Abstract
Cardiac Troponin I-interacting kinase (TNNI3K) is a cardiac specific kinase whose biological function remains largely unknown. We have recently shown that TNNI3K expression greatly accelerates cardiac dysfunction in mouse models of cardiomyopathy, indicating an important role in modulating disease progression. To further investigate TNNI3K kinase activity in vivo, we have generated transgenic mice expressing both wild-type and kinase-dead versions of the human TNNI3K protein. Importantly, we show that the increased TNNI3K kinase activity induces mouse cardiac remodeling, and its kinase activity promotes accelerated disease progression in a left-ventricular pressure overload model of mouse cardiomyopathy. Using an in vitro kinase assay and proteomics analysis, we show that TNNI3K is a dual-function kinase with Tyr and Ser/Thr kinase activity. TNNI3K expression induces a series of cellular and molecular changes, including a reduction of sarcomere length and changes in titin isoform composition, which are indicative of cardiac remodeling. Using antisera to TNNI3K, we show that TNNI3K protein is located at the sarcomere Z disc. These combined data suggest that TNNI3K mediates cell signaling to modulate cardiac response to stress.
Keywords: TNNI3K, kinase, cardiac remodeling
1. Introduction
Cardiac Troponin I-interacting kinase (TNNI3K) is a cardiac specific kinase [1]. It contains three recognizable domains/motifs: ten copies of an N-terminal ankyrin repeat followed by a protein kinase domain and a C-terminal Ser-rich domain. The overall domain structure of TNNI3K is similar to integrin-linked kinase (ILK) that regulates signaling pathways controlling cardiac growth, contractility and repair [2, 3]. To date, downstream targets of TNNI3K have not been identified, and no connection between TNNI3K and other signaling pathways has been established. A yeast two-hybrid screen with a C-terminal fragment of TNNI3K suggested interaction with several proteins, including cardiac Troponin I [1], the latter leading to the name troponin I-interacting kinase. But these putative protein-protein interactions have yet to be validated in the appropriate cellular context of mammalian cardiomyocytes. Although TNNI3K has been proposed as a potential pharmaceutical target of kinase inhibitors in the context of heart disease [4, 5], the role of the TNNI3K kinase activity in vivo remains unknown.
We recently determined that expression of Tnni3k modulates the course of disease progression in mouse cardiomyopathy models [6]. We found that levels of Tnni3k mRNA and protein vary widely among inbred mouse stains. Robust levels of TNNI3K protein are detected in C57BL/6J and other strains, but due to a mRNA splicing mutation, little or no protein is made in DBA/2J or other strains that effectively represent naturally-occurring Tnni3k-null mice [6]. In these healthy inbred mouse strains, neither extreme of Tnni3k expression results in cardiomyopathy. However, in the context of the calsequestrin (Csq) transgenic model of dilated cardiomyopathy, high expression of Tnni3k greatly accelerates the disease course, leading to premature heart failure and death. Tnni3k expression also accelerates disease progression in another mouse model of heart disease due to left ventricular pressure overload induced by transverse aortic constriction (TAC) [6].
By contrast, in a murine model of cardiac ischemia, intramyocardial transplantation of Tnni3k-overexpressing P19CL6 cells promotes cardiomyogenesis and improves cardiac function [7]. Additionally, the Tnni3k locus has also been mapped as a quantitative trait locus (QTL) for electrocardiographic parameters [8, 9] and for susceptibility to viral myocarditis [8, 9]. Thus, expression of Tnni3k may be detrimental in certain pathological conditions such as aberrant calcium regulation or pressure overload, but protective in other cardiomyopathic disease contexts. TNNI3K expression was recently found to be 6-fold higher in heart tissue from end-stage idiopathic dilated cardiomyopathy (DCM) patients as compared to healthy controls [10]. Although protein levels and activity have yet to be measured in human disease cohorts, these data are consistent with a potential role in modulating disease progression and outcome in human heart disease.
Given the pivotal role of TNNI3K in multiple forms of cardiomyopathy, the characterization of this protein is critical to a clear understanding of its in vivo function. Unfortunately, to date, there are few studies on the biochemical properties of TNNI3K protein [1, 7, 11–13]. Some clues to its properties can be gleaned from its domain structure suggesting that TNNI3K may be a dual-function kinase [1, 11]. However, TNNI3K’s closest relative in the kinome, ILK, has been the subject of debate, and is thought by some to be a “pseudo” kinase and serve primarily as an adapter protein [14]. Here, to further investigate TNNI3K kinase activity in vivo, we have generated transgenic mice expressing both wild-type [6] and here, kinase-dead versions of the human TNNI3K protein. Importantly, we show that the increased TNNI3K kinase activity induces cardiac remodeling, and the kinase activity promotes accelerated disease progression in a left-ventricular pressure overload model of mouse cardiomyopathy. Using an in vitro kinase assay and proteomics analysis, we show that TNNI3K is a dual-function kinase with Tyr and Ser/Thr kinase activity. Phosphorylation of TNNI3K increases after pressure overload stress. We propose that activated TNNI3K might induce cardiac remodeling by modulating sarcomere length and titin isoform composition.
2. Material and methods
2.1. Animal care and handling
All mice were handled according to approved protocols and animal welfare regulations of the Institutional Review Board at Duke University Medical Center. All inbred mouse strains used in the course of this study were obtained from Jackson Laboratory (Bar Harbor, ME). Transgenic miceTNNI3Ktg and TNKDtg were created as previously described [6] and bred and maintained on a DBA/2J genetic background that expresses almost no endogenous Tnni3k [6].
2.2. Cloning of TNNI3K constructs, cell culture and transfection
A full-length 2.5 kb human TNNI3K cDNA was amplified from normal human heart RNA following RT-PCR. Site-directed mutagenesis was used to change a single base in the hTNNI3K cDNA construct. The mutation, an ‘a’ to ‘g’, changed the AAA Lysine codon to an AGA Arginine codon at nucleotide position 1469/aa position 490. hTNNI3K cDNA was cloned into pRK5 with a Flag tag at the amino terminus. FLAG-TNNI3K 5’:GGGAATTCATGGACTACAAGGACGAC GACGACCAA GGAAATTATAAATCTAGACC; FLAG-TNNI3K 3’: GGGAATT CCGCCGAATGCT GTCAGC. Human embryonic kidney HEK293T (293T) cells (ATCC, Manassas, VA) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) containing 10% fetal bovine serum at 37°C in 5% CO2. HL-1 cardiomyocytes were cultured in Claycomb Medium (SAFC Laboratories, Lenexa, KS) supplemented with Fetal Bovine Serum at 10%, 2 mM L-Glutamine, 100mg/ml Penicillin/Streptomycin, and 100mM fungizone. Cells were grown on 35 mm2 plates and transfected with 1µg plasmid DNA using FuGene reagent (Roche, Indianapolis, IN) according to the manufacturer’s protocol.
2.3. Immunoblotting and gel electrophoresis
Whole heart protein lysates were prepared using flash-frozen heart tissue resuspended in lysis buffer with protease and phosphatase inhibitors. Lysates were analyzed by SDS-PAGE and western blotting was performed using standard methods. A polyclonal peptide antiserum (Bethyl Laboratories, Montgomery, TX) was raised against a human C-terminal TNNI3K peptide (FHSCRNSSSFEDSS). The antiserum was purified on a Protein A column (GenScript, Piscataway, NJ). TNNI3K antiserum was used at a 1:1000 dilution in TBST with 5% dry milk. Other primary antibodies were obtained from commercial sources; Mouse anti-Flag M2 (1:1000, sigma); PY99 (1:500, Santa Cruz); mouse anti-phosphoserine PSR-45 (1:200, Sigma); mouse anti-phosphothreonine-HRP (1:100, abcam); mouse anti-alpha tubulin (1:500, DSHB, U. of Iowa). Protein bands were visualized using secondary antibodies conjugated to HRP (1:3000, BioRad) followed by incubation with Pierce SuperSignal West Pico Chemiluminescant Substrate (Thermo Fisher Scientific, Rockford, IL) and exposure to X-OMAT film (Kodak). For titin protein analysis, SDS–agarose electrophoresis was performed as previously described [15]. Briefly, muscle samples were solubilized in a urea and glycerol buffer and analyzed by vertical SDS–agarose electrophoresis. The 1% agarose gels were run at 10 mA per gel for 4 hr. The gels were stained with Silver stain plus kit (BioRad).
2.4. Immunoprecipitation and in vitro kinase assay
Cell lysates were incubated with antibodies overnight at 4 °C, then with protein A/G conjugated agarose beads (30 µl, Santa Cruz) for 2 hours. The pellet was washed three times with lysis buffer. For the in vitro kinase assay, the pellet was washed with kinase buffer, then incubated in kinase buffer with 200µM ATP at 30°C for 30 min.
2.5. Creation of a TNNI3K kinase dead (TNKD) transgenic mouse
Two TNKD lines were generated in the identical manner as the previously-described wild-type TNNI3K transgenic lines [6]. Briefly, the TNKD cDNA was cloned downstream of the murine α-myosin heavy chain (αMHC) promoter. An artificial minx intron was inserted upstream of the TNNI3K start codon. The construct was linearized and an 8 kb fragment containing the αMHC promoter, the TNKD cDNA, and an SV40 polyadenylation sequence was purified and used for microinjection. B6SJLF1/J blastocysts were injected with the linearized transgene and subsequently implanted into surrogate mice. The resulting founder animals were genotyped for presence of the TNKD transgene using a 5’ primer in the αMHC promoter and a 3’ primer in the TNKD transgene. Two robustly-expressing transgenic lines were chosen for backcrossing to the DBA/2J mouse strain for 5 generations before the lines were used in the experiments described here. SYBR green qRT-PCR analysis of transgenic transcripts showed that levels of TNNI3K transgene expression in both wild-type and kinase dead transgenic mice ranged from 5 to 20-fold higher than endogenous murine Tnni3k measured in heart tissue from C57BL/6, a strain that expresses robust levels of the endogenous murine transcript [6].
