Abstract
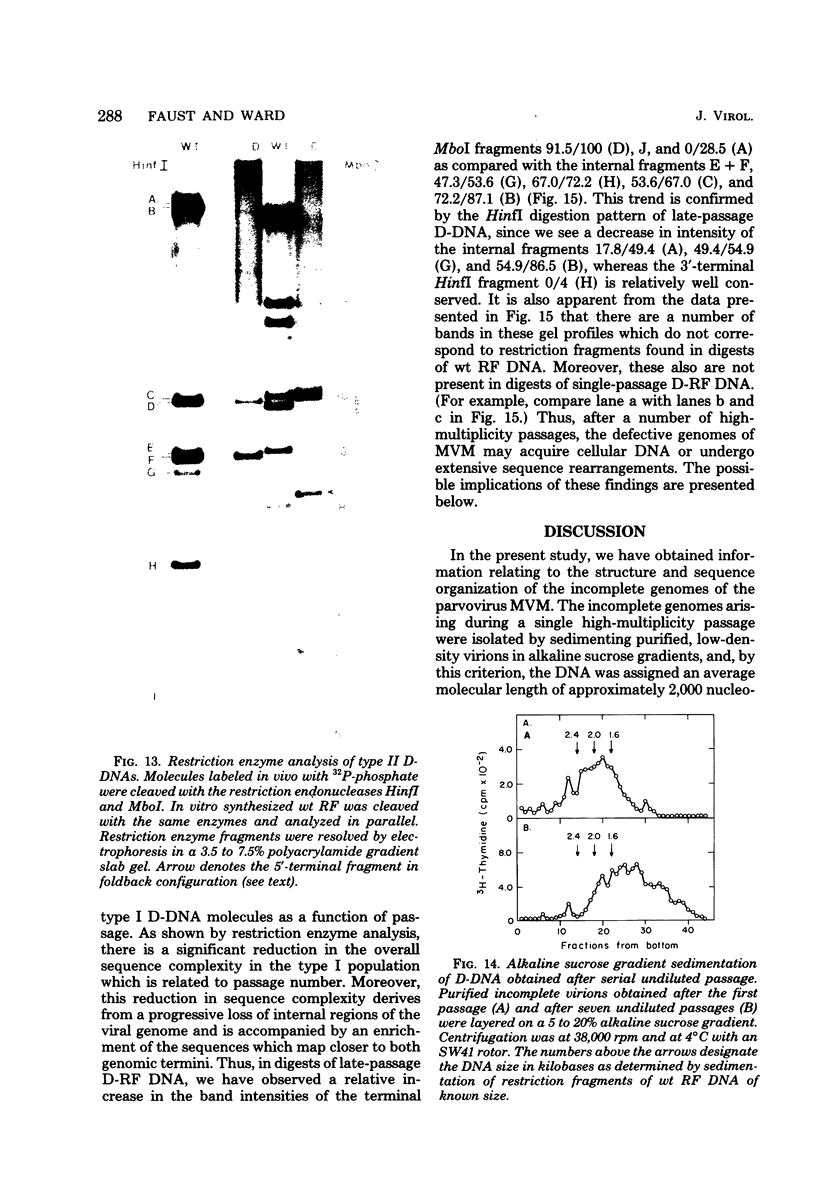
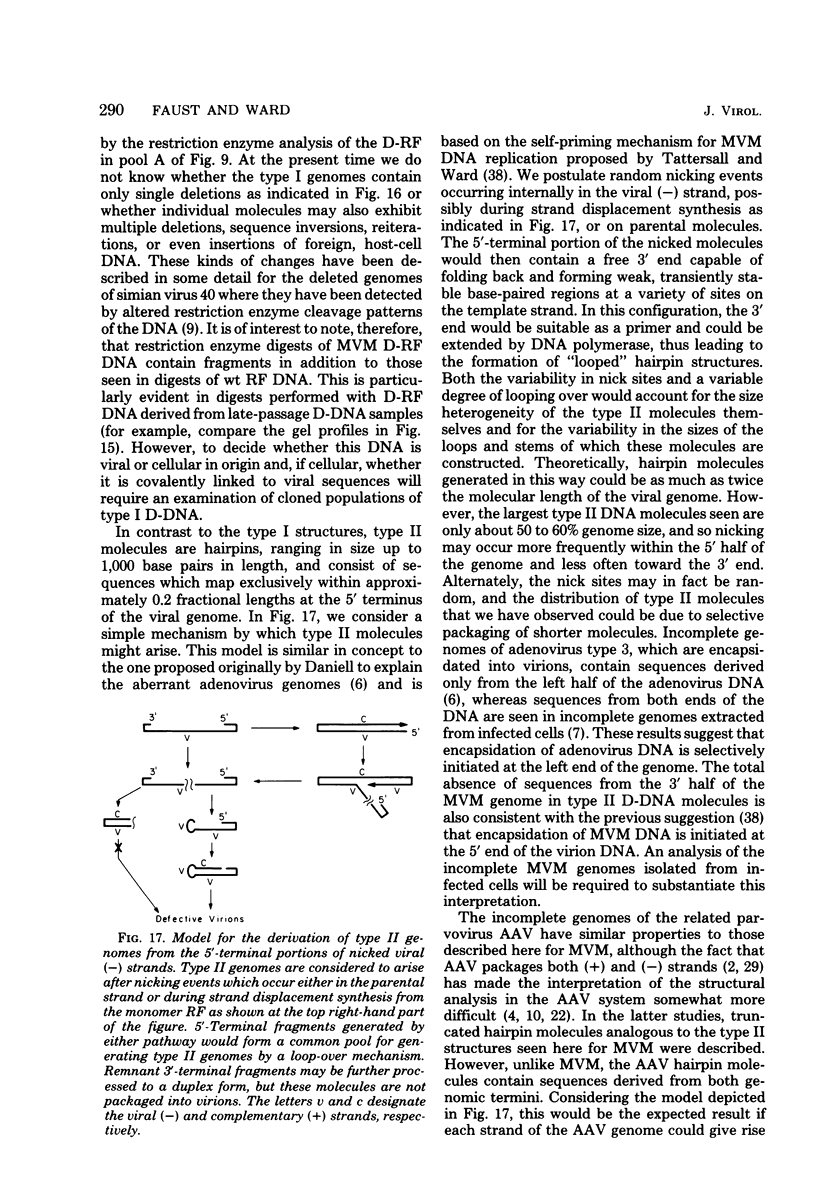
Deletion mutants of minute virus of mice arising during a single high-multiplicity passage and after serial undiluted passage have been isolated, and the incomplete viral genomes contained therein have been analyzed. The DNA isolated from incomplete virions derived from a single high-multiplicity passage was heterogeneous, ranging in size from 15 to 70% of the intact viral genome, with an average molecular length of approximately, 2,000 nucleotides. Two distinct types of molecules, designated as type I D-DNA and type II D-DNA, could be distinguished on the basis of their degree of secondary structure, and these were present in roughly equal amounts. Type I D-DNAs were predominantly single-stranded, recombinant molecules in which the self-complementary sequences derived from both genomic termini were conserved. The 5' terminus was modified relative to the analogous wild-type structure. Although virtually all of the wild-type genome sequence was seen in the total type I D-DNA population, sequences which map between coordinates 47.3 and 87.1 were clearly underrepresented. However, the extent and position of the deletions in individual molecules varied significantly. The shortest molecules in the population lacked between 90 and 95% of the internal wild-type genome sequence and consisted of sequences derived almost exclusively from within 5.0 map units (250 nucleotides) at both ends of the viral genome. Moreover, these miniature recombinant molecules were selectively amplified during serial undiluted passage and were therefore believed to contain all of the critical recognition sites necessary for the replication of minute virus of mice viral DNA. Type II D-DNAs were virus-specific, double-stranded hairpin molecules whose complementary strands were covalently continuous at variable sites distal to the 5' end of the viral minus strand. In sharp contrast to the type I genomes, these hairpin molecules consisted of sequences which mapped entirely at the 5' end of the viral genome between positions 85.0 and 100. Furthermore, type II molecules were gradually lost from the total D-DNA population during serial undiluted passage, suggesting that these molecules are not competent for DNA Replication but arise as the result of fatal replication errors. Deletion mutants of the type described here for minute virus of mice should be valuable generally as aids to future studies on parvovirus DNA replication, transcription, and cell-virus interactions.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Astell C. R., Smith M., Chow M. B., Ward D. C. Sequence of the 3' terminus of the genome from Kilham rat virus, a nondefective parvovirus. Virology. 1979 Jul 30;96(2):669–674. doi: 10.1016/0042-6822(79)90127-2. [DOI] [PubMed] [Google Scholar]
