Abstract
Background
The rs3818361 single nucleotide polymorphism in CR1 is associated with increased risk of Alzheimer's disease (AD). Although this novel variant is associated with a small effect size and, is unlikely to be useful as a predictor of AD risk, it may provide insights into AD pathogenesis. We examined the association between rs3818361 and brain amyloid deposition in non-demented older individuals.
Methods
We used 11C-Pittsburgh Compound-B (PiB) PET to quantify brain amyloid burden in 57 non-demented older individuals (mean age 78.5 years) in the neuroimaging substudy of the Baltimore Longitudinal Study of Aging. In a replication study, we analyzed 11C-PiB PET data from 22 cognitively normal older individuals (mean age 77.1 years) in the Alzheimer's disease neuroimaging initiative (ADNI) dataset.
Results
Risk allele carriers of rs3818361 have lower brain amyloid burden relative to non-carriers. There is a strikingly greater variability in brain amyloid deposition in the non-carrier group relative to risk carriers, an effect explained partly by APOE genotype. In non-carriers of the CR1 risk allele, APOE ε4 individuals showed significantly higher brain amyloid burden relative to APOE ε4 non-carriers. We also independently replicate our observation of lower brain amyloid burden in risk allele carriers of rs3818361 in the ADNI sample.
Conclusions
Our findings suggest complex mechanisms underlying the interaction of CR1, APOE and brain amyloid pathways in AD. Our results are relevant to treatments targeting brain Aβ in non-demented individuals at risk for AD and suggest that clinical outcomes of such treatments may be influenced by complex gene-gene interactions.
Keywords: CR1, APOE, single nucleotide polymorphism, Alzheimer's disease, amyloid, 11C-PiB PET
Introduction
Recent large-scale Genome Wide Association Studies (GWAS) have identified novel risk variants for sporadic Alzheimer's disease (AD) (1, 2). These findings have since been independently replicated (3, 4). Although the identification of novel genetic risk factors for AD is a significant advance, these variants occur commonly in the general population and are associated with small effect sizes. Moreover, they are believed to be merely proxies for true AD risk variants. Their clinical utility as stand-alone predictors of disease risk is therefore likely to be limited (5). They may however be invaluable in the delineation of pathways intrinsic to disease mechanisms or their modifiers in at-risk older individuals. Single nucleotide polymorphisms (SNPs) in the complement component (3b/4b) receptor-1 (CR1) were reported to be associated with greater risk of AD (2-4). More recently, the rs6656401A risk allele of CR1 was also related to greater cognitive decline over time as well as with the extent of neuritic plaque burden at autopsy in older individuals who were non-demented at baseline (6). Together with a large body of evidence supporting a role for the complement system in modulating AD pathogenesis (7), these findings suggest that the AD risk variant of CR1 might influence pathways related to brain Aβ clearance and/or deposition.
The aim of the present study was to investigate the association between the AD risk variant rs3818361 SNP in CR1 and brain amyloid burden in non-demented older individuals within the neuroimaging substudy of the Baltimore Longitudinal Study of Aging (BLSA-NI) (8). In light of the findings by Lambert and colleagues in their original GWAS study demonstrating an interaction between this SNP and APOE genotype in influencing risk for AD (2), it was also of interest to examine the effect of APOE genotype in modifying associations between CR1 and brain amyloid during aging.
Subjects and Methods
The Baltimore Longitudinal Study of Aging (BLSA) is one of the largest and longest-running longitudinal studies of aging in the United States (8). The community dwelling unpaid volunteer participants are predominantly white, of upper-middle socioeconomic status, and with an above average educational level. In general, at the time of entry into the study, participants have no physical and cognitive impairment (i.e. Mini-Mental State Examination (MMSE) score ≤ 24) and no chronic medical condition with the exception of well-controlled hypertension.
The BLSA Neuroimaging sub-study (BLSA-NI) began in 1994. BLSA participants were initially prioritized for admission to the neuroimaging study based on health considerations and the amount of prior cognitive data available for each individual (8). At enrollment, participants were free of central nervous system disease (e.g. epilepsy, stroke, bipolar illness, dementia), severe cardiac disease (e.g. myocardial infarction, coronary artery disease requiring angioplasty or coronary artery bypass surgery), pulmonary disease or metastatic cancer.
Participants in the current report were 57 (mean age; 78.5±6.3 years) non-demented individuals in the neuroimaging substudy of the BLSA, who underwent 11C-PiB PET amyloid imaging scans and genome-wide genotyping. They were ascertained from the initial 61 BLSA-NI participants consecutively assessed with 11C-PiB from June 2005 to March 2007 and were representative of the entire BLSA-NI with respect to baseline age, sex, race, and education. We excluded individuals with clinical strokes, brain trauma, and those meeting consensus criteria for AD (NINCDS-ADRDA) and mild cognitive impairment (MCI) as determined by consensus case conference (9, 10). This study was approved by the local institutional review board. All participants provided written informed consent prior to each assessment. Previous studies utilizing 11C-PiB PET data from these BLSA-NI participants have reported on the association of in vivo brain amyloid deposition with cognitive decline during aging (11), brain atrophy (12) and resting state regional cerebral blood flow (13).
The Alzheimer's disease neuroimaging initiative (ADNI) is a multi-center longitudinal study initiated in 2003 by the National Institute on Aging (NIA) (http://www.adni-info.org; PI Michael M. Weiner) (supplemental information). The principal goal of ADNI is to test whether neuroimaging and other biomarkers, together with clinical assessments can better detect and measure the progression of AD. Data used in the current report were derived from 22 cognitively normal ADNI participants (mean age; 77.1±6.2 years) who underwent 11C-PiB PET imaging and genome-wide genotyping.