2.6. Trypsin digestion and phosphopeptide enrichment
Flag-TNNI3K and Flag-TNKD protein were immunoprecipitated from transfected 293T cell lysates. Immunoprecipitated TNNI3K/TNKD protein was separated by SDS-PAGE. The corresponding protein bands on the SDS-PAGE gel were excised, chopped into small pieces and subjected to in-gel trypsin digestion. In brief, the gel pieces were destained by 25 mM ammonium bicarbonate in 50% acetonitrile. The proteins in the gel were reduced by dithiothreitol (DTT), alkylated by iodoacetamide (IAA), and then subjected to overnight trypsin digestion. Extracted peptides were lyophilized and resuspended in 150 µL of immobilized metal ion affinity chromatography (IMAC) wash/equilibration buffer (25mM formic acid, 40% acetonitrile). 25 µL of a 1:1 slurry of precharged IMAC resin [Fe(III)-loaded IMAC slurry; Phos-Select iron affinity gel] (Sigma-Aldrich) were added to the resuspended peptide sample [16, 17]. The IMAC resin was prewashed three times in 1mL of wash/equilibration buffer. Samples were agitated for 90 min at room temperature and washed three times with 150 µL of wash/equilibration buffer. Bound phosphopeptides were eluted twice with 45 µL of 50 mM KH2PO4/NH3 (pH 10.0) and acidified with 45 µL of 5% formic acid, 5% acetonitrile. The enriched phosphopeptides were resuspended in 40 µL of 5% formic acid and desalted on C18 resin, using handmade StageTips [18]. Peptides were eluted with 5% formic acid, 50% acetonitrile, lyophilized with a speed-vac, reconstituted in 0.1% trifluoroacetic acid, 2% acetonitrile, 25 mM citrate, and subjected to LC-MS/MS analysis.
2.7. LC/MS/MS analyses
LC/MS/MS analyses were performed on a Thermo Scientific LTQ Orbitrap XL (Thermo Scientific) with a Finnigan Nanospray II electrospray ionization source. Enriched phosphopeptides were injected onto a 75 µm × 150 mm BEH C18 column (particle size 1.7 µm, Waters) and separated using a Waters nano ACQUITY Ultra Performance LC™ (UPLC™) System (Waters, Milford, MA). The LTQ Orbitrap XL was operated in the data dependent mode using the TOP10 strategy [19]. In brief, each scan cycle was initiated with a full MS scan of high mass accuracy [375 –1,800 m/z; acquired in the Orbitrap XL at 6 × 104 resolution setting and automatic gain control (AGC) target of 106], which was followed by MS/MS scans (AGC target 5,000; threshold 3,000) in the linear ion trap on the 10 most abundant precursor ions. Selected ions were dynamically excluded for 30 s. Singly charged ions were excluded from MS/MS analysis. MS/MS spectra were searched against a composite database containing the human TNNI3K sequence using the SEQUEST algorithm. Search parameters allowed for three missed tryptic cleavages, a mass tolerance of ± 80 ppm, a static modification of 57.02146 Daltons (carboxyamidomethylation) on cysteine, and up to six total dynamic modifications: 79.96633 Dalton (phosphorylation) on serine, threonine, and tyrosine, 15.99491 Dalton (oxidation) on methionine. Matches for tryptic phosphopeptides were validated manually with special consideration of intense fragment ions formed through cleavage N-terminal to proline residues and neutral losses of phosphoric acid.
2.8. Phosphorylation site localization
The probability of the assignment of the correct position for each phosphorylation site was determined using an Ascore algorithm [20]. This algorithm considers all phosphoforms of a peptide and uses the presence or absence of experimental fragment ions unique to each to create an ambiguity score (Ascore). Parameters included a window size of 100-m/z units and a fragment ion tolerance of +/− 0.6 m/z units. Sites with an Ascore ≥13 (P ≤ 0.05) were considered to be confidently localized and those with an Ascore ≥19 (P ≤ 0.01) were considered to be localized with near certainty. If the same peptide was detected multiple times with the same site locations, the highest Ascore was selected for each site and reported.
2.9. Mouse RNA isolation and qRT-PCR
Whole hearts were removed from age- and sex-matched animals. Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA). cDNA was synthesized from 1µg total RNA using iScript (BioRad) and used as the template for qRT-PCR. GAPDH was used as the endogenous control. All amplifications were carried out in triplicate on an ABI Prism 7000 Real Time PCR system and analyzed with ABI software. All statistical analyses were done using an unpaired, one-tailed T-test. Nppb 5’ primer GAGGTCACTCCTATCCTCTGG; 3’ GCCATTTC CTCCGACTTTTCTC. Nppa 5’ GCTTCCAGGCCATATTGGAG; 3’ GGGGGCATGACCTCATCTT.
2.10. M-mode echocardiography
Transthoracic two-dimensional M-mode echocardiography was performed between 10 and 18 weeks of age in conscious mice using either a Vevo 770 echocardiograph (Visual Sonics, Toronto, Canada) or an HDI 5000 echocardiograph with a 15-MHz frequency probe (Phillips Electronics, Bothell, WA) as previously described [21]. Measurements of cardiac function included heart rate, posterior and septal wall thickness, left-ventricular end diastolic diameter (LVEDD) and left-ventricular end systolic diameter (LVESD). Fractional shortening (FS) was calculated with the formula: FS= (LVEDD-LVESD)/LVEDD, as previously described [22].
2.11. Transverse aortic constriction
Mice were anesthetized with a mixture of ketamine (100 mg/kg) and xylazine (2.5 mg/kg), and transverse aortic constriction (TAC) was performed as originally described[23]. TAC was performed on animals at 10 weeks of age. The mice were analyzed by echocardiography (as described above) just prior to surgery and at 4 and 8 weeks following surgery.
2.12. Histology and transmission electron microscopy
Hearts were perfused with 300 µl of 6.7% KCl and then fixed in 10% neutral buffered formalin, dehydrated in 75%, 90% and 100% ethanol, and embedded in paraffin. Sections were cut at 5 mm thickness and then stained with Masson’s trichrome stain. The percentage of fibrosis was quantified using the Frida image analysis software (http://bui3.win.ad.jhu.edu/frida/). For transmission electron microscopy, hearts were fixed overnight with 4% glutaraldehyde in 100 mM cacodylic acid (pH 7.4) at 4°C and post-fixed with OsO4 in cacodylate buffer. They were then dehydrated in a graded series of ethanol followed by propylene oxide, and embedded in PolyBed 812 resin (Polyscience, Warrington, PA). Ultrathin sections were cut on a Reichert-Jung ultramicrotome (Leica, Buffalo Grove, IL) and stained with uranyl acetate and lead citrate. Sections were examined in a Philips CM 12 transmission electron microscope (FEI, Hillsboro, OR). Images were collected digitally on an AMT XR100-2Vu camera (Advanced Microscopy Techniques, Woburn, MA).
2.13. Immunocytochemistry
Hearts were dissected and fixed overnight in 4% PFA-PBS. Antisera used included Rabbit anti-human TNNI3K (1: 50); Mouse anti-desmin (1:50, clone D33, DAKO); Mouse anti-myosin (1:50, DSHB, U. of Iowa). These were added to the blocking solution and were incubated by rocking at 4°C overnight. Samples were rinsed three times for 30 min in PBT (PBS and 0.1% Triton X-100) with 5% BSA and 0.1% heat-inactivated goat serum, and incubated overnight at 4 °C in blocking solution with Alexa Fluor 594 phalloidin, Alexa Fluor 488 and 594 secondary antibodies (1:500; Invitrogen). Samples were washed three times for 30 min in PBT then mounted in ProLong Gold antifade reagent with DAPI (Invitrogen) and imaged on a Zeiss LSM420 confocal microscope.
3. Results
3.1. TNNI3K kinase activity induces cardiac remodeling
We had previously generated transgenic mouse lines, TNNI3Ktg, expressing wild-type human TNNI3K from a cardiac-specific promoter [6]. To further investigate the role of TNNI3K kinase function in vivo, we have generated similar transgenic mouse lines expressing the kinase-dead version of the human TNNI3K, TNKDtg, which has one residue change at the canonical ATP binding site (490, Lys>Arg) [11]. As determined by quantitation of protein immunoblots of heart lysates over multiple generations (n>10) of each lineage, there is no significant difference in protein expression levels between the TNNI3Ktg and TNKDtg lines (FigureS1, p=0.46). Each of the TNNI3Ktg and TNKDtg transgenes have been introgressed into the DBA/2J inbred strain background that shows no detectable murine TNNI3K protein [6]. Thus, TNNI3K protein in these lines derives solely from the exogenous human transgene.
