Abstract
The steady state concentration of lactate was compared in an inducible Her2/nue transgenic breast cancer mouse model and in tumors from the same Her2/neu model grown orthotopically. In vivo lactate was detected by MRS utilizing the Hadamard encoded selective multiple quantum coherence pulse sequence (HadSelMQC) recently developed by our laboratory (1). A lower lactate signal was observed in the inducible tumors than in the orthotopic tumors in vivo, while ex vivo analysis of perchloric acid extracts revealed similar amounts of this metabolite in both models. Histological staining of mammary tumor specimens demonstrated a much higher level of fat tissue in inducible tumors compared to the orthotopic model. Phantom studies with [3-13C] lactate indicated that the lipid environment can significantly reduce the T2 of lactate and impede its detection. The transgenic inducible model for breast cancer not only better recapitulates the biological aspects of the human disease, but also provides additional characteristics related to the in vivo detection of lactate that are not available in orthotopic or xenograft models. This study suggests that the level of lactate measured by the HadSelMQC pulse sequence may be underestimated in human patients in the presence of high lipid levels, which are typically encountered in the breast.
Keywords: Lactate, NMR-invisible lactate, breast cancer, transgenic breast cancer model, inducible oncogene
Introduction
Xenograft models of breast cancer are extensively used but only partially reflect critical aspects of tumor biology and cannot be used to investigate several important properties of cancer, such as the natural tumor microenvironment. Orthotopic and transgenic tumor models have been utilized. The orthotopic model is initiated by injection of cancer cells into a target organ in which the spontaneously growing tumor naturally occurs. Growth characteristics of the orthotopic tumor are more realistic than obtained by subcutaneous injection to produce xenografts, but still differ considerably from those of a spontaneous tumor. Transgenic mouse models yield spontaneous tumors that recapitulate key features of the naturally occurring human disease. We have developed a number of inducible transgenic mouse models for human breast cancer (2–8)
The Her2/neu oncogene is amplified in approximately 20% of human breast cancers and is associated with aggressive tumor behavior and poor clinical outcome (9,10). MMTV-rtTA; TetO-neu (MTB/TAN) transgenic mice conditionally express the oncogene Her2/neu in response to doxycycline administration in the drinking water (7). Mammary tumor cells from MTB/TAN mice can also be cultured in vitro and implanted either as orthotopic tumors in the mouse mammary gland or as subcutaneous tumors (i.e., xenografts) in the flanks of immunocompromised mice (8). Xenografts were not included in this study because they show the weakest resemblance to the microenvironment of naturally occurring breast cancer.
Glycolysis is a key energy source for cancer cells that also provides building blocks for cellular replication (11–14). Lactate is the end-product of glycolysis and its concentration can serve as a biological marker of glycolytic activity, although other pathways such as glutaminolysis and alanine metabolism can also produce lactate. Enhanced glycolysis of cancer cells and the undeveloped vasculature of tumor tissues often result in higher steady-state lactate levels in tumors (15). Elevated lactate levels have been proposed as a biomarker of cancer and have been utilized for tumor diagnosis (16) as well as the detection of therapeutic response (17–20).
In vivo 1H magnetic resonance spectroscopy (MRS) enables measurement of concentrations of cellular metabolites (19–22). However, in vivo MRS detection of lactate is difficult due to the overlap of the proton resonances emanating from the CH3-groups of lactate and the CH2 groups of lipids. A double frequency selective multiple quantum coherence filter (SelMQC) has been shown to detect lactate in vivo with no interference from lipid signals (23,24). Hadamard slice encoding (HadSelMQC) was subsequently added to SelMQC sequence in order to obtain the lactate signal from particular locations within large tumors and to facilitate lactate quantification (1,25). This technique has been implemented to measure steady state lactate levels in xenograft models of lymphoma, breast and prostate cancers (20,26,27), and decreases of lactate have been demonstrated to serve as early biomarkers of response to chemotherapy in lymphoma tumor models and in human patients with non-Hodgkin’s lymphoma (28).
Quantum filter techniques utilize differences in evolution times required for refocusing of proton signals from lactate and lipids. The scalar vicinal coupling constant for the lactate methyl group is approximately 7 Hz; therefore, echo times of ~140 msec are required for refocusing of the lactate methyl resonance. This time limit restricts application of the quantum filtering technique to metabolites with sufficiently long T2s. Measurement of lactate concentration with the double quantum filter technique in muscle tissue has been reported, and a significantly lower lactate concentration was detected in vivo than in extracts (29,30). With a double quantum filter technique having an effective TE of 150 ms, only 20–30 % of the endogenous lactate was NMR detectable. The low “visibility of lactate” in muscle was attributed to the presence of a rapidly relaxing lactate pool with a T2 less than 50 ms. The existence of the rapidly relaxing lactate pool was confirmed in experiments with a shorter echo time (30).
In this study, we implemented the HadSelMQC pulse sequence for measuring steady state lactate levels in inducible and orthotopic Her2/neu models of breast cancer in mice. We found that in the inducible tumor model in vivo levels of lactate were lower and the detection of this metabolite was more difficult than in the orthotopic models. However, analysis of perchloric acid extracts from the same tissues demonstrated that the actual levels of lactate in inducible and orthotopic Her2/neu tumors were essentially the same. This finding has important implications for the use of lactate as an indicator of human breast cancer and as a marker of tumor response to chemotherapy. The objective of this study was to determine why the detection level of lactate by the HadSelMQC pulse sequence was inappropriately low in inducible tumor models of breast cancer.
Methods
Animal model
A detailed description of the inducible transgenic Her2/neu mouse model (MTB/TAN) has been published elsewhere (7). Briefly, mice conditionally expressing the Her2/neu oncogene were generated using the tetracycline regulatory system by cloning the coding sequence of activated neu downstream from the minimal tet operator. Mice harboring this TetO-neu transgene, referred to as TAN, were mated with transgenic MMTV-rtTA mice (MTB) to yield bitransgenic MTB/TAN offspring (3,7). Beginning at 6 weeks of age, MTB/TAN mice (n=7) were administered doxycycline 0.1 mg/mL in their drinking water. This treatment induced Her2/neu oncogene expression in a mammary-specific manner. Mice were monitored weekly for tumor formation. Palpable tumors were detected approximately 15 weeks after initial induction. MR experiments were begun when tumors reached dimensions of approximately 10 mm in diameter.
