluorescent intensity at each location fit well with the prediction, with a slope of 14.6 (Fig. S3). These data indicate that consecutive seeding processes can be used to overcome cell adhesion issues associated with large tilt angles, where cell rolling across the surface becomes an issue.
Figure 3.
Fluorescence micrographs showing gradients in cell density generated on glass slides at tilt angles of (a) 75 and (b) 29 degrees, respectively. (c) Cumulative gradients in cell density generated after two consecutive seeding processes at 29 degrees each along the same direction of gradient. The cells were stained using a live/dead kit immediately after seeding to give live and dead cells green and red colors, respectively.
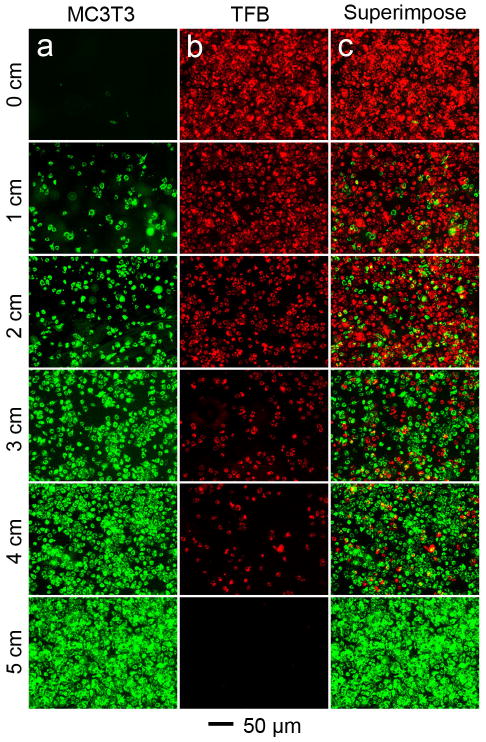
We also generated opposite gradients in cell density on the same glass slide to demonstrate the power of this method for applications such as tendon-to-bone tissue engineering. Specifically, MC3T3 preosteoblasts and tendon fibroblasts (TFB) were used to generate reverse gradients for the densities of these two cell types. MC3T3 and TFB were pre-labeled with DiO (green) and DiI (red), respectively, prior to sedimentation. The MC3T3 preosteoblasts were seeded onto the glass slide at a tilt angle of 30 degrees for 2 h. The glass slide was briefly rinsed with culture medium to wash off loosely attached cells and observed under a fluorescent microscope (Fig. 4a). The glass slide was then horizontally rotated 180 degrees and inserted into a suspension of TFB cells at a tilt angle of 30 degrees for 2 h. After washing off loosely attached TFB cells, the glass slide was imaged again. As shown in Figure 4b, a similar gradient in the density of TFB cells was generated along the glass slide but in the reverse direction of MC3T3 preosteoblasts. Figure 4c shows a superimposed image of the images in 4a and 4b, indicating a transition zone at the center of the scaffold where MC3T3 preosteoblasts and TFB were brought into close proximity.
Figure 4.
Fluorescence micrographs showing the generation of reverse gradients in cell density for MC3T3 preosteoblasts and TFB on the same glass slide after two consecutive sedimentation processes along opposite directions. The two types of cells were labeled with different membrane dyes prior to seeding.
The gradient in cell density could also be fabricated on a scaffold of electrospun nanofibers using the same procedure (Fig. S5). The nanofibers were fabricated according to our previously published protocols.[17] The scaffold was attached to a glass slide prior to seeding and the tilt angle was set to 30 degrees. The scaffold was designed to have two different types of structures with uniaxially aligned nanofibers on one region and random nanofibers on the other. When placed in the cell suspension, the random nanofibers were kept at the bottom whereas the aligned end was kept at the top. A clear gradient in cell density can be observed on the scaffold with no obvious difference in cell attachment between the aligned region and the random region. This observation indicates that both aligned and random nanofibers are suitable substrates for the generation of gradients in cell density using this simple method.
There are several advantages associated with this method for generating gradients in cell density as compared to previously published approaches. First, the experimental setup is simple and cell-friendly compared to the printing techniques for live cells. Maintaining sterility is trivial for the method presented in this study, but can be difficult for the cell printing approaches. Due to minimal cell processing, it is expected that the cells will display similar behaviors to the cells plated using the conventional methods. Maintaining cell function is especially critical for studies involving stem cell differentiation or primary cell types that must maintain their phenotypes. Techniques involving chemical, mechanical, or electrical stimulation for the formation of cell gradients may result in unwanted or uncontrollable differentiation of stem cells or de-differentiation of primary cells. A second advantage of the method presented here is the predictability of the gradient based on the tilt angle. A simple linear relationship can be used to reproducibly generate cell gradients in a particular density. A third advantage of the present method is the ability to easily produce complex gradients in terms of patterns and cell types via sequential seeding processes, many of which would be difficult or impossible to be fabricated using the previously reported techniques.
In summary, a simple and robust method for generating gradients in cell density has been demonstrated. In this method, a substrate is inserted into a homogenous suspension of cells at a pre-set angle and a cell density gradient will be naturally created due to the variation in the number of cells available for sedimentation above the substrate. By adjusting the tilt angle, various gradient patterns can be easily obtained in a controllable and reproducible fashion. Even reverse gradients of different cell types can also be fabricated on the same substrate by taking advantage of multiple sequential seeding processes.