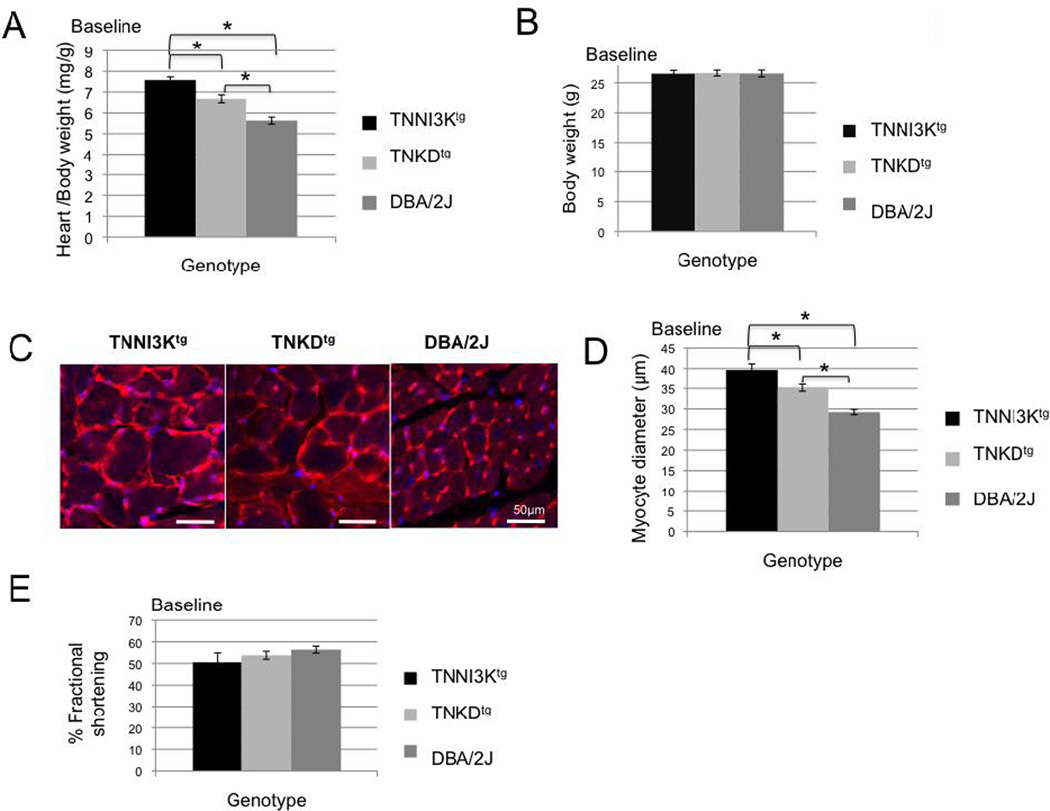
To study the in vivo effect of increased TNNI3K kinase activity, we examined the hearts from TNNI3Ktg, TNKDtg and DBA mice under normal physiological conditions, without any cardiac stress. We first measured the heart-to-body weight ratio of mice for all three strains. As we previously reported [6], we have not observed significant phenotypic differences between male and female mice. However, the measurements shown here were carried out on male mice at 2 months of age. The relative cardiac mass is significantly higher in the adult TNNI3Ktg mice compared to DBA and TNKDtg mice (Figure 1A, P<0.0001), although all the animals exhibit a similar body weight (Figure 1B, P>0.8). As seen in Figure 1C and D, cardiomyocytes from the TNNI3Ktg mice are significantly enlarged compared with that of the TNKDtg and DBA/2J mice (p<0.001). Cardiac function measured by echocardiography shows slight, but not statistically significant, decline in adult TNNI3Ktg mice (Figure 1E). Although transgene expression can vary among individual mice, the heart-to-body weight ratio is not significantly different between mice with low and high levels of the transgene expression (Figure S1). Instead, the differences in the resulting cardiac phenotypes correlate with genotype of the transgene (wild-type or kinase-dead).
Figure 1. TNNI3K kinase activity induces an increase of cardiac mass.
Heart weight and body weight of adult male DBA/2J (n=11), TNKDtg (n=11) and TNNI3Ktg (n=11) mice were measured at 2 months of age. Heart weight to body weight ratio of TNNI3Ktg mice is significantly higher than in the other strains (A, * p<0.0001), while body weight is similar among the three strains (B, p=0.97). (C, D) Heart sections from TNNI3Ktg, TNKDtg, and DBA/2J mice were stained with wheat germ agglutinin (C, red and counter stained with DAPI (blue)) and the diameter of the myocytes was measured (D). The diameter was significantly increased in the TNNI3Ktg mice. (n=60 measurements of 3 mice from each genotype, * P <0.001) Each bar represents 50 µm. (E) Echocardiography was performed for TNNI3Ktg mice (n=8), TNKDtg (n=7) and wild type littermate control male mice (n=7) at 2 months of age. There was no significant difference in contractile function in any of the lines, as shown by similar shortening (FS) among three groups (p=0.25 TNNI3Ktg vs DBA; p=0.53, TNNI3Ktg vs TNKDtg). Each data point is shown as mean± SEM.
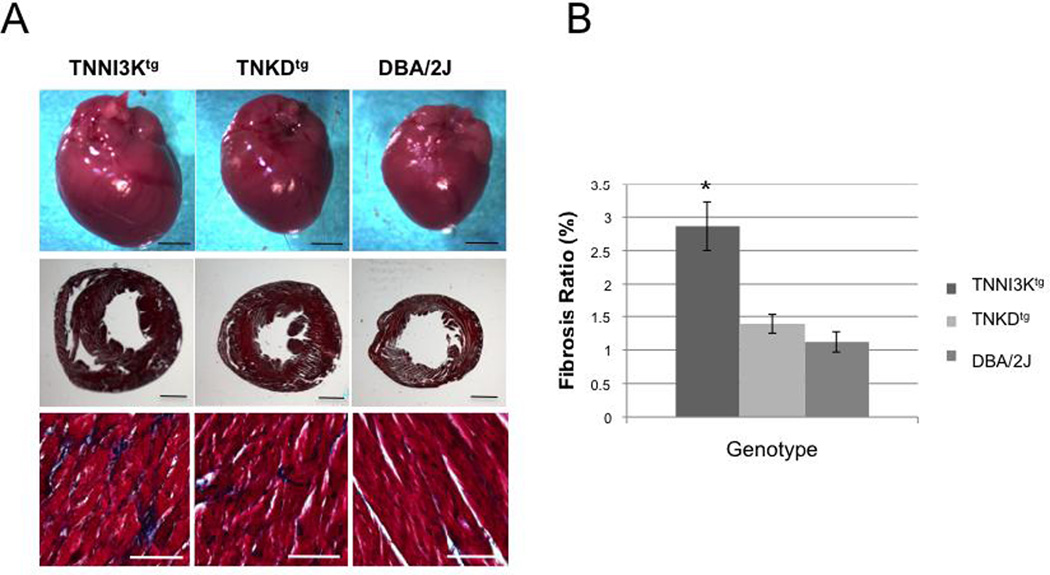
Consistent with the heart-to-body weight ratio, the gross morphology of whole hearts of adult TNNI3Ktg mice shows an increase in size relative to hearts from DBA and TNKDtg mice (Figure 2). In humans under persistent cardiac stress, cardiac hypertrophy frequently progresses to ventricular dilatation. Similarly, adult TNNI3Ktg mice show a slight dilatation of the ventricular chambers (Figure 2). One of the hallmarks of heart failure is fibrosis of the ventricular wall. As revealed by Masson Trichrome staining, the hearts of TNNI3Ktg mice contained more deposits of collagen than the other two strains (Figure 2, P<0.001).
Figure 2. TNNI3K kinase activity results in cardiac remodeling and increased fibrosis.
(A) Upper row: Representative gross morphology of whole hearts (upper rows) of adult (age 2 months, male) wild-type DBA/2J, TNKDtg and TNNI3Ktg mice. Middle row: Masson trichrome staining of transverse sections of left ventricular wall tissue of hearts from the three strains. Bottom row: High magnification of the tissue reveals excess collagen (blue) in the TNNI3Ktg mice. Each bar represents 2 mm in the upper two rows and 200 µm in the bottom row of photographs. (B) The percentage of area exhibiting fibrosis in the left ventricle is shown (* p<0.001).
Interestingly, TNKDtg mice also show a relative cardiac mass increase and slightly increased fibrosis compared to DBA/2J mice (Figure 1A–D, P<0.0001, and Figure 2), suggesting that other domains of TNNI3K, such as the ankyrin repeats, might also play a role in cardiac remodeling.
3.2. TNNI3K is an active dual-function kinase
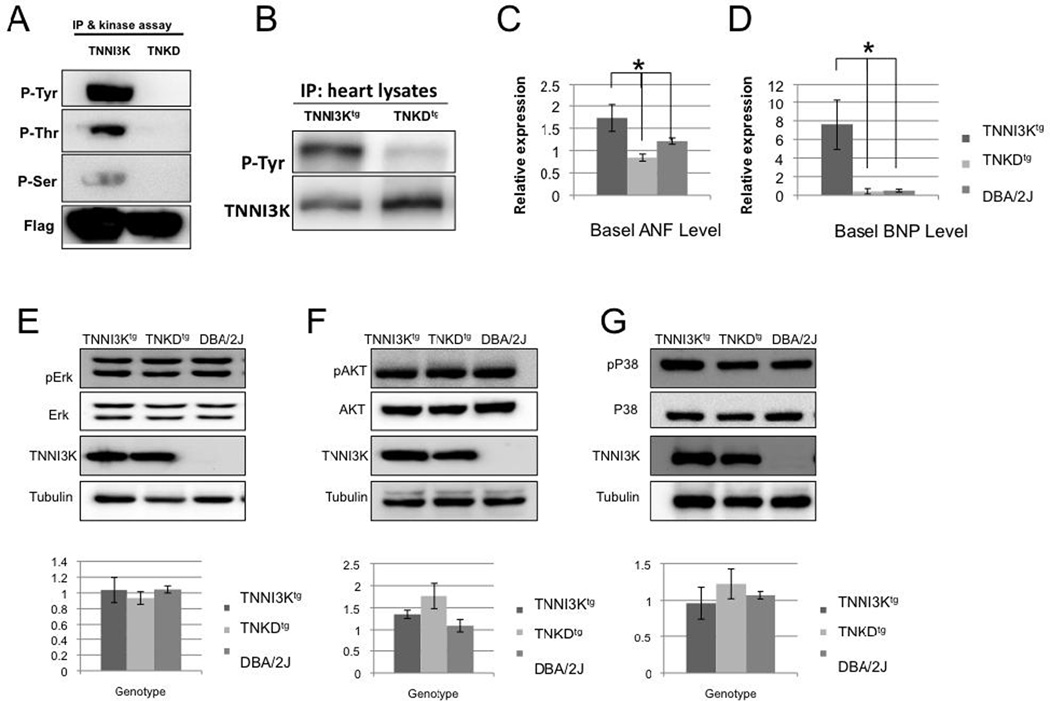
Based on a homology search against the mammalian “kinome” database, the sequence of the TNNI3K kinase domain places this novel kinase in the MAPKKK family [1, 24]. This suggests that like other family members, TNNI3K might be a dual-function kinase with both Tyr and Ser/Thr kinase activities [1]. In the absence of a validated downstream target of TNNI3K kinase activity, we investigated its kinase function using an in vitro kinase assay based on the previously described autophosphorylation of TNNI3K [11]. In vitro, wild type TNNI3K auto-phosphorylates at Tyr and also at Thr and Ser residues, indicating that it is a dual-function kinase (Figure 3A). The kinase domain includes an invariant residue at the canonical ATP binding site that allows the creation of a kinase-dead (490, Lys>Arg, TNKD) version of the protein[11]. All kinase activities are abolished in this kinase-dead version of human TNNI3K (Figure 3A).