The orthotopic model was initiated by injecting 2 × 106 of 54074 MTB/TAN tumor cells into the mammary fat pads of NCr/nu:nu nude mice (n = 5). After cancer cell implantation, animals were treated with 2.0 mg/ml of doxycycline. After 6–8 weeks, palpable tumors developed, and MRI/MRS studies were performed when tumors reached 10 mm in diameter. Thus, both groups of animals produced tumors in their mammary glands driven by the same oncogene and with the same genetic background, but one group developed these tumors spontaneously whereas the other group had tumors exogenously implanted.
In vivo MRS
All MRS experiments were performed on a 9.4 T, 8.9 cm diameter bore Varian spectrometer equipped with 55 mm ID diameter gradient coils having a maximal strength of 100 G/cm and interfaced to a Direct Drive console (Agilent, Palo Alto, CA). Animals were anesthetized by continuous inhalation of 1–1.5% isofluorine in oxygen. Tumors were positioned in a home-built loop-gap resonator, which provided a uniform B1 field over most of the tumor. Shimming of the B0 field was performed on each animal. Lactate spectroscopy was performed after shimming to a line-width of ~ 100 Hz on the water proton signal from the entire tumor. Core body temperature, monitored during data collection with an MRI-compatible fibrooptic probe (Luxtron, LumaSense Technologies, Inc. Santa Clara, CA), was maintained at 37°C by blowing warm air through the magnet bore. Anatomical images were acquired using a multi-slice gradient echo 1H MRI sequence (TR/TE/flip angle = 150 ms/5 ms/20°, slice thickness 1 mm).
Lactate was detected utilizing the HadSelMQC pulse sequence (1). This sequence can detect lactate in selected regions of multiple slices, but in this study to maximize the signal-to-noise ratio we used only one slice covering the entire tumor. The slice thickness for all MRS studies was 10 mm. Gaussian pulses of 8192 μs were used as frequency selective pulses. For selection of the coherence pathway, shaped gradients with amplitude ratios of 0:−1.5:3 Gauss/cm and duration of 3 ms were applied. Other acquisition parameters were: TR = 4 s, spectral width SW = 4 kHz, number of data points np = 512 for in vivo experiments and 2048 for phantoms studies, number of averages NA = 128 for in vivo and 4 for phantoms. Receiver gain for HadSelMQC experiments was set to 60 dB. For quantitation a Hadamard-encoded spin echo sequence was used to detect water in the same slice (TE= 140 ms, TR = 8 s and NA = 4). The lactate/water ratios were calculated for each in vivo experiment. To demonstrate inability of conventional spectroscopy methods to detect lactate and demonstrate the efficacy of the quantum filter technique for eliminating lipid signals, spectra were acquired using a single voxel STEAM sequence with CHESS water suppression pulses. Three CHESS pulses were used before the start of the STEAM sequence and one CHESS pulse was used during the mixing time (TM). The following sequence parameters were used: TR=8 s, TE = 20 ms, TM = 40 ms, voxel dimensions 10 × 10 × 10 mm3, number of averages = 128. A 1000 mm3 voxel was utilized to encompass the entire tumor volume as it was performed with HadSelMQC sequence for lactate detection.
PCA tissue extracts
At the end of the in vivo experiments, tumor metabolites were extracted with perchloric acid and analyzed by high resolution NMR spectroscopy. To avoid lactate generation in isolated tissue, we excised the tumor from the living animal and froze it as quickly as possible. The animal was anesthetized by ip injection of ketamine/xylazine, placed on a surgical table; a skin incision was made and the tumor tissue was carefully isolated from connected skin and non-tumor tissue. Special care was taken to avoid any disruptions of blood vessels feeding the tumor. When the tissue was isolated, the tumor was lifted with forceps and isolated with scissors, freeze clamped with a pair of tongs dipped in liquid nitrogen and finally immersed into liquid nitrogen. The entire procedure from tumor excision to dropping it into liquid nitrogenous was performed in 3–4 sec. A similar procedure was applied to excised leg muscle tissue to measure lactate in a resting muscle. After excision, animals were euthanized under CO2.
Frozen tissues (~ 200–600 mg) were ground at liquid nitrogen temperature with 6% perchloric acid (3.25 ml per 1 g of tissue) using a method described previously (22). The homogenate was centrifuged at 13,000 rpm for 30 min at 4°C. The supernatant was neutralized to pH 7.0 ± 0.2 with 100 mM of KOH. The precipitated salt was removed by centrifugation at 1,000 rpm for 10 min, and the supernatant was stored at −20°C until further processing.
Liposome preparation
In order to mimic the effects of high levels of adoposites (fat tissue) in mammary tumors on the lactate NMR signal, we performed NMR measurements on lactate dissolved in liposomes. Unilamellar liposomes were prepared as described elsewhere (31). Briefly, chloroform or methanol solutions of 7.34 mg of dipalmitoyl phosphatidylcholine, 3.67 mg of phosphatidylcholine, 1.16 mg of cholesterol and 1.48 mg of phosphatidylglycerol were evaporated under nitrogen flow in a spherical vial to which 900μl of vegetable oil and 500 μl of 10 mM of regular or [3-13C] lactate were added and mixed vigorously with a vortex shaker. The solution was extruded 10 times through a 200 nm membrane. The resulting ~120 nm liposomes were stored at 4° C. The liposome diameter was measured with dynamic light scattering method.
High resolution NMR spectroscopy
The tissue extract was concentrated by lyophilization; the resulting powder was re-dissolved in 0.6 ml D2O, and the pH (calibrated with standard buffers and uncorrected for the deuterium isotope effect) was adjusted to 7.0 ± 0.1 with 1 mM KOH. The solution was then transferred to a 5-mm NMR tube, and a capillary tube containing 0.56 mM of sodium 3-(trimethylsilyl)-[2,2,3,3,-2H4]-1-propionate (TSP) was inserted as an external reference standard and metabolite concentrations were calculated as described previously (32). High-resolution 1H NMR spectra were acquired with a 9.4 T superconducting magnet interfaced to a Bruker DMX400 spectrometer (Bruker BioSpin, Billerica, MA). The following parameters were used: 45° flip angle, TR 8.8 s, and 128 averages. A pre-saturation pulse was used to suppress the water signal. The Fourier transformed spectra were analyzed using the XWin NMR program (Bruker, BioSpin, Billerica, MA).