- Astell C. R., Smith M., Chow M. B., Ward D. C. Structure of the 3' hairpin termini of four rodent parvovirus genomes: nucleotide sequence homology at origins of DNA replication. Cell. 1979 Jul;17(3):691–703. doi: 10.1016/0092-8674(79)90276-9. [DOI] [PubMed] [Google Scholar]
- Berns K. I., Adler S. Separation of two types of adeno-associated virus particles containing complementary polynucleotide chains. J Virol. 1972 Feb;9(2):394–396. doi: 10.1128/jvi.9.2.394-396.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon G. J., Tattersall P. J., Ward D. C. DNA of minute virus of mice: self-priming, nonpermuted, single-stranded genome with a 5'-terminal hairpin duplex. J Virol. 1976 Oct;20(1):290–306. doi: 10.1128/jvi.20.1.290-306.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B. J., Laughlin C. A., de la Maza L. M., Myers M. Adeno-associated virus autointerference. Virology. 1979 Jan 30;92(2):449–462. doi: 10.1016/0042-6822(79)90149-1. [DOI] [PubMed] [Google Scholar]
- Daniell E. Genome structure of incomplete particles of adenovirus. J Virol. 1976 Aug;19(2):685–708. doi: 10.1128/jvi.19.2.685-708.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell E., Mullenbach T. Synthesis of defective viral DNA in HeLa cells infected with adenovirus type 3. J Virol. 1978 Apr;26(1):61–70. doi: 10.1128/jvi.26.1.61-70.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli D., Ganem D., Nussbaum A. L., Fareed G., Howley P. M., Khoury G., Martin M. A. Genome Structures of reiteration mutants of simian virus 40. Virology. 1977 Mar;77(1):110–124. doi: 10.1016/0042-6822(77)90411-1. [DOI] [PubMed] [Google Scholar]
- Fife K. H., Berns K. I., Murray K. Structure and nucleotide sequence of the terminal regions of adeno-associated virus DNA. Virology. 1977 May 15;78(2):475–477. doi: 10.1016/0042-6822(77)90124-6. [DOI] [PubMed] [Google Scholar]
- Ganem D., Nussbaum A. L., Davoli D., Fareed G. C. Propagation of a segment of bacteriophage lamda-DNA in monkey cells after covalent linkage to a defective simian virus 40 genome. Cell. 1976 Mar;7(3):349–359. doi: 10.1016/0092-8674(76)90164-1. [DOI] [PubMed] [Google Scholar]