Figure 3. TNNI3K is a dual-function kinase, and cardiac remodeling resulting from TNNI3K kinase activity is not mediated via the MAPK or AKT signaling pathways.
(A) An in vitro kinase assay monitoring autophosphorylation of TNNI3K indicates that TNNI3K is a dual-function (Tyr and Ser/Thr) kinase. Flag-tagged hTNNI3K or the flag-tagged kinase-dead mutant of hTNNI3K (TNKD) was transfected into 293T cells. Cell lysates were immunoprecipitated using an anti-Flag antibody and the lysate used in an in vitro kinase assay. Wild type TNNI3K exhibited autophosphorylation at Tyr, Ser, and Thr residues, as detected by the three distinct anti-phosphorylation antibodies. By contrast, autophosphorylation of TNKD was barely detectable. (B) TNNI3K is phosphorylated in vivo, and the kinase-dead mutation abolishes TNNI3K autophosphorylation at Tyr residues. The heart lystates from the TNNI3Ktg and TNKDtg transgenic mice were immunoprecipitated using an anti-human TNNI3K antibody and then immunoblotted with a pan phosphotyrosine antibody. Phospho-Tyr antiserum detects strong signals in the TNNI3Ktg heart lysates, but not in lysates from TNKDtg hearts. (C, D) Expression of markers of cardiac hypertrophy is increased in TNNI3Ktg mice relative to TNKDtg and wild type DBA/2J mice. Message levels were determined by qRT-PCR of RNA from hearts (n=3 for each genotype) of the indicated genotypes for (C), ANF and (D), BNP. Both ANF (p=0.023) and BNP (p=0.029) were increased in the TNNI3Ktg when compared to the TNKDtg transgenic animals (*P < 0.05). (E)TNNI3K kinase activity does not activate ERK, AKT or p38 pathways. Heart lysates of TNNI3Ktg, TNKDtg and wild type DBA/2J mice were immunoblotted with antisera against total and phosphorylated ERK, AKT and p38 [25]. No significant phosphorylation increase was detected for any of the three effectors (p>0.05, n>=3 hearts from each strains). Each data point is shown as mean± SEM.
Paralleling the in vitro data, heart lysates from adult TNNI3Ktg mice demonstrate increased TNNI3K phosphorylation at Tyr residues when compared to the non-transgenic DBA/2J strain, suggesting that autophosphorylation also occurs in vivo. This interpretation is supported by lower TNNI3K phosphorylation at Tyr residues in the TNKDtg mice (Figure 3B).
To further identify autophosphorylation sites of TNNI3K, we analyzed in vitro phosphorylated TNNI3K protein by mass spectrometry. Flag-TNNI3K and Flag-TNKD protein were immunoprecipitated from transfected 293T cell lysates followed by the in vitro kinase assay. After Trypsin digestion, extracted peptides from both proteins were analyzed by Liquid chromatography–tandem mass spectrometry (LC/MS/MS). In the Flag-TNNI3K sample, multiple phosphorylated peptides were identified. The phosphorylated residues (Y24, T399, Y416, Y425, T622, S737, S739, S741, Y771, Y804, T805, Y812) are enriched in the ankyrin repeats and Ser-rich tail domain (Table 1, Figure S2), which are predicted to be involved in protein-protein interaction and kinase activity. These sites may be the targets of other kinases/phosphatases and/or involved in binding with other proteins (Table S2). By contrast, in the Flag-TNKD protein sample, only three phosphorylated peptides were identified (S427, S430, S737). Phosphorylated residues that are only detected in the wild type TNNI3K protein are potential autophosphorylation sites.
TABLE 1. The phosphoryltion sites of TNNI3K identified by phophoproteomic analysis.
Flag-TNNI3K and Flag-TNKD protein were immunoprecipitated from transfected 293T cell lysates followed by the in vitro kinase assay. After Trypsin digestion, extracted peptides from both proteins were analyzed by Liquid chromatography–tandem mass spectrometry (LC/MS/MS).
| Ascore Seq | Ascore | Site | |
|---|---|---|---|
| TNKD | RPQDELPCNEYSQPGGDGSYVS#VPSPLGK | 31.9 | S427 |
| TNKD | RPQDELPCNEYSQPGGDGSYVSVPS#PLGK | 33.5 | S430 |
| TNKD | LEECLCNIELMSPASSNSS#GSLSPSSSSDCLVNR | 8.7 | S737 |
| TNKD | LEECLCNIELMSPASSNSS#GSLSPSSSSDCLVNR | 9.4 | S737 |
| TNKD | LEECLCNIELMSPASSNSS#GSLSPSSSSDCLVNR | 20.9 | S737 |
| TNKD | RSLQY#T#PIDK | 5.1/4.1 4 | Y804/T805 |
| TNNI3K | VSESY#VITIER | 19.3 | Y24 |
| TNNI3K | KVSESY#VITIER | 32.4 | Y24 |
| TNNI3K | GHDAIVT#LLK | 1000 | T399 |
| TNNI3K | RPQDELPCNEY#SQPGGDGSY#VSVPSPLGK | 13.3/27.6 | Y416/Y425 |
| TNNI3K | RPQDELPCNEY#SQPGGDGSYVSVPSPLGK | 15.9 | Y416 |
| TNNI3K | RPQDELPCNEY#SQPGGDGSY#VSVPSPLGK | 28.5/6.4 | Y416/Y425 |
| TNNI3K | RPQDELPCNEYSQPGGDGSY#VSVPSPLGK | 12.3 | Y425 |
| TNNI3K | RPQDELPCNEYSQPGGDGSY#VSVPSPLGK | 14 | Y425 |
| TNNI3K | RPQDELPCNEYSQPGGDGSY#VSVPSPLGK | 27.7 | Y425 |
| TNNI3K | FLQSLDEDNMT#KQPGNLR | 33.5 | T622 |
| TNNI3K | LEECLCNIELMSPASSNSS#GSLSPSSSSDCLVNR | 34.5 | S737 |
| TNNI3K | LEECLCNIELMSPASSNSSGS#LSPSSSSDCLVNR | 26.8 | S739 |
| TNNI3K | LEECLCNIELMSPASSNSSGSLS#PSSSSDCLVNR | 19.4 | S741 |
| TNNI3K | SRFELEY#ALNAR | 5.2 | Y771 |
| TNNI3K | SRFELEY#ALNAR | 16.5 | Y771 |
| TNNI3K | SRFELEY#ALNAR | 88.6 | Y771 |
| TNNI3K | SRFELEY#ALNAR | 107.7 | Y771 |
| TNNI3K | FELEY#ALNAR | 1000 | Y771 |
| TNNI3K | SY#AALSQSAGQYSSQGLSLEEMKR | 0 | Y778 |
| TNNI3K | SLQY#TPIDK | 32.4 | Y804 |
| TNNI3K | SLQYT#PIDK | 28.9 | T805 |
| TNNI3K | Y#GYVSDPMSSMHFHSCR | 11 | Y810 |
| TNNI3K | Y#GYVSDPMSSMHFHSCR | 11.2 | Y810 |
| TNNI3K | YGY#VSDPMSSMHFHSCR | 26.8 | Y812 |
| TNNI3K | YGY#VSDPMSSMHFHSCR | 27 | Y812 |
| TNNI3K | SLQYTPIDKYGY#VSDPMSSMHFHSCR | 27 | Y812 |
| TNNI3K | YGY#VSDPMSSMHFHSCR | 35.8 | Y812 |
| TNNI3K | YGY#VSDPMSSMHFHSCR | 44.7 | Y812 |
# Phosphorylation sites at Y24, T399, Y416, Y425, T622, S737, S739, S741, Y771, Y804, T805, Y812 were identified in wild type TNNI3K protein; Phosphorylation sites at S427, S430 and S737 were identified in TNKD protein.
Each phosphorylation site was measured using an Ascore algorithm. Ascore ≥13 (P ≤ 0.05) is considered confident, and Ascore ≥19 (P ≤ 0.01) is considered near certainty.
3.3. TNNI3K overexpression induces a series of molecular and cellular changes in the cardiomyocyte that leads to cardiac remodeling
We next sought to determine whether increased TNNI3K kinase activity induces cardiac remodeling by modulating one of the known cardiac intracellular signaling pathways. We first examined expression of two prototypical markers of hypertrophy, Nppa (which encodes atrial natriuretic factor - ANF) and Nppb (which encodes brain natriuretic peptide - BNP). Consistent with the hypertrophic phenotype, ANF and BNP are both elevated only in the TNNI3Ktg line (Figure 3C,D). In the heart, p38, ERKs and AKT are important effectors transducing cardiac remodeling signals [25]. We analyzed the phosphorylation status of p38, ERK 1/2 and AKT from heart lysates of TNNI3Ktg, TNKDtg and DBA mice (Figure 3E). However, the phosphorylation states of these three effectors did not show significant changes in hearts from TNNI3Ktg mice compared to the other two strains (p>0.1), suggesting that TNNI3K may not directly regulate these effectors.