In order to evaluate the efficiency of the HadSelMQC sequence to detect lactate in a lipid environment, several experiments were performed on phantoms. Three phantoms were prepared as follows: 1) Phantom 1: 5 mm NMR tube filled with 500 μl of 10 mM lactate solution in H2O was inserted into a 10 mm NMR tube filled with D2O; 2) Phantom 2: a 5 mm NMR tube filled with 10 mM lactate in H2O was inserted into a 10 mm NMR tube containing vegetable oil; 3) Phantom 3: a 5 mm NMR tube with 10 mM lactate containing a liposomal suspension in H2O was inserted into a 10 mm NMR tube filled with D2O. Two types of NMR spectra were acquired with a 10 mm high-resolution NMR probe for each of these phantoms. First, a water suppressed pulse-acquire sequence was utilized to demonstrate overlapping of lipid and lactate signals. Parameters for this sequence were: TR=8 sec, 26 μs hard RF pulse with 90° flip angle, number of repetitions 4, receiver gain set to 2 dB. A 1.5 sec saturation pulse was utilized for water suppression. Second, the HadSelMQC sequence was implemented to detect lactate signal. The same acquisition protocol as for in vivo experiments was utilized with NA = 4. Quantitation of the HadSelMQC spectrum was performed offline with Mnova NMR data processing and analysis software (Mestrelab Research SL). The lactate methyl peak was integrated, and areas were compared in the phantoms or in vivo experiments. T2 measurements of an aqueous lactate solution were performed using a standard CPMG pulse sequence: 90 and 180 RF pulses separated by time τ/2 = 14 ms. The echo time was increased by increasing the number of 180 pulses and varied from 14 to 2988 ms with 14ms increments, np=16000, TR=20 sec, 4 averages. Single exponential curve fitting was performed to analyze experimental data and measure T2. To compare the T2 of lactate in aqueous solution with its T2 in a lipid environment, [3-C13] labeled lactate was utilized. T2 of [3-C13] labeled lactate was measured in the liposomal suspension with the same parameters that were used in the aqueous lactate solution but at carbon resonance frequency.
Histological staining
Histological studies were performed on a separate matched cohort of animals with the same size tumors as those employed in the NMR studies; these animals (n=5) were sacrificed, and their tumors surgically excised and fixed in 4% paraformaldehyde. The tissue was cryo-protected by incubation in 5 M sucrose solution, embedded in OCT and frozen on a bed of crushed dry ice. Cryo-sections (5 μm-thick) were prepared and subjected to Haematoxylin and Eosin staining (H&E). Oil Red staining was performed to detect the lipid/fat content of tumor specimens. Briefly, slides containing tumor tissues were immersed in an Oil Red working solution (30ml of 0.5% of oil red and 20 ml of distilled water) for 15 min, rinsed with 60% isopropanol, dipped into alum haematoxylin for 15–20 sec, rinsed with water and coverslipped for microscopic examination. Five images of five different sections were taken from induced and orthotopic tumors tissues. The total numbers of Oil Red-positive and negative pixels were counted. The average percentages of Oil Red positive pixels in inducible and orthotopic models were compared. Image analysis was performed with ImagePro software (Media Cybernetics, Inc., Bethesda, MD).
Results
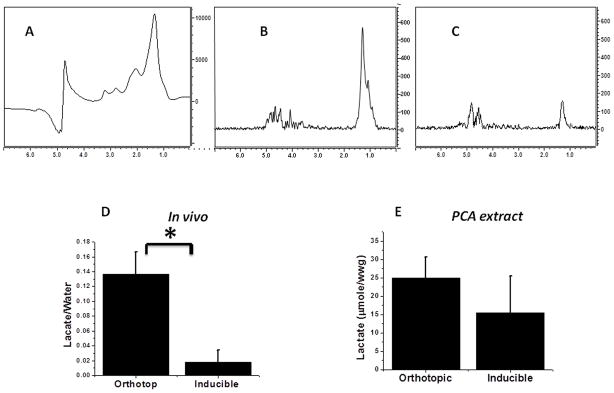
Figure 1(A) shows a typical in vivo MR spectrum acquired from an orthotopic mammary tumor utilizing the STEAM pulse sequence. The large peak at 1.3 ppm originates from an unresolved mixture of lactate and lipid signals. Implementation of the quantum filtering technique reveals the hidden lactate NMR signal. The HadSelMQC pulse sequence utilized in this study exhibits very high efficiency in filtering out all the tissue NMR signals except the lactate methyl proton resonance. Figure 1(B) shows typical in vivo HadSelMQC lactate spectra acquired from the orthotopic (B) and induced mammary gland (C) tumors. The peak on the right (high field) is the CH3 resonance of lactic acid, and the peak on the left (low field) is the residual water signal. Much stronger lactate signals were detected from the orthotopic tumor (Lactate/Water ratio 0.136 ± 0.03) compared to the inducible transgenic tumor (0.018 ± 0.016) (Figure 1D, p < 0.01). However, the PCA extracts obtained from the same snap-frozen tissues revealed similar high lactate concentrations in both the orthotopic and inducible tumors (24.9 ± 5.6 and 15.3 ± 10 μg/ww respectively; there was no statistical differences between these two groups, p >0.05). Note that in spite of the significant variation in lactate concentration in tumor tissue specimens, the concentration of lactate in the tumor was consistently higher than the lactate concentration in resting muscle tissue (1±0.3 μg/ww). An integration of the lactate signals revealed that only ~12% of the signal in the orthotopic model was visible in the inducible model in vivo.
Figure 1.
(A) In vivo NMR spectrum of inducible Her2/neu mammary tumor acquired with STEAM sequence. Large peak at 1.3 ppm presents unresolved signals from lipids and lactate. In vivo lactate spectra acquired with HadSelMQC pulse sequence of orthotopic (B) and inducible (C) Her2/neu transgenic mammary tumors. The lactate level in the inducible model was lower and less reproducible than that of the orthotopic model. (C) Quantitation of in vivo data. (D) Concentration of lactate in tumor PCA extracts of orthotopic and inducible transgenic models. The same high concentration of lactate was found in both tumor models ex vivo. Errors bars are the standard deviation of 7 and 5 experiments respectively.