- Hamer D. H., Davoli D., Thomas C. A., Jr, Fareed G. C. Simian virus 40 carrying an Escherichia coli suppressor gene. J Mol Biol. 1977 May 15;112(2):155–182. doi: 10.1016/s0022-2836(77)80137-x. [DOI] [PubMed] [Google Scholar]
- Hauswirth W. W., Berns K. I. Adeno-associated virus DNA replication: nonunit-length molecules. Virology. 1979 Feb;93(1):57–68. doi: 10.1016/0042-6822(79)90275-7. [DOI] [PubMed] [Google Scholar]
- Hauswirth W. W., Berns K. I. Origin and termination of adeno-associated virus DNA replication. Virology. 1977 May 15;78(2):488–499. doi: 10.1016/0042-6822(77)90125-8. [DOI] [PubMed] [Google Scholar]
- Jeppesen P. G. A method for separating DNA fragments by electrophoresis in polyacrylamide concentration gradient slab gels. Anal Biochem. 1974 Mar;58(1):195–207. doi: 10.1016/0003-2697(74)90458-8. [DOI] [PubMed] [Google Scholar]
- Kowalski D., Kroeker W. D., Laskowski M., Sr Mung bean nuclease I. Physical, chemical, and catalytic properties. Biochemistry. 1976 Oct 5;15(20):4457–4463. doi: 10.1021/bi00665a019. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Khoury G. Deletion mutants of simian virus 40 defective in biosynthesis of late viral mRNA. Proc Natl Acad Sci U S A. 1979 Jan;76(1):71–75. doi: 10.1073/pnas.76.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeveld S. A., van Mansfeld A. D., Baas P. D., Jansz H. S., van Arkel G. A., Weisbeek P. J. Nucleotide sequence of the origin of replication in bacteriophage phiX174 RF DNA. Nature. 1978 Feb 2;271(5644):417–420. doi: 10.1038/271417a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Laughlin C. A., Myers M. W., Risin D. L., Carter B. J. Defective-interfering particles of the human parvovirus adeno-associated virus. Virology. 1979 Apr 15;94(1):162–174. doi: 10.1016/0042-6822(79)90446-x. [DOI] [PubMed] [Google Scholar]
- Little J. W. An exonuclease induced by bacteriophage lambda. II. Nature of the enzymatic reaction. J Biol Chem. 1967 Feb 25;242(4):679–686. [PubMed] [Google Scholar]
- Mulligan R. C., Howard B. H., Berg P. Synthesis of rabbit beta-globin in cultured monkey kidney cells following infection with a SV40 beta-globin recombinant genome. Nature. 1979 Jan 11;277(5692):108–114. doi: 10.1038/277108a0. [DOI] [PubMed] [Google Scholar]