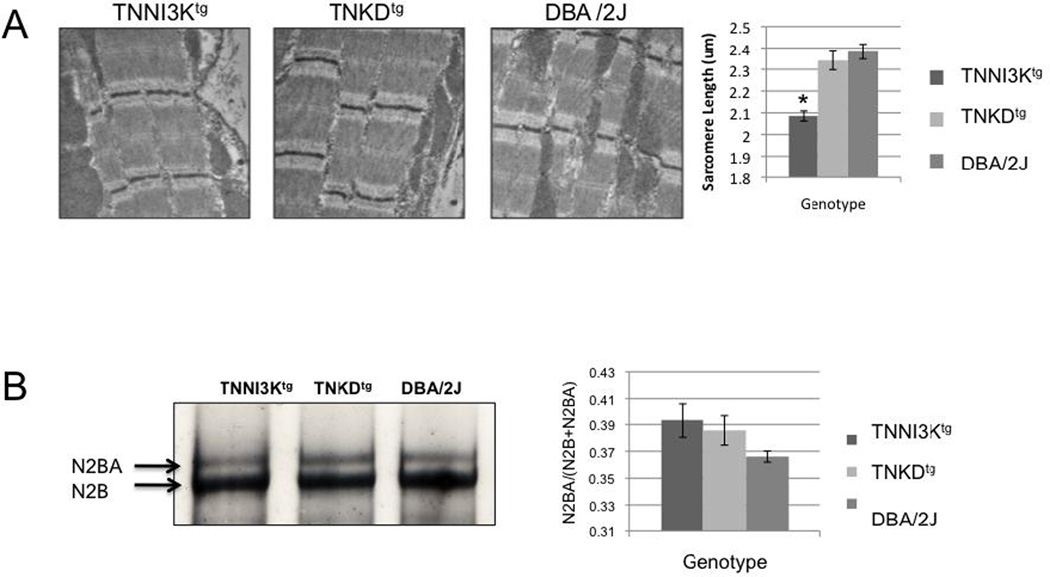
We further examined the ultrastructure of cardiomyocytes using electron microscopy to determine whether overexpressed TNNI3K protein would interrupt the structure of the contractile unit. No gross sarcomeric defect was observed in the hearts of any of the three strains (Figure 4A), suggesting that neither the protein, nor its kinase activity is required for the integrity of the sarcomeric structure. However, the resting sarcomere length is significantly reduced in TNNI3Ktg compared to TNKDtg and DBA/2J mice (p<0.0001). Reduced sarcomere length has been observed in other mouse models of cardiomyopathy [26, 27].
Figure 4. TNNI3K expression reduces sarcomere length and changes titin isoform composition.
(A) Representative images of transmission electron microscopy in resting cardiomyocytes from TNNI3Ktg, TNKDtg and DBA mice showing normal sarcomeric integrity. However, the resting sarcomere length of TNNI3Ktg (n=27 measurements) is significantly shorter than TNKDtg (n=31) and DBA/2J mice (n=28) (* p<0.0001). (B) Titin isoform expression. 1% vertical agarose protein gel of lysates from the TNNI3Ktg, TNKDtg transgenic and DBA/2J mouse hearts reveal the N2BA and N2B titin isoforms. Increased N2BA to N2B isoform ratios are seen in both transgenic animals (n=4 mice for each strain and repeat 3 times, p=0.09 and 0.15) compared to the DBA control mice.
In light of the above, we examined the expression pattern of the protein isoforms of titin in the context of TNNI3K genotypes. Titin functions as a molecular spring that develops passive force in sarcomeres during mechanical stretch [28]. Two main isoforms are co-expressed in the adult heart, the N2B isoform that is stiff (3.0 MDa) and the larger N2BA isoform that is more compliant (3.4 MDa). Changes in titin stiffness occur during heart disease via a shift in the expression ratio of the two main titin isoforms [29–32]. We found a trend towards an increase of the N2BA ratio in TNNI3Ktg and TNKDtg mice when compared to DBA/2J mice, although the difference was not statistically significant (Figure 4B, p=0.09 and 0.15 respectively). This shift is consistent with results from human cardiomyopathy patients and the pressure overload mouse cardiomyopathy model [29–31].
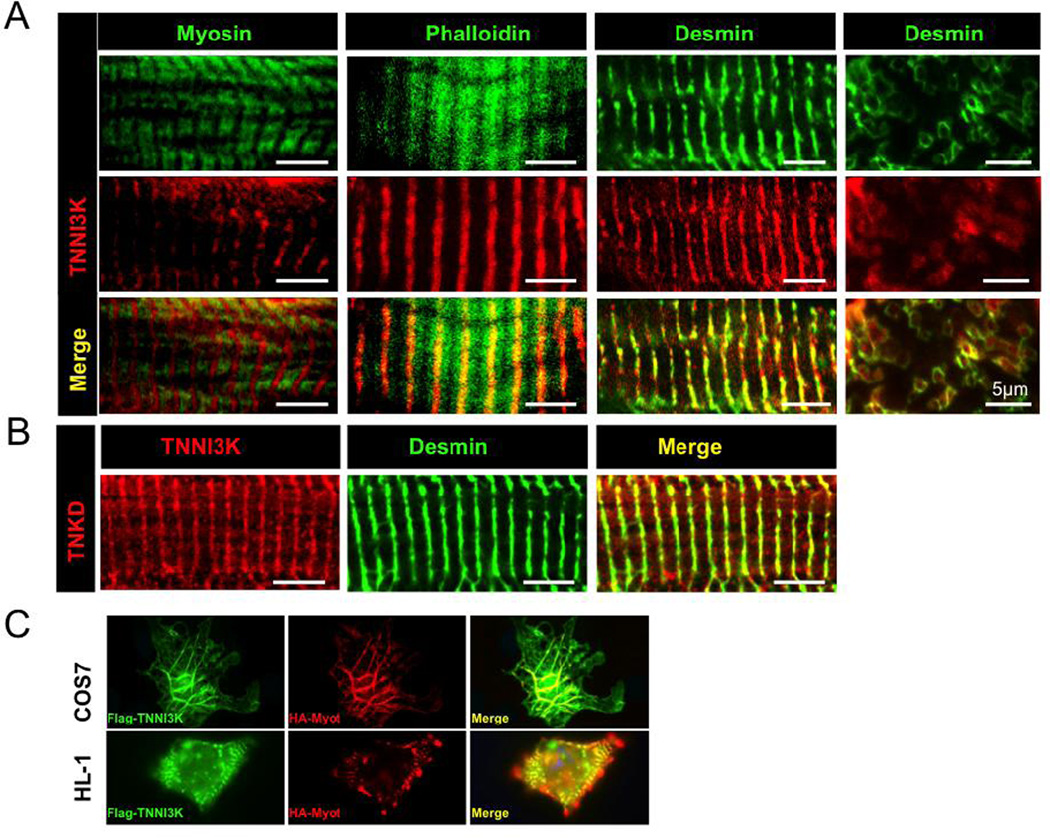
3.4. TNNI3K localizes to the sarcomeric Z disc independent of its kinase activity
To determine the intracellular localization of TNNI3K in cardiomyocytes from heart tissue, we performed immunostaining on cryosections of TNNI3Ktg adult mouse hearts using antibodies against human C-terminal TNNI3K and markers for the various sarcomeric components (Figure 5A). TNNI3K shows a reciprocal (out-of-register) staining pattern with myosin that forms the sarcomere thick filaments, is centrally distributed along the actin thin filaments, and nearly perfectly overlaps with desmin, the intermediate filament protein surrounding the Z disc [33]. In cross-section, TNNI3K localizes inside the desmin ring structures. Endogenous mouse TNNI3K exhibits the same striated expression pattern in C57BL/6J mouse heart tissue; a pattern not seen in hearts from strain DBA/2J that express no TNNI3K protein [6] (Figure S3). Using TNKDtg mouse hearts, we found that the kinase-dead protein shows an identical staining pattern to the wild-type transgene (Figure 5B), suggesting that the kinase activity of TNNI3K is not required for its Z disc localization. The striated staining pattern in cardiac muscle tissue suggested to us that TNNI3K might also be present in striated skeletal muscle, but this was not the case (Figure S3), consistent with previous reports that the protein is cardiac specific [1].
Figure 5. TNNI3K localizes to the sarcomeric Z disc in cardiomyocytes, and its kinase activity is not required for its localization.
(A) TNNI3K localizes to the sarcomeric Z disc. TNNI3Ktg transgenic mouse heart sections were co-immunostained with antisera against human TNNI3K (red) and other sarcomeric proteins (green). In longitudinal sections of sarcomeres, TNNI3K shows a reciprocal staining pattern with myosin (green), partially overlaps with F-actin (phalloidin, green), and co-localizes with desmin (green), the intermediate filaments surrounding the Z disc. In cross-section, TNNI3K localizes inside the desmin ring structures. (B) TNNI3K kinase activity is not required for Z disc localization. TNKDtg transgenic mouse heart sections were co-immunostained using antisera against human TNNI3K (red) and desmin (green). The lack of TNNI3K kinase activity does not alter its localization. Each bar represents 5µm. (C) Co-localization of Flag-hTNNI3K and HA-hMyotilin in transfected cells grown in culture. COS7 or HL-1 cells were transfected with Flag-hTNNI3K and HA-hMyotilin. Immunostaining with anti-Flag (green) and anti-HA (red) antisera shows the co-localization of Flag-hTNNI3K and HA-hMyotilin in transfected cells (100× magnification).
In order to further investigate the cellular location of TNNI3K, we co-expressed flag-tagged TNNI3K and HA-tagged myotilin (a component of the sarcomere Z disc) in either COS7 cells or HL-1 cells, the latter a cardiac muscle cell line. Using antisera to the epitope tags for visualization, both proteins co-localized in the cells with a filamentous (COS7) or striated (HL-1) staining pattern (Figure 5C). These data obtained with epitope-tagged protein in cell culture are consistent with our data obtained with TNNI3K-specific antisera in mouse heart tissue.