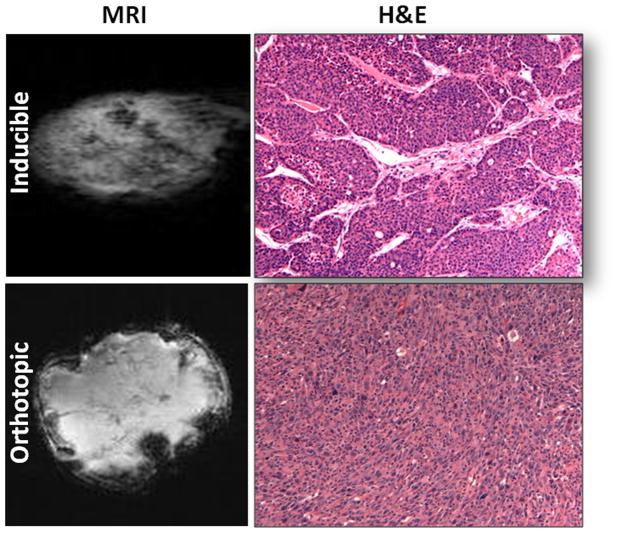
To determine why the in vivo lactate signal was suppressed in inducible tumors of MTB/TAN mice, we conducted histological studies. Figure 2 shows MRI and H&E staining of tumor tissues. Inducible tumors proved to be considerably more heterogeneous as indicated by the MR and H&E images, in part due to the greater amount of stroma present compared to orthotopic tumors (Figure 2).
Figure 2.
In vivo MR images and H&E staining (200× magnification) of inducible (top) and orthotopic (bottom) tumor tissue. More heterogeneity was observed in inducible tumors.
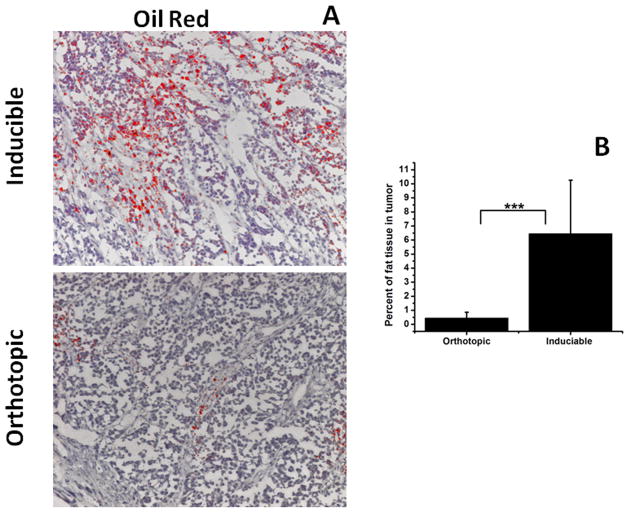
We also compared the amount of fat tissue in these two models using Oil Red staining. Higher levels of fat (red) within adipocytes were detected in inducible (6.46 ± 3.8) compared to orthotopic (0.46 ± 0.4, p < 0.001) tumors (Figure 3). Significant variations in the amount of fatty tissue in inducible tumors were observed, which is reflected in the large standard deviation of these measurements.
Figure 3.
A - Oil Red (fat) staining of inducible (top) and orthotopic (bottom) tumor tissue (200× magnification). B – Quantification of histological data. Errors bars are the standard deviation of 50 measurements, p < 0.001. Higher levels of fat (red) were observed in inducible tumors.
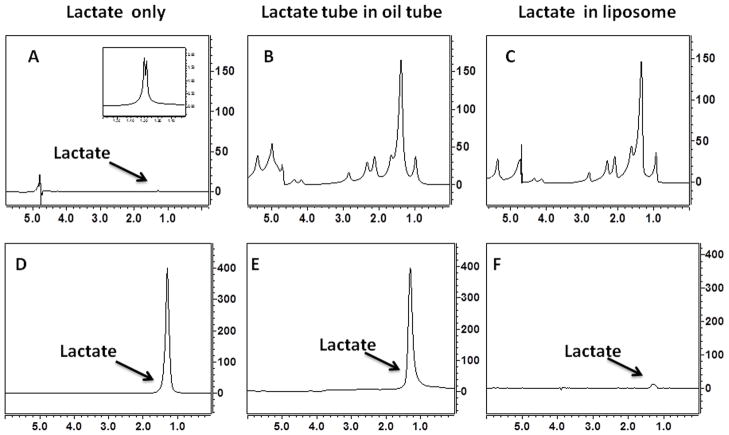
To understand the discrepancy in lactate levels in vivo and ex vivo, we assessed the ability of the HadSelMQC sequence to detect lactate in a high lipid environment. As a reference point, we recorded NMR spectrum from phantom 1 (10 mM aqueous lactate solution) utilizing a simple water suppressed pulse-acquire sequence (Figure 4A) and the HadSelMQC sequence (Figure 4D). Well-defined signals were detected with both sequences. A difference in signal intensity is due to different receiver gains, 2 and 60 dB for spectra A, B, C and D, E, F, respectively. To demonstrate the efficiency of HadSelMQC sequence to detect a lactate signal in the presence of high amounts of lipid, we conducted an experiment with phantom 2 (5 mm NMR tube with 10 mM of lactate contained in a 10 mm NMR tube with vegetable oil). The water-suppressed NMR spectrum of phantom 2 is shown on Figure 4B, and the corresponding HadSelMQC spectrum appears in Figure 4E. Large and unresolved peaks at 1.3 ppm and several others (1.6, 2.1, 2.3, 2.8 ppm) from vegetable oil dominate the unedited spectrum and completely obscure the lactate signal. Meanwhile, efficient suppression of all NMR signals except the lactate methyl resonance was achieved with the HadSelMQC sequence. Nearly complete water suppression and strong lactate signals were detected in the aqueous lactate solutions in the presence or absence of external lipids (panels D and E). Significant reduction of the lactate methyl signal was observed in phantom 3 (10 mM of lactate in liposomal suspension) Figure 4F. Similar to our in vivo studies, a weaker signal (~3 % of the signal intensity of the original signal (panel D)) was observed from lactate in the liposomal suspension.
Figure 4.