- Rhode S. L., 3rd Defective interfering particles of parvovirus H-1. J Virol. 1978 Aug;27(2):347–356. doi: 10.1128/jvi.27.2.347-356.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode S. L., 3rd Replication process of the parvovirus H-1. VI. Characterization of a replication terminus of H-1 replicative-form DNA. J Virol. 1977 Feb;21(2):694–712. doi: 10.1128/jvi.21.2.694-712.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. J., Bellett J. D. A circular DNA-protein complex adenoviruses and its possible role in DNA replication. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):523–531. doi: 10.1101/sqb.1974.039.01.064. [DOI] [PubMed] [Google Scholar]
- Rose J. A., Berns K. I., Hoggan M. D., Koczot F. J. Evidence for a single-stranded adenovirus-associated virus genome: formation of a DNA density hybrid on release of viral DNA. Proc Natl Acad Sci U S A. 1969 Nov;64(3):863–869. doi: 10.1073/pnas.64.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Singer I. I., Rhode S. L., 3rd Replication process of the parvovirus H-1. VII. Electron microscopy of replicative-form DNA synthesis. J Virol. 1977 Feb;21(2):713–723. doi: 10.1128/jvi.21.2.713-723.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda E., Kimura G., Miura K. Similarity of nucleotide sequences around the origin of DNA replication in mouse polyoma virus and simian virus 40. Proc Natl Acad Sci U S A. 1978 Jan;75(1):162–166. doi: 10.1073/pnas.75.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian K. N., Dhar R., Weissman S. M. Nucleotide sequence of a fragment of SV40 DNA that contains the origin of DNA replication and specifies the 5' ends of "early" and "late" viral RNA. III. Construction of the total sequence of EcoRII-G fragment of SV40 DNA. J Biol Chem. 1977 Jan 10;252(1):355–367. [PubMed] [Google Scholar]
- Tattersall P., Cawte P. J., Shatkin A. J., Ward D. C. Three structural polypeptides coded for by minite virus of mice, a parvovirus. J Virol. 1976 Oct;20(1):273–289. doi: 10.1128/jvi.20.1.273-289.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall P. Replication of the parvovirus MVM. I. Dependence of virus multiplication and plaque formation on cell growth. J Virol. 1972 Oct;10(4):586–590. doi: 10.1128/jvi.10.4.586-590.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall P., Ward D. C. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature. 1976 Sep 9;263(5573):106–109. doi: 10.1038/263106a0. [DOI] [PubMed] [Google Scholar]
- Van Ventrooij W. J., Henshaw E. C., Hirsch C. A. Nutritional effects on the polyribosome distribution and rate of protein synthesis in Ehrlich ascites tumor cells in culture. J Biol Chem. 1970 Nov 25;245(22):5947–5953. [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]