Peri-nuclear staining was seen in C57BL/6J mouse heart tissue (Figure S3), but not in TNNI3Ktg transgenic mice using the antibody against human TNNI3K, or in transfected cell lines using an anti-Flag anitibody (Figure 5C). This suggests that perinuclear staining may represent non-specific background. We do not see any staining within the nucleus (Figure S3), which contrasts with a previous report [1]. We conclude that TNNI3K is located at the sarcomere Z disc of the cardiomyocyte.
3.5. TNNKI3K kinase activity is essential to accelerating mechanical stress-induced cardiomyopathic disease progression
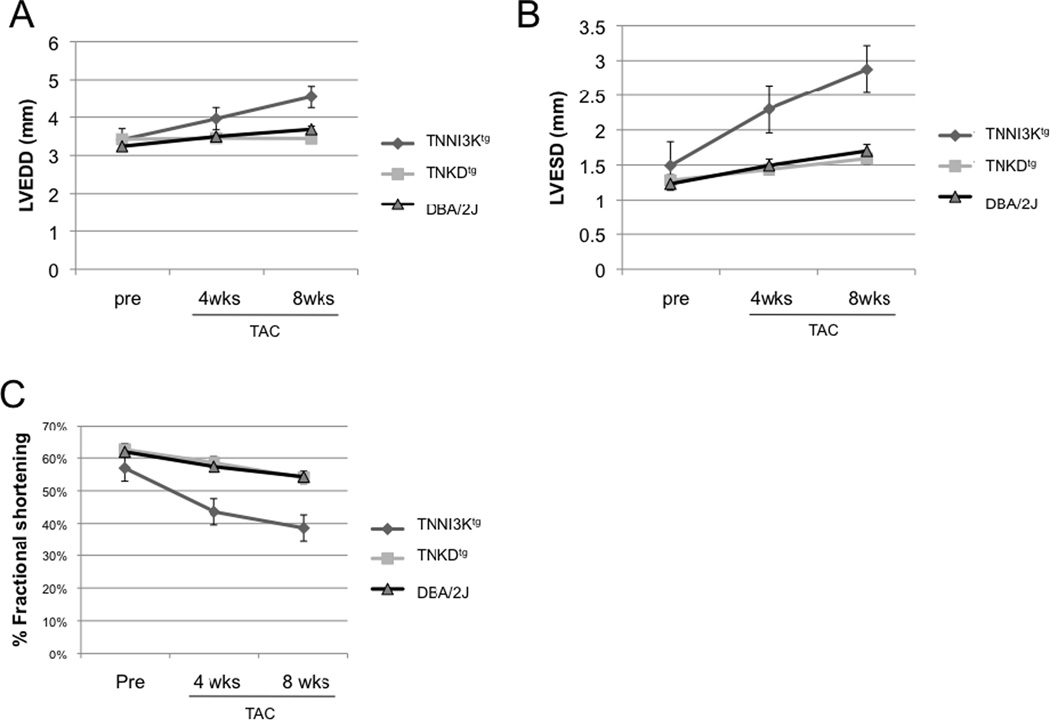
We had previously shown that transgenic expression of TNNI3K accelerates disease progression in two different mouse models of cardiomyopathy [6]. To determine whether TNNI3K kinase activity is important for this disease-accelerating effect, we compared the effects of left-ventricular pressure overload on adult TNNI3Ktg and TNKDtg mice. We employed surgical transthoracic aortic constriction (TAC) at 10 weeks of age to induce pressure overload and monitored cardiac function by echocardiography prior to and at 4 and 8 weeks post surgery. As expected [34], cardiac function in all TAC animals declined 4 and 8 weeks post-surgery (Figure 6, S4, Table S1). However, in mice expressing wild-type TNNI3K, left-ventricular dysfunction progressed more rapidly than non-transgenic controls (p=0.012 at 4 weeks; p=0.006 at 8 weeks), while left-ventricular dysfunction in TNKDtg mice mirrored that of the non-transgenic controls (p=0.597 at 4 weeks; p=0.402 at 8 weeks). These results demonstrate that TNNI3K kinase activity is essential to the disease accelerating properties of TNNI3K in biomechanical stress-induced cardiomyopathy.
Figure 6. TNNI3K kinase activity accelerates left-ventricular dysfunction in the transthoracic aortic constriction model of cardiomyopathy.
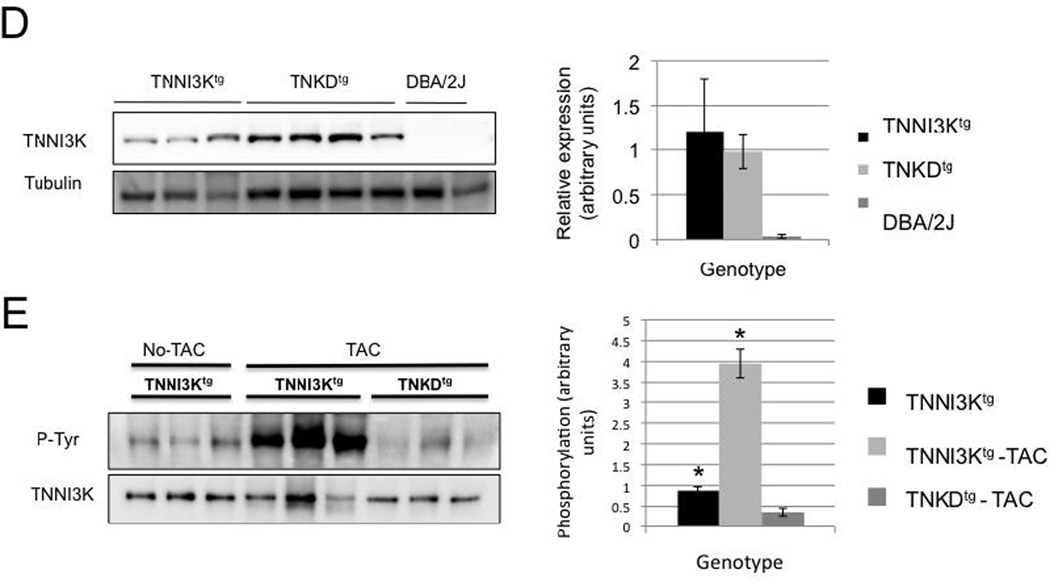
(A–C) Echocardiography was performed prior to transverse aortic constriction (TAC) and at 4 and 8 weeks post TAC surgery for TNNI3Ktg mice (n=12), TNKDtg (n=15) and wild type littermate control mice (n=27 in two groups of 13 and 14). The surgeries for the two transgenic genotypes were staggered over the course of many months. In order to control for unforeseen variables in surgical technique, each line of transgenic mice was compared to a new control group that underwent surgery at the same time. As expected, there was no evidence of decreased contractile function in any of the lines prior to surgery, as shown by similar fractional shortening (FS) among three groups (p=0.21 TNNI3Ktg vs DBA; p=0.71, TNKDtg vs DBA). After surgery, the TNNI3Ktg mice showed a more rapid decline in fractional shortening than non-transgenic controls (p=0.012 at 4 weeks; p=0.006 at 8 weeks). By contrast, disease progression in the TNKDtg mice was identical to non-transgenic littermate control mice (p=0.597 at 4 weeks; p=0.402 at 8 weeks). Each data point is shown as mean± SEM. (D) Representative immmunoblots of the heart lystates from the TNNI3Ktg, TNKDtg transgenic and DBA/2J mice are shown. Each lane represents heart lysate from a different individual animal performed with TAC surgery, and was immunoblotted with an anti human TNNI3K antibody and an anti tubulin antibody. A quantitative result of relative transgenic protein expression is shown. (E) The heart lysates of the TNNI3Ktg and TNKDtg mice at 8 weeks post-TAC surgery and non-surgery TNNI3Ktg control mice were immunoprecipitated using an anti-human TNNI3K antibody and then immunoblotted with a pan phosphotyrosine antibody. Phosphorylation of Tyr is significantly higher in post-TAC TNNI3Ktg mice compared to TNKDtg mice and non-surgery control mice (p<0.001). Each lane represents the heart lysate from a different individual animal. TNNI3K transgene protein was measured by anti-TNNI3K antiserum.
Importantly, TAC-induced cardiomyopathy was associated with increased TNNI3K phosphorylation levels of TNNI3Ktg mice compared to non-surgical controls or TNKDtg mice (Figure 6D,E). This result indicates that the kinase activity of TNNI3K is elevated by signals induced by mechanical stress, and that TNNI3K further accelerates the cardiac dysfunction in this pressure overload model of disease.
4. Discussion
Although TNNI3K was first identified as a cardiac specific kinase in 2003 [1], and recent data from mouse models of cardiomyopathy demonstrates a critical role in disease progression [6], its biochemical characteristics and biological function remain largely unknown. This study reveals the importance of the kinase activity of TNNI3K, and suggests testable hypothesis concerning its function in vivo.
First, we have demonstrated that TNNI3K kinase activity induces cardiac remodeling under normal physiological conditions, and accelerates disease progression in a left-ventricular pressure overload model of mouse cardiomyopathy. We also showed that TNNI3K is a dual function kinase both in vitro and in vivo. These data suggest that the kinase activity is essential for TNNI3K function, but that in many cases, high level of kinase activity leads to cardiac pathology. This further supports the potential of TNNI3K as a pharmaceutical target of kinase inhibitors in heart disease.