Water suppressed pulse acquire (A, B, C) and HadSelMQC (D, E, F) spectra of 10 mM lactate phantoms. (A) Phantom 1: lactate in aqueous solution, (B) Phantom 2: an aqueous solution of lactate inserted into a tube with vegetable oil, (C) Phantom 3: lactate in a liposomal suspension. Large lipid signals in phantoms 2 and 3 (panel B and C) completely obscure the lactate signal (A). The HadSelMQC sequence filtered out the lipids signals and revealed the underlying lactate resonance (panels E and D). A difference in signal intensity in top and bottom panels is due to differences in receiver gain: 2 and 60 dB for spectrum A, B, C and D, E, F respectively. No differences in signals intensity were detected in experiments in phantoms containing only lactate (D) or in lactate in the presence of oil but not in direct physical contact with it (E). The liposome provides a large surface area for integration between lactate and lipid head groups. Such an interaction leads to reduction of lactate T2 and reduction of NMR visibility of lactate (panel F).
We hypothesize that the weak signal of lactate in inducible transgenic mammary tumors and in the liposome suspension was due to the reduction of lactate T2 caused by interactions with lipids. To test this hypothesis, we measured the T2 of lactate in water and in a liposomal suspension. The T2 of lactate in saline solution at the proton frequency was 1.24 ± 0.039 sec. Determination of the lactate T2 at the proton frequency in liposomal suspension was not possible due to the overlap of lipid and lactate signals. To overcome this problem, we measured the T2 of [3-13C] lactate at the carbon frequency in which lipid and lactate produce distinctly resolvable resonances. The T2 of [3-13C] lactate in saline was 1.75 ± 0.041 sec while in the liposomal suspension it was 0.33 ± 0.013 sec, (p <0.001).
Discussion
Promising preclinical data demonstrating the utility of lactate levels as a marker of therapy response have been reported (1,17–21). The HadSelMQC pulse sequence is currently being utilized by our laboratory as a robust method for the in vivo detection of lactate by NMR in mice and in human patients with non-Hodgkin’s lymphoma (19–21,25,33). However this technique was not capable of reliably detecting lactate in an inducible transgenic mouse model of breast cancer. In vivo detection of lactate in the inducible model was difficult and not reproducible. Sometimes lactate was completely undetectable in vivo despite the fact that high levels of lactate were demonstrated by analysis of tumor extracts.
Several parameters control lactate NMR signal intensity. The most critical factors that can reduce the NMR visibility of the lactate in vivo are B0 and B1 field homogeneities and the transverse relaxation time of lactate. The following considerations indicate that either B0 or B1 fields were the cause of the dramatic decrease of lactate signal intensity in inducible tumors. First, both tumor models were examined with the same RF coil and, therefore, experienced similar B1 field. Second, after global shimming, the line-widths of the water signal in both tumor models were the same (~100 Hz) indicating comparable B0 field homogeneity. Third, in our phantom experiments, we found that the HadSelMQC sequence exhibited relatively low sensitivity to both B1 and B0 field in homogeneity (data not shown).
We believe, however, that the major reason for the lactate signal suppression in inducible tumor model was a reduction of the lactate T2 due to an interaction with high levels of fat found in mammary glands of transgenic animals (Figure 3). Oil red staining of tumor tissues arising in transgenic MTB/TAN mice clearly showed higher amounts of intratumoral fat (Figure 3). The distribution of oil red positive tissue was patchy and not uniform, which may explain the significant variation of the lactate detected in the transgenic animals. In contrast, orthotopic tumors exhibited very little fat infiltration.
In vivo MRS detection of lactate is difficult due to overlap of the lactate methyl and lipid methylene signals. Several techniques have been implemented by other investigators to suppress lipid in order to reveal the underlying lactate signals in various tissues (34–41). These methods can be broadly divided into two categories. The first category encompasses techniques using the difference method. This approach strongly depends on a stability of the specimen (patient) and the instrument between acquisitions, which is hard to achieve in experiments with live animals. The second category of methods to suppress lipid signals utilizes modified versions of the quantum filter technique. These methods suffer from the same problem: inability to detect metabolites with short T2. Nevertheless, each of these sophisticated methods observed significant discrepancies between the amounts of lactate detected in vivo and ex vivo (in extracts). Substantially higher concentrations of the lactate were detected in tissue extracts compared to in vivo measurements (30, 42–44). Several explanations were suggested to clarify this so-called “NMR-invisible lactate” phenomenon. Nonspecific binding of lactate to large molecules such as albumin in blood or muscles has been demonstrated (44–46). The presence of rapidly and slowly relaxing (intra- and extracellular) populations of the lactate in muscle tissue has been suggested as an alternative explanation (29, 43).
In our study, we found another factor that can reduce NMR visibility of the lactate. We attributed the low lactate signal in inducible transgenic mouse models of human breast cancer to high levels of adipocytes within these tumors. As a first step, we verified the ability of our sequence to detect lactate in the presence of high lipid content. No differences in signal intensity were detected in experiments with lactate only and lactate with a high amount of vegetable oil separated by a barrier (Figure 4D and E). Thus, the diminution in lactate detectability is not simply a dynamic range limitation of the pulse sequence. Second, we compared the lactate signal in aqueous solution with liposomal suspensions. Liposomes were chosen for the following reasons: Aqueous lactate solution practically is not soluble in lipids, and interactions between these two types of molecules are limited by a thin layer of interaction between the lipid and aqueous phases. However, emulsification drastically increases the surface of interaction between these phases and therefore significantly facilitates lactate-lipid interactions. Adipocytes detected in inducible tumor tissues provide a large surface for interaction of lipids with extracellular lactate. The spherical shape of adipocytes was mimicked by the liposomal vesicles. The HadSelMQC spectrum of the lactate in the liposomal suspension was only 3% of the aqueous lactate signal. Third, a five-fold decrease of the lactate T2 in the liposomal suspension compared to the aqueous solution was detected in our phantom studies. All these facts allow us to conclude that the interaction between lactate and adipocytes in breast tissue of transgenic mice occurs. However, we are not arguing that the liposome is providing an environment similar to that produced in adipocytes for interaction of lactate methyl protons with lipids. In liposomes only the head group of the phospholipids will be exposed to lactate molecules on the inner and outer surfaces of the liposomes, whereas adipocytes are believed to contain large quantities of fatty acids whose methylene protons are accessible to lactic acid. The cationic head groups of phospholipids are the predominant binding sites for the anionic lactate molecules that interact with lysosomes. Much more extensive interactions with the interior methylenes of fatty acids are likely to occur in adipocytes. Therefore, the experiments with the liposomes are intended to demonstrate that exposure to fatty acids even in the form of head groups of liposomes provides a very efficient relaxation pathway for lactate methyl protons.