Supplementary Material
Footnotes
This work was supported in part by a grant from the NIH (1R01 AR060820) and startup funds from Georgia Institute of Technology.
Supporting information for this article is available on the WWW under http://dx.doi.org/.
References
- 1.Moffat KL, Wang I-NE, Rodeo SA, Lu HH. Clin Sport Med. 2009;28:157–176. doi: 10.1016/j.csm.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang PJ, Temenoff JS. Tissue Eng. 2009;15:127–141. doi: 10.1089/ten.teb.2008.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacNeil S. Nature. 2007;455:874–880. doi: 10.1038/nature05664. [DOI] [PubMed] [Google Scholar]
- 4.a) Rosso F, Giordano A, Barbarisi M, Barbarisi A. J Cell Physiol. 2004;199:174–180. doi: 10.1002/jcp.10471. [DOI] [PubMed] [Google Scholar]; b) Bhatia SN, Balis UJ, Yarmush ML, Toner M. FASEB J. 1999;13:1883–1900. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- 5.Nelson CM, Liu WF, Chen CS. In: Comprehensive Supramolecular Chemistry. Coutts AS, editor. Vol. 370. Springer; Berlin: 2007. pp. 1–9. [Google Scholar]
- 6.Rosenkilde MM, Schwartz TW. APMIS. 2004;112:481–495. doi: 10.1111/j.1600-0463.2004.apm11207-0808.x. [DOI] [PubMed] [Google Scholar]
- 7.a) Pennisi DJ, Mikawa T. Dev Biol. 2005;279:378–390. doi: 10.1016/j.ydbio.2004.12.028. [DOI] [PubMed] [Google Scholar]; b) Liu L, Ratner BD, Sage EH, Jiang S. Langmuir. 2007;23:11168–11173. doi: 10.1021/la701435x. [DOI] [PubMed] [Google Scholar]
- 8.a) Helm CE, Fleury ME, Zisch AH, Boschetti F, Schwartz MA. Proc Natl Acad Sci U S A. 2005;102:15779–15784. doi: 10.1073/pnas.0503681102. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Li X, Xie J, Lipner J, Yuan X, Thomopoulos S, Xia Y. Nano Lett. 2009;9:2763–2768. doi: 10.1021/nl901582f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng SY, Heilman S, Wasserman M, Archer S, Shuler ML, Wu M. Lab Chip. 2007;7:763–769. doi: 10.1039/b618463d. [DOI] [PubMed] [Google Scholar]
- 10.Wan AMD, Brooks DJ, Gumus A, Fischbach C, Malliaras GG. Chem Commun. 2009;35:5278–5280. doi: 10.1039/b911130a. [DOI] [PubMed] [Google Scholar]
- 11.Webb K, Hlady V, Tresco PA. J Biomed Mater Res. 2000;49:362–368. doi: 10.1002/(sici)1097-4636(20000305)49:3<362::aid-jbm9>3.0.co;2-s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Ruardy TG, Schakenraad JM, van der Mei HC, Busscher HJ. J Biomed Res. 1995:29,1415–1423. doi: 10.1002/jbm.820291113. [DOI] [PubMed] [Google Scholar]; b) Schakenraad JM, Busscher HJ, Wildevuur CRH, Arends J. J Biomed Mater Res. 1986;20:773–784. doi: 10.1002/jbm.820200609. [DOI] [PubMed] [Google Scholar]; c) Altankov G, Grinnell F, Groth T. J Biomed Mater Res. 1996;30:385–391. doi: 10.1002/(SICI)1097-4636(199603)30:3<385::AID-JBM13>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 13.a) Huttenlocher A, Sanborg RR, Horwitz AF. Curr Opin Cell Biol. 1995;7:697–706. doi: 10.1016/0955-0674(95)80112-x. [DOI] [PubMed] [Google Scholar]; b) Lauffenburger DA, Horowitz AF. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]; c) Dimilla PA, Stone JA, Quinn JA, Albelda SM, Lauffenburger DA. J Cell Biol. 1993;122:729–737. doi: 10.1083/jcb.122.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barron JA, Wu P, Ladouceur HD, Ringeisen BR. Biomed Microdevices. 2004;6:139–147. doi: 10.1023/b:bmmd.0000031751.67267.9f. [DOI] [PubMed] [Google Scholar]
- 15.Xu T, Jin J, Gregory C, Hickman JJ, Boland T. Biomaterials. 2004;26:93–99. doi: 10.1016/j.biomaterials.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 16.Barron JA, Krizman DB, Ringeisen BR. Ann Biomed Eng. 2005;33:121–130. doi: 10.1007/s10439-005-8971-x. [DOI] [PubMed] [Google Scholar]
- 17.a) Liu W, Thomopoulos S, Xia Y. Adv Healthcare Mater. 2012;1:10–25. doi: 10.1002/adhm.201100021. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Xie J, Liu W, MacEwan MR, Yeh Y-C, Thomopoulos S, Xia Y. Small. 2011;3:293–297. doi: 10.1002/smll.201001446. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Xie J, Li X, Lipner J, Manning CN, Schwartz AG, Thomopoulos S, Xia Y. Nanoscale. 2010;2:923–926. doi: 10.1039/c0nr00192a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.