Although the downstream target(s) of TNNI3K kinase activity remain to be identified, using mass spectrometry, we have identified several potential autophosphorylation sites. Several sites are located near the end of the ankyrin repeat domain. Phosphorylation in the ankyrin repeat domain might interfere with protein-protein interaction [35]. Recent studies reveal that cardiac proteins with ankyrin repeat domains, such as CARP and ILK [2, 36, 37], play important roles in cardiac function through interaction with other proteins. Our TNKDtg transgenic mice, which lack TNNI3K kinase activity, also show signs of cardiomyopathy, though less severe compared to the TNNI3Ktg mice. These include an increase in cardiac mass and a possible change in titin isoform composition, suggesting that TNNI3K’s ankyrin repeat domain may likewise form a complex with other proteins to modulate sarcomere function. Several other sites are clustered in the C-terminal Serine-rich tail of the protein. In other kinases, C-terminal Ser-rich tails are negative regulatory domains [38]. In support of a similar role for this domain in TNNI3K, deletion of the Ser-rich tail of TNNI3K has been shown to augment autophosphorylation in vitro [11]. Additionally, one potential autophosphorylation site (T622) is located in a region that by sequence conservation would be predicted to be located within the activation loop [39]. By analogy with other kinases, phosphorylation at this site might also critically regulate TNNI3K kinase activity. The identification of these putative autophosphorylation sites provides the molecular groundwork for future studies on its biological role in normal heart function, and in cardiomyopathy. We propose that TNNI3K kinase function may be activated by cardiac stress signals via an upstream kinase/phosphatase. Additionally, TNNI3K auto-phosphorylation activity in vivo suggests a feedback regulatory loop that might modulate its function. The phosphorylation states of these sites might be altered under different physiological conditions. Thus, TNNI3K activation may be critical to its role in regulating downstream signaling pathways related to the stress response.
To date, the up-stream and down-stream signaling pathways of TNNI3K remain unknown. TNNI3K shares an overall domain order and structure with ILK, and its kinase domain belongs to the mitogen-activated protein kinase kinase kinase (MAPKKK) family [1]. In the heart, MAPK pathways play critical roles in stress-induced cardiac remodeling [25]. Importantly, our data on overexpressed TNNI3K and TNKD transgenic mice suggests that TNNI3K kinase activity might be involved in a hypertrophic response pathway. However, activities of three important hypertrophic effecters, ERK, p38 and AKT, do not show significant change with increased TNNI3K kinase activity in vivo, suggesting that the hypertrophic program induced by TNNI3K overexpression does not rely heavily on these effectors. Recently, Lai et al. have also shown that ERK phosphorylation is not significantly changed, while p38 phosphorylation is slightly decreased in a P19CL6-derived cardiomyocyte cell line with overexpression of TNNI3K [7].
In TNNI3Ktg mice, we observed a series of molecular and cellular changes that are related to sarcomere dysfunction. Although the underlying mechanism remains unknown, sarcomere length reduction has also been reported in two other cardiomyopathy transgenic mouse models that express specific Troponin T or myosin heavy chain mutations originally identified in patients [26, 27]. Interestingly, we observed an increase of the N2BA/N2B titin isoform ratio in both TNKDtg and TNNI3Ktg cardiomyocytes, relative to DBA controls. This shift is consistent with findings in human dilated cardiomyopathy and ischemic heart disease patients as well as the TAC mouse model [29–31]. Increased expression of N2BA-isoforms reduces the passive force of the sarcomere and depresses titin-based stiffness in cardiomopathic hearts [29–31]. Altogether, these data suggest that TNNI3K induces cellular and molecular changes in the cardiomyocyte that alter sarcomere function eventually leading to cardiac remodeling and an acceleration of cardiac dysfunction under stress.
Since TNNI3K kinase activity accelerates disease progression in a left-ventricular pressure overload model of mouse cardiomyopathy, TNNI3K may also respond to biomechanical stretch signals. Such a role is consistent with our data showing that TNNI3K is localized at the cardiac sarcomere Z disc, which not only anchors the actin thin filaments, but also “senses” mechanical stretch [40]. LIM-domain protein MLP (muscle LIM protein), which is anchored to the Z-disc, is thought to function as an internal stretch sensor through a complex with telethonin and titin [40, 41]. Additionally, the integrin-interacting molecule melusin has been implicated as a sensor of mechanical stress in cardiomyocytes [42]. Thus, TNNI3K might phosphorylate components of this pathway to modulate and amplify the stress signals.
TNNI3K expression is increased six-fold over healthy controls in heart tissue from severe idiopathic dilated cardiomyopathy patients [10], providing additional evidence that increased TNNI3K kinase activity is associated with heart disease. This clinical data is consistent with its disease-accelerating properties in the murine models of cardiomyopathy, where the naturally occurring null allele is the protective sequence variant, and the risk allele is the variant that expresses robust levels of TNNI3K protein [6]. To date however, TNNI3K mutations have not been reported in patients with heart disease. Extrapolating from the murine and human data, loss-of-function mutations in the gene would be predicted to have minimal or possibly no phenotypic consequences under normal physiological conditions. Gain-of-function mutations, particularly those that increase or constitutively-activate TNNI3K kinase function, would be predicted to induce cardiomyopathy or accelerate disease progression. Idiopathic cardiomyopathy patients exhibiting a precipitous decline in cardiac dysfunction might reveal such mutations. Additionally, TNNI3K activity might play different roles in other disease contexts [8, 9], such as ischemic cardiac injury [7].
Despite the advent of significant therapeutic advances for cardiomyopathy in recent decades, heart disease remains the leading cause of mortality in developed countries [43]. Thus, there is a major unmet need for new therapies. We have previously shown that TNNI3K is cardiac-specific kinase that modulates disease progression in different mouse models of cardiomyopathy. Given that TNNI3K kinase activity is critical for its disease-accelerating capacity in these murine models, further study of this protein in cardiomyopathy patients may reveal its relevance to human heart disease.
Supplementary Material
Highlights.
The increased TNNI3K kinase activity induces mouse cardiac remodeling;
Its kinase activity promotes accelerated disease progression in a model of mouse cardiomyopathy;
TNNI3K is a dual-function kinase, and is located at the sarcomere Z disc.
Acknowledgements
We thank Christopher Clayton and Barbara Williams for animal husbandry and technical assistance. We thank Dr. Steven P Gygi at Harvard Medical School for the use of Ascore software and for useful discussion. We thank Dr. Teng-Yi Huang for technical assistance with proteomics sample preparation. This work was supported by NIH grants HL083155 to DAM, HL056687 to HAR, 2P01-HL-075443 to HAR and KX, and AHA 11POST7600109 to HT.
Abbreviations
- TNNI3K
Cardiac Troponin I-interacting kinase
- TAC
transverse aortic constriction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
H.A.R is cofounder of Trevena INC., a company developing drugs targeting G protein-coupled receptors.
References
- 1.Zhao Y, Meng XM, Wei YJ, Zhao XW, Liu DQ, Cao HQ, Liew CC, Ding JF. Cloning and characterization of a novel cardiac-specific kinase that interacts specifically with cardiac troponin I. J Mol Med. 2003 May;81(5):297–304. doi: 10.1007/s00109-003-0427-x. [DOI] [PubMed] [Google Scholar]
- 2.Lu H, Fedak PW, Dai X, Du C, Zhou YQ, Henkelman M, Mongroo PS, Lau A, Yamabi H, Hinek A, Husain M, Hannigan G, Coles JG. Integrin-linked kinase expression is elevated in human cardiac hypertrophy and induces hypertrophy in transgenic mice. Circulation. 2006 Nov 21;114(21):2271–2279. doi: 10.1161/CIRCULATIONAHA.106.642330. [DOI] [PubMed] [Google Scholar]
- 3.Hannigan GE, Coles JG, Dedhar S. Integrin-linked kinase at the heart of cardiac contractility, repair, and disease. Circ Res. 2007 May 25;100(10):1408–1414. doi: 10.1161/01.RES.0000265233.40455.62. [DOI] [PubMed] [Google Scholar]
- 4.Luft FC. Hearts of this ILK rely on TNNI3K, a MAPKKK that regulated TNNI3. J Mol Med. 2003 May;81(5):279–280. doi: 10.1007/s00109-003-0436-9. [DOI] [PubMed] [Google Scholar]
- 5.Lai ZF. TNNI3K could be a novel molecular target for the treatment of cardiac diseases. Recent Pat Cardiovasc Drug Discov. 2009 Nov;4(3):203–210. doi: 10.2174/157489009789152285. [DOI] [PubMed] [Google Scholar]
- 6.Wheeler FC, Tang H, Marks OA, Hadnott TN, Chu PL, Mao L, Rockman HA, Marchuk DA. Tnni3k modifies disease progression in murine models of cardiomyopathy. PLoS Genet. 2009 Sep;5(9):e1000647. doi: 10.1371/journal.pgen.1000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai ZF, Chen YZ, Feng LP, Meng XM, Ding JF, Wang LY, Ye J, Li P, Cheng XS, Kitamoto Y, Monzen K, Komuro I, Sakaguchi N, Kim-Mitsuyama S. Overexpression of TNNI3K, a cardiac-specific MAP kinase, promotes P19CL6-derived cardiac myogenesis and prevents myocardial infarction-induced injury. Am J Physiol Heart Circ Physiol. 2008 Aug;295(2):H708–H716. doi: 10.1152/ajpheart.00252.2008. [DOI] [PubMed] [Google Scholar]