It is reasonable to assume that the presence of high concentrations of adipocytes can reduce the T2 of the lactate in breast tissue of transgenic mice in a similar manner as it did in the liposomal phantom. The T2 relaxation time of lactate in different tissue has been measured and exhibits a wide range: T2 ~ 180–140 ms in muscle and brain at 3 and 4.7 T (30,46,47). The T2 values for the MCa and Colon-38 tumors were 68 ms and 117 ms, respectively at 4.7 T (48). Rat C6-glioma reported a value of T2 = 200 ms for lactate at the same magnetic field (49). Interaction with adipocytes will lead to reduction of the lactate T2 below 50 ms and formation of two fractions of lactate molecules present in transgenic tumor tissue. First, a rapidly relaxing fraction that is interacting with fat tissue and, therefore, is NMR invisible. Second, a solid tissue-associated fraction of lactate with a long T2 that is NMR visible. The fraction of the rapidly relaxing subpopulation of lactate depends on the amount of fat tissue in a tumor and varies significantly from animal to animal. Based on our in vivo experiments, this fraction can reach up to 78% of the total lactate (only ~12% of lactate detected in orthotopic transgenic model was visible in inducible transgenic tumors while the concentration of lactate in extracts was essentially the same).
The experiments with inducible and orthotopic mouse model of breast cancer demonstrated the presence of a large fraction of NMR invisible lactate in mouse breast tissue. We were able to demonstrate a correlation between a disappearance of the lactate signal with the presence of high levels of adipocytes in mammary glands and a reduction of lactate T2 in lipid environments. However we still have not delineated the molecular mechanism of lactate and lipid interaction.
The inducible mouse model for breast cancer more realistically reflects important aspects of the tumor microenvironment observed in the human disease that are relevant to the lactate detection in the clinical setting. The very heterogeneous structure of the inducible tumors was shown in the inducible model. High tissue heterogeneity was also observed in human breast cancers in clinical studies. In contrast, orthotopic models revealed relatively uniform cell distributions and homogeneous structure that avoid such technical problems which may be encountered in breast cancer patients.
The HadSelMQC sequence has distinct limitations in detection of rapidly relaxing lactate. A lipid-rich microenvironment can cause a significant reduction of the lactate T2 and decrease the NMR visibility of this important metabolite. We anticipate that the same problem of lactate detection is likely to occur in human breast cancer, but this has to be verified in clinical studies. This may limit the utility of HadSelMQC sequence for studies tumors of the breast and perhaps other tissues that exhibit high lipid content.
Acknowledgments
All in vivo and ex vivo studies performed in the Small Animal Imaging Facilities of University of Pennsylvania. We thank Suzanne Wehrli from the Children’s Hospital of Philadelphia for her assistance with high resolution NMR spectroscopy. We also thank Michael Garwood (Department of Radiology, University of Minnesota) for useful discussions. This work was supported by Small Animal Imaging Resource NIH grant 5U24CA08315-07 (JDG and LAC) and U01CA105490, CA98371, CA143296 and 148774 (LAC).
Abbreviations used
- HadSelMQC
Hadamard slice selective-multiple quantum coherence
- MRI
magnetic resonance imaging
- MRS
magnetic resonance spectroscopy
- T2
spin-spin relaxation time
- TSP
3-(trimethylsilyl)-[2,2,3,3,-2H4]-1-propionate
References
- 1.Pickup S, Lee SC, Mancuso A, Glickson JD. Lactate imaging with Hadamard-encoded slice-selective multiple quantum coherence chemical-shift imaging. Magn Reson Med. 2008;60(2):299–305. doi: 10.1002/mrm.21659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gunther EJ, Belka GK, Wertheim GB, Wang J, Hartman JL, Boxer RB, Chodosh LA. A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. Faseb J. 2002;16(3):283–292. doi: 10.1096/fj.01-0551com. [DOI] [PubMed] [Google Scholar]
- 3.D’Cruz CM, Gunther EJ, Boxer RB, Hartman JL, Sintasath L, Moody SE, Cox JD, Ha SI, Belka GK, Golant A, Cardiff RD, Chodosh LA. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nature medicine. 2001;7(2):235–239. doi: 10.1038/84691. [DOI] [PubMed] [Google Scholar]
- 4.Boxer RB, Stairs DB, Dugan KD, Notarfrancesco KL, Portocarrero CP, Keister BA, Belka GK, Cho H, Rathmell JC, Thompson CB, Birnbaum MJ, Chodosh LA. Isoform-specific requirement for Akt1 in the developmental regulation of cellular metabolism during lactation. Cell metabolism. 2006;4(6):475–490. doi: 10.1016/j.cmet.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Jang JW, Boxer RB, Chodosh LA. Isoform-specific ras activation and oncogene dependence during MYC- and Wnt-induced mammary tumorigenesis. Molecular and cellular biology. 2006;26(21):8109–8121. doi: 10.1128/MCB.00404-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boxer RB, Jang JW, Sintasath L, Chodosh LA. Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer cell. 2004;6(6):577–586. doi: 10.1016/j.ccr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, Innocent N, Cardiff RD, Schnall MD, Chodosh LA. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer cell. 2002;2(6):451–461. doi: 10.1016/s1535-6108(02)00212-x. [DOI] [PubMed] [Google Scholar]
- 8.Sarkisian CJ, Keister BA, Stairs DB, Boxer RB, Moody SE, Chodosh LA. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nature cell biology. 2007;9(5):493–505. doi: 10.1038/ncb1567. [DOI] [PubMed] [Google Scholar]
- 9.Berger MS, Locher GW, Saurer S, Gullick WJ, Waterfield MD, Groner B, Hynes NE. Correlation of c-erbB-2 gene amplification and protein expression in human breast carcinoma with nodal status and nuclear grading. Cancer research. 1988;48(5):1238–1243. [PubMed] [Google Scholar]
- 10.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science (New York, NY. 1987;235(4785):177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 11.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science (New York, NY. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson CB. Attacking cancer at its root. Cell. 2009;138(6):1051–1054. doi: 10.1016/j.cell.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Tong X, Zhao F, Mancuso A, Gruber JJ, Thompson CB. The glucose-responsive transcription factor ChREBP contributes to glucose-dependent anabolic synthesis and cell proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(51):21660–21665. doi: 10.1073/pnas.0911316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson CB. Metabolic enzymes as oncogenes or tumor suppressors. The New England journal of medicine. 2009;360(8):813–815. doi: 10.1056/NEJMe0810213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aboagye EO, Bhujwalla ZM, He Q, Glickson JD. Evaluation of lactate as a 1H nuclear magnetic resonance spectroscopy index for noninvasive prediction and early detection of tumor response to radiation therapy in EMT6 tumors. Radiation research. 1998;150(1):38–42. [PubMed] [Google Scholar]
- 16.Czernicki Z, Horsztynski D, Jankowski W, Grieb P, Walecki J. Malignancy of brain tumors evaluated by proton magnetic resonance spectroscopy (1H-MRS) in vitro. Acta neurochirurgica. 2000;76:17–20. doi: 10.1007/978-3-7091-6346-7_4. [DOI] [PubMed] [Google Scholar]
- 17.Huang MQ, Nelson DS, Pickup S, Qiao H, Delikatny EJ, Poptani H, Glickson JD. In vivo monitoring response to chemotherapy of human diffuse large B-cell lymphoma xenografts in SCID mice by 1H and 31P MRS. Academic radiology. 2007;14(12):1531–1539. doi: 10.1016/j.acra.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shungu DC, Bhujwalla ZM, Wehrle JP, Glickson JD. 1H NMR spectroscopy of subcutaneous tumors in mice: preliminary studies of effects of growth, chemotherapy and blood flow reduction. NMR in biomedicine. 1992;5(5):296–302. doi: 10.1002/nbm.1940050517. [DOI] [PubMed] [Google Scholar]
- 19.Lee SC, Delikatny EJ, Poptani H, Pickup S, Glickson JD. In vivo (1)H MRS of WSU-DLCL2 human non-Hodgkin’s lymphoma xenografts: response to rituximab and rituximab plus CHOP. NMR in biomedicine. 2009;22(3):259–265. doi: 10.1002/nbm.1316. [DOI] [PubMed] [Google Scholar]
- 20.Lee SC, Huang MQ, Nelson DS, Pickup S, Wehrli S, Adegbola O, Poptani H, Delikatny EJ, Glickson JD. In vivo MRS markers of response to CHOP chemotherapy in the WSU-DLCL2 human diffuse large B-cell lymphoma xenograft. NMR in biomedicine. 2008;21(7):723–733. doi: 10.1002/nbm.1250. [DOI] [PubMed] [Google Scholar]
- 21.Lee SC, Poptani H, Pickup S, Jenkins WT, Kim S, Koch CJ, Delikatny EJ, Glickson JD. Early detection of radiation therapy response in non-Hodgkin’s lymphoma xenografts by in vivo 1H magnetic resonance spectroscopy and imaging. NMR in biomedicine. 2010;23(6):624–632. doi: 10.1002/nbm.1505. [DOI] [PubMed] [Google Scholar]
- 22.Magnitsky S, Vite CH, Delikatny EJ, Pickup S, Wehrli S, Wolfe JH, Poptani H. Magnetic resonance spectroscopy of the occipital cortex and the cerebellar vermis distinguishes individual cats affected with alpha-mannosidosis from normal cats. NMR in biomedicine. 2010;23(1):74–79. doi: 10.1002/nbm.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Q, Shungu DC, van Zijl PC, Bhujwalla ZM, Glickson JD. Single-scan in vivo lactate editing with complete lipid and water suppression by selective multiple-quantum-coherence transfer (Sel-MQC) with application to tumors. Journal of magnetic resonance. 1995;106(3):203–211. doi: 10.1006/jmrb.1995.1035. [DOI] [PubMed] [Google Scholar]
- 24.He Q, Bhujwalla ZM, Maxwell RJ, Griffiths JR, Glickson JD. Proton NMR observation of the antineoplastic agent Iproplatin in vivo by selective multiple quantum coherence transfer (Sel-MQC) Magn Reson Med. 1995;33(3):414–416. doi: 10.1002/mrm.1910330315. [DOI] [PubMed] [Google Scholar]
- 25.Mellon EA, Lee SC, Pickup S, Kim S, Goldstein SC, Floyd TF, Poptani H, Delikatny EJ, Reddy R, Glickson JD. Detection of lactate with a hadamard slice selected, selective multiple quantum coherence, chemical shift imaging sequence (HDMD-SelMQC-CSI) on a clinical MRI scanner: Application to tumors and muscle ischemia. Magn Reson Med. 2009;62(6):1404–1413. doi: 10.1002/mrm.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glunde K, Artemov D, Penet MF, Jacobs MA, Bhujwalla ZM. Magnetic resonance spectroscopy in metabolic and molecular imaging and diagnosis of cancer. Chemical reviews. 2010;110(5):3043–3059. doi: 10.1021/cr9004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Artemov D, Pilatus U, Chu S, Mori N, Nelson JB, Bhujwalla ZM. Dynamics of prostate cancer cell invasion studied in vitro by NMR microscopy. Magn Reson Med. 1999;42(2):277–282. doi: 10.1002/(sici)1522-2594(199908)42:2<277::aid-mrm9>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 28.Lee SC, Poptani H, Delikatny EJ, Pickup S, Nelson DS, Schuster SJ, Nasta SD, Svoboda J, Goldstein SC, Wallace SG, Loevner LA, Mellon EA, Reddy R, Glickson JD. NMR metabolic and physiological markers of therapeutic response. Advances in experimental medicine and biology. 2011;701:129–135. doi: 10.1007/978-1-4419-7756-4_18. [DOI] [PubMed] [Google Scholar]