- 8.Scicluna BP, Tanck MW, Remme CA, Beekman L, Coronel R, Wilde AA, Bezzina CR. Quantitative trait loci for electrocardiographic parameters and arrhythmia in the mouse. J Mol Cell Cardiol. 2011 Mar;50(3):380–389. doi: 10.1016/j.yjmcc.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Wiltshire SA, Leiva-Torres GA, Vidal SM. Quantitative trait locus analysis, pathway analysis, and consomic mapping show genetic variants of Tnni3k, Fpgt, or H28 control susceptibility to viral myocarditis. J Immunol. 2011 Jun 1;186(11):6398–6405. doi: 10.4049/jimmunol.1100159. [DOI] [PubMed] [Google Scholar]
- 10.Colak D, Kaya N, Al-Zahrani J, Al Bakheet A, Muiya P, Andres E, Quackenbush J, Dzimiri N. Left ventricular global transcriptional profiling in human end-stage dilated cardiomyopathy. Genomics. 2009 Jul;94(1):20–31. doi: 10.1016/j.ygeno.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y, Cao HQ, Liu Z, Ding JF, Meng XM. Identification of the dual specificity and the functional domains of the cardiac-specific protein kinase TNNI3K. Gen Physiol Biophys. 2007 Jun;26(2):104–109. [PubMed] [Google Scholar]
- 12.Feng Y, Liu DQ, Wang Z, Liu Z, Cao HQ, Wang LY, Shi N, Meng XM. AOP-1 interacts with cardiac-specific protein kinase TNNI3K and down-regulates its kinase activity. Biochemistry (Mosc) 2007 Nov;72(11):1199–1204. doi: 10.1134/s0006297907110053. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Wang H, Ye J, Xu RX, Song L, Shi N, Zhang YW, Chen X, Meng XM. Adenovirus-mediated overexpression of cardiac troponin I-interacting kinase promotes cardiomyocyte hypertrophy. Clin Exp Pharmacol Physiol. 2011 Apr;38(4):278–284. doi: 10.1111/j.1440-1681.2011.05499.x. [DOI] [PubMed] [Google Scholar]
- 14.Lange A, Wickstrom SA, Jakobson M, Zent R, Sainio K, Fassler R. Integrin-linked kinase is an adaptor with essential functions during mouse development. Nature. 2009 Oct 15;461(7266):1002–1006. doi: 10.1038/nature08468. [DOI] [PubMed] [Google Scholar]
- 15.Warren CM, Krzesinski PR, Greaser ML. Vertical agarose gel electrophoresis and electroblotting of high-molecular-weight proteins. Electrophoresis. 2003 Jun;24(11):1695–1702. doi: 10.1002/elps.200305392. [DOI] [PubMed] [Google Scholar]
- 16.Xiao K, Sun J, Kim J, Rajagopal S, Zhai B, Villen J, Haas W, Kovacs JJ, Shukla AK, Hara MR, Hernandez M, Lachmann A, Zhao S, Lin Y, Cheng Y, Mizuno K, Ma'ayan A, Gygi SP, Lefkowitz RJ. Global phosphorylation analysis of beta-arrestin-mediated signaling downstream of a seven transmembrane receptor (7TMR) Proc Natl Acad Sci U S A. 2010 Aug 24;107(34):15299–15304. doi: 10.1073/pnas.1008461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nobles KN, Xiao K, Ahn S, Shukla AK, Lam CM, Rajagopal S, Strachan RT, Huang TY, Bressler EA, Hara MR, Shenoy SK, Gygi SP, Lefkowitz RJ. Distinct Phosphorylation Sites on the {beta}2-Adrenergic Receptor Establish a Barcode That Encodes Differential Functions of {beta}-Arrestin. Sci Signal. 2011 Aug 9;4(185):ra51. doi: 10.1126/scisignal.2001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003 Feb 1;75(3):663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 19.Haas W, Faherty BK, Gerber SA, Elias JE, Beausoleil SA, Bakalarski CE, Li X, Villen J, Gygi SP. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol Cell Proteomics. 2006 Jul;5(7):1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006 Oct;24(10):1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 21.Cho MC, Rapacciuolo A, Koch WJ, Kobayashi Y, Jones LR, Rockman HA. Defective beta-adrenergic receptor signaling precedes the development of dilated cardiomyopathy in transgenic mice with calsequestrin overexpression. J Biol Chem. 1999 Aug 6;274(32):22251–22256. doi: 10.1074/jbc.274.32.22251. [DOI] [PubMed] [Google Scholar]
- 22.Esposito G, Santana LF, Dilly K, Cruz JD, Mao L, Lederer WJ, Rockman HA. Cellular and functional defects in a mouse model of heart failure. Am J Physiol Heart Circ Physiol. 2000 Dec;279(6):H3101–H3112. doi: 10.1152/ajpheart.2000.279.6.H3101. [DOI] [PubMed] [Google Scholar]
- 23.Rockman HA, Ross RS, Harris AN, Knowlton KU, Steinhelper ME, Field LJ, Ross J, Jr, Chien KR. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8277–8281. doi: 10.1073/pnas.88.18.8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002 Dec 6;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 25.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006 Aug;7(8):589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 26.Tardiff JC, Hewett TE, Palmer BM, Olsson C, Factor SM, Moore RL, Robbins J, Leinwand LA. Cardiac troponin T mutations result in allele-specific phenotypes in a mouse model for hypertrophic cardiomyopathy. J Clin Invest. 1999 Aug;104(4):469–481. doi: 10.1172/JCI6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsson MC, Palmer BM, Stauffer BL, Leinwand LA, Moore RL. Morphological and functional alterations in ventricular myocytes from male transgenic mice with hypertrophic cardiomyopathy. Circ Res. 2004 Feb 6;94(2):201–207. doi: 10.1161/01.RES.0000111521.40760.18. [DOI] [PubMed] [Google Scholar]
- 28.Fukuda N, Terui T, Ishiwata S, Kurihara S. Titin-based regulations of diastolic and systolic functions of mammalian cardiac muscle. J Mol Cell Cardiol. 2010 May;48(5):876–881. doi: 10.1016/j.yjmcc.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Neagoe C, Kulke M, del Monte F, Gwathmey JK, de Tombe PP, Hajjar RJ, Linke WA. Titin isoform switch in ischemic human heart disease. Circulation. 2002 Sep 10;106(11):1333–1341. doi: 10.1161/01.cir.0000029803.93022.93. [DOI] [PubMed] [Google Scholar]
- 30.Hudson B, Hidalgo C, Saripalli C, Granzier H. Hyperphosphorylation of mouse cardiac titin contributes to transverse aortic constriction-induced diastolic dysfunction. Circ Res. 2011 Sep 30;109(8):858–866. doi: 10.1161/CIRCRESAHA.111.246819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makarenko I, Opitz CA, Leake MC, Neagoe C, Kulke M, Gwathmey JK, del Monte F, Hajjar RJ, Linke WA. Passive stiffness changes caused by upregulation of compliant titin isoforms in human dilated cardiomyopathy hearts. Circ Res. 2004 Oct 1;95(7):708–716. doi: 10.1161/01.RES.0000143901.37063.2f. [DOI] [PubMed] [Google Scholar]
- 32.Nagueh SF, Shah G, Wu Y, Torre-Amione G, King NM, Lahmers S, Witt CC, Becker K, Labeit S, Granzier HL. Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation. 2004 Jul 13;110(2):155–162. doi: 10.1161/01.CIR.0000135591.37759.AF. [DOI] [PubMed] [Google Scholar]
- 33.Frank D, Kuhn C, Katus HA, Frey N. The sarcomeric Z-disc: a nodal point in signalling and disease. J Mol Med. 2006 Jun;84(6):446–468. doi: 10.1007/s00109-005-0033-1. [DOI] [PubMed] [Google Scholar]
- 34.Rockman HA, Wachhorst SP, Mao L, Ross J., Jr ANG II receptor blockade prevents ventricular hypertrophy and ANF gene expression with pressure overload in mice. Am J Physiol. 1994 Jun;266(6 Pt 2):H2468–H2475. doi: 10.1152/ajpheart.1994.266.6.H2468. [DOI] [PubMed] [Google Scholar]
- 35.Ranganathan P, Vasquez-Del Carpio R, Kaplan FM, Wang H, Gupta A, Vanwye JD, Capobianco AJ. Hierarchical Phosphorylation within the Ankyrin Repeat Domain Defines a Phosphoregulatory Loop That Regulates Notch Transcriptional Activity. J Biol Chem. 2011 Aug 19;286(33):28844–28857. doi: 10.1074/jbc.M111.243600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller MK, Bang ML, Witt CC, Labeit D, Trombitas C, Watanabe K, Granzier H, McElhinny AS, Gregorio CC, Labeit S. The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J Mol Biol. 2003 Nov 7;333(5):951–964. doi: 10.1016/j.jmb.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 37.Duboscq-Bidot L, Charron P, Ruppert V, Fauchier L, Richter A, Tavazzi L, Arbustini E, Wichter T, Maisch B, Komajda M, Isnard R, Villard E. Mutations in the ANKRD1 gene encoding CARP are responsible for human dilated cardiomyopathy. Eur Heart J. 2009 Sep;30(17):2128–2136. doi: 10.1093/eurheartj/ehp225. [DOI] [PubMed] [Google Scholar]
- 38.Raveh T, Berissi H, Eisenstein M, Spivak T, Kimchi A. A functional genetic screen identifies regions at the C-terminal tail and death-domain of death-associated protein kinase that are critical for its proapoptotic activity. Proc Natl Acad Sci U S A. 2000 Feb 15;97(4):1572–1577. doi: 10.1073/pnas.020519497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor SS, Radzio-Andzelm E. Three protein kinase structures define a common motif. Structure. 1994 May 15;2(5):345–355. doi: 10.1016/s0969-2126(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 40.Knoll R, Hoshijima M, Hoffman HM, Person V, Lorenzen-Schmidt I, Bang ML, Hayashi T, Shiga N, Yasukawa H, Schaper W, McKenna W, Yokoyama M, Schork NJ, Omens JH, McCulloch AD, Kimura A, Gregorio CC, Poller W, Schaper J, Schultheiss HP, Chien KR. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002 Dec 27;111(7):943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- 41.Arber S, Hunter JJ, Ross J, Jr, Hongo M, Sansig G, Borg J, Perriard JC, Chien KR, Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997 Feb 7;88(3):393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- 42.Brancaccio M, Fratta L, Notte A, Hirsch E, Poulet R, Guazzone S, De Acetis M, Vecchione C, Marino G, Altruda F, Silengo L, Tarone G, Lembo G. Melusin, a muscle-specific integrin beta1-interacting protein, is required to prevent cardiac failure in response to chronic pressure overload. Nat Med. 2003 Jan;9(1):68–75. doi: 10.1038/nm805. [DOI] [PubMed] [Google Scholar]
- 43.Mudd JO, Kass DA. Tackling heart failure in the twenty-first century. Nature. 2008 Feb 21;451(7181):919–928. doi: 10.1038/nature06798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.