- 29.Shen D, Gregory CD, Dawson MJ. Observation and quantitation of lactate in oxidative and glycolytic fibers of skeletal muscles. Magn Reson Med. 1996;36(1):30–38. doi: 10.1002/mrm.1910360107. [DOI] [PubMed] [Google Scholar]
- 30.Jouvensal L, Carlier PG, Bloch G. Low visibility of lactate in excised rat muscle using double quantum proton spectroscopy. Magn Reson Med. 1997;38(5):706–711. doi: 10.1002/mrm.1910380505. [DOI] [PubMed] [Google Scholar]
- 31.Manevich Y, Reddy KS, Shuvaeva T, Feinstein SI, Fisher AB. Structure and phospholipase function of peroxiredoxin 6: identification of the catalytic triad and its role in phospholipid substrate binding. Journal of lipid research. 2007;48(10):2306–2318. doi: 10.1194/jlr.M700299-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Bottomley PA, Hardy CJ. Rapid, reliable in vivo assays of human phosphate metabolites by nuclear magnetic resonance. Clinical chemistry. 1989;35(3):392–395. [PubMed] [Google Scholar]
- 33.Lee SC, Poptani H, Delikatny EJ, Pickup S, Nelson DS, Schuster SJ, Nasta SD, Svoboda J, Goldstein SC, Wallace SG, Loevner LA, Mellon EA, Reddy R, Glickson JD. Molecular imaging of cancer: Prediction and early detection of response by NMR spectroscopy and imaging. Advances in experimental medicine and biology. 2010;701:129–135. doi: 10.1007/978-1-4419-7756-4_18. [DOI] [PubMed] [Google Scholar]
- 34.Keller AM, Sorce DJ, Sciacca RR, Barr ML, Cannon PJ. Very rapid lactate measurement in ischemic perfused hearts using 1H MRS continuous negative echo acquisition during steady-state frequency selective excitation. Magn Reson Med. 1988;7(1):65–78. doi: 10.1002/mrm.1910070108. [DOI] [PubMed] [Google Scholar]
- 35.McKinnon GC, Boesiger P. A one-shot lactate-editing sequence for localized whole-body spectroscopy. Magn Reson Med. 1988;8(3):355–361. doi: 10.1002/mrm.1910080313. [DOI] [PubMed] [Google Scholar]
- 36.Meyer RA. Echo acquisition during frequency-selective pulse trains for proton spectroscopy of metabolites in vivo. Magn Reson Med. 1987;4(3):297–301. doi: 10.1002/mrm.1910040311. [DOI] [PubMed] [Google Scholar]
- 37.Ugurbil K, Petein M, Maidan R, Michurski S, Cohn JN, From AH. High resolution proton NMR studies of perfused rat hearts. FEBS letters. 1984;167(1):73–78. doi: 10.1016/0014-5793(84)80835-2. [DOI] [PubMed] [Google Scholar]
- 38.Reddy R, Subramanian VH, Clark BJ, Leigh JS. Longitudinal spin-order-based pulse sequence for lactate editing. Magn Reson Med. 1991;19(2):477–482. doi: 10.1002/mrm.1910190241. [DOI] [PubMed] [Google Scholar]
- 39.Rothman DL, Behar KL, Hetherington HP, den Hollander JA, Bendall MR, Petroff OA, Shulman RG. 1H-Observe/13C-decouple spectroscopic measurements of lactate and glutamate in the rat brain in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(6):1633–1637. doi: 10.1073/pnas.82.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freeman DM, Sotak CH, Muller HH, Young SW, Hurd RE. A double quantum coherence transfer proton NMR spectroscopy technique for monitoring steady-state tumor lactic acid levels in vivo. Magn Reson Med. 1990;14(2):321–329. doi: 10.1002/mrm.1910140217. [DOI] [PubMed] [Google Scholar]
- 41.Thakur SB, Yaligar J, Koutcher JA. In vivo lactate signal enhancement using binomial spectral-selective pulses in selective MQ coherence (SS-SelMQC) spectroscopy. Magn Reson Med. 2009;62(3):591–598. doi: 10.1002/mrm.22065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao P, Storey CJ, Babcock EE, Malloy CR, Sherry AD. 1H NMR detection of lactate and alanine in perfused rat hearts during global and low pressure ischemia. Magn Reson Med. 1995;33(1):53–60. doi: 10.1002/mrm.1910330108. [DOI] [PubMed] [Google Scholar]
- 43.Zhao P, Sherry AD, Malloy CR, Babcock EE. Direct observation of lactate and alanine by proton double quantum spectroscopy in rat hearts supplied with [3-13C]pyruvate. FEBS letters. 1992;303(2–3):247–250. doi: 10.1016/0014-5793(92)80530-t. [DOI] [PubMed] [Google Scholar]
- 44.Chatham JC, Forder JR. Lactic acid and protein interactions: implications for the NMR visibility of lactate in biological systems. Biochimica et biophysica acta. 1999;1426(1):177–184. doi: 10.1016/s0304-4165(98)00154-8. [DOI] [PubMed] [Google Scholar]
- 45.Bell JD, Brown JC, Kubal G, Sadler PJ. NMR-invisible lactate in blood plasma. FEBS letters. 1988;235(1–2):81–86. doi: 10.1016/0014-5793(88)81238-9. [DOI] [PubMed] [Google Scholar]
- 46.Luo Y, Rydzewski J, de Graaf RA, Gruetter R, Garwood M, Schleich T. In vivo observation of lactate methyl proton magnetization transfer in rat C6 glioma. Magn Reson Med. 1999;41(4):676–685. doi: 10.1002/(sici)1522-2594(199904)41:4<676::aid-mrm5>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 47.Annarao S, Thomas K, Pillarsetty N, Koutcher JA, Thakur SB. In vivo lactate T1 and T2 relaxation measurements in ER-positive breast tumors using SS-SelMQC editing sequence. ISMRM proceeding; 2011. [Google Scholar]
- 48.Muruganandham M, Koutcher JA, Pizzorno G, He Q. In vivo tumor lactate relaxation measurements by selective multiple-quantum-coherence (Sel-MQC) transfer. Magn Reson Med. 2004;52(4):902–906. doi: 10.1002/mrm.20206. [DOI] [PubMed] [Google Scholar]
- 49.Terpstra M, High WB, Luo Y, de Graaf RA, Merkle H, Garwood M. Relationships among lactate concentration, blood flow and histopathologic profiles in rat C6 glioma. NMR in biomedicine. 1996;9(5):185–194. doi: 10.1002/(SICI)1099-1492(199608)9:5<185::AID-NBM414>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]