Genotyping
Genome-wide genotyping procedures in BLSA and ADNI have been described previously (14-16) (supplemental information).
11C-PiB studies
Dynamic 11C-PiB PET studies were performed in the BLSA participants as described previously (13). PET scanning started immediately after an intravenous bolus injection of 540.2±33.3 MBq (14.6 ± 0.9 mCi) of 11C-PiB with a specific activity of 208.68 ± 111GBq/μmol (range, 36.26– 540.94 GBq/μmol). PiB-PET data in ADNI were collected as described previously (17) (supplemental information).
MRI-based Region-of-Interest (ROI) definition
In the BLSA PiB-PET study, T1-weighted volumetric MRI scans were co-registered to the mean of the first 20min dynamic PET images using the mutual information method in the Statistical Parametric Mapping software (SPM 2; Wellcome Department of Imaging Neuroscience, London, U.K.). Besides the cerebellum, which was used as a reference region, 15 ROIs (caudate, putamen, thalamus, lateral temporal, medial temporal, orbitofrontal, prefrontal, occipital, superior frontal, parietal, anterior cingulate, posterior cingulate, pons, midbrain, and white matter) were manually drawn on the co-registered MR images (18).
Quantification of distribution volume ratios (DVRs) in the BLSA PiB-PET study
Reference tissue model (RTM) is a compartmental modeling approach that uses a reference tissue, such as cerebellum, time activity curve (TAC) as input for quantification of ligand-receptor dynamic PET without blood sampling. The distribution volume ratio (DVR) of [11C]PiB binding can be estimated directly by reference tissue models using the reference tissue TAC as input (19). DVR parametric images were estimated by simultaneous fitting of a simplified reference tissue model using linear regression with spatial constraints (SRTM-LRSC) and the cerebellum as reference tissue (19) (supplemental information).
Methods for the estimation of global amyloid burden in the ADNI dataset have been described previously (17) (supplemental information).
Neuropsychological testing
BLSA participants completed a battery of twelve neuropsychological tests evaluating six cognitive domains concurrent with the 11C-PiB PET scans (supplemental information). A similar battery of neuropsychological tests was also administered to the ADNI participants who underwent 11C-PiB PET imaging (20).
Statistical analyses
Our main aim was to investigate inter-group differences in brain amyloid burden between risk (AG/AA) and non-risk (GG) carriers of the AD variant rs3818361 SNP in CR1. All the analyses were conducted in SAS 9.2 (Cary, NC). During initial exploratory analyses plotting values of PiB DVR across different brain regions, we observed a striking difference in the variability of PiB distribution between the two groups (i.e. AA/AG versus GG) in most brain regions.
We therefore used generalized least square regression models which allowed us to investigate the differences in variability of PiB distribution and differences in mean levels of brain amyloid burden between risk (AG/AA) and non-risk (GG) carriers of the AD variant SNP in CR1 in one unified model. Mean cortical and regional PiB DVRs were used as dependent variables. We used the group variable (coded 0 for GG and 1 for AG/AA) as the main predictor, and included age, sex and race as covariates to adjust for their effects. We first used two separate residual error variance terms (one for each group) and then used likelihood ratio tests to test if the residual variances were equal between two groups. One residual error variance (pooled) was used for regions that showed statistically non-significant differences in variance and two residual error variances were used for regions that showed statistically significant differences (p<0.05) in variance. Once the residual variance terms were determined, the differences in mean levels of brain amyloid burden were then estimated. In the light of previous reports including our own that have shown robust effects of age and APOE ε4 status on brain amyloid deposition (11, 21-23), we conducted targeted analyses examining whether the effects of age and APOE ε4 status on PiB DVRs were different between risk (AG/AA) and non-risk (GG) groups. In this regression model, the predictors included age, APOE ε4 status (ε4-positive or ε4-negative), CR1 group (AA/AG or GG), interaction between age and CR1 group, and interaction between APOE ε4 status and CR1 group. Sex and race were included in the model as covariates. Significant interactions indicate whether the effects of age or APOE ε4 status on brain amyloid burden differed between CR1-risk (AG/AA) and non-risk (GG) groups. To control for potential type 1 error due to multiple comparisons, we report False Discovery Rate (FDR) adjusted p-values (padj) based on the method described by Benjamini and Hochberg (24).
In our replication study in the ADNI 11C-PiB dataset, our main aim was to confirm our findings of differences in brain amyloid burden between CR1 risk and non-risk groups among BLSA participants. Our replication analyses used a measure of global brain amyloid burden that has been previously validated by ADNI investigators both as a quantitative phenotype in genetic analyses as well as to derive cut-off measures to establish PiB positivity/negativity (17, 25). In restricting the replication study to a single validated measure of global amyloid burden, we avoided making multiple comparisons across several brain regions in the much smaller ADNI dataset. In the replication analysis, the null hypothesis tested was that our original observation of lower brain amyloid in CR1 risk-carriers was a false positive finding. The p-value reported for the replication analysis is therefore for a one-sided t-test comparing mean values of global brain amyloid burden between the CR1 risk and non-risk groups.
Results
Sample characteristics
The two groups (risk carriers; AA/AG and non-risk carriers; GG) did not differ significantly in age, sex, number of years of education or APOE ε4 status. Their Mini Mental State Examination (MMSE) scores and domain-specific (memory, language, executive function, visuospatial function and attention) cognitive performance did not differ significantly. There were a significantly higher number of African American participants in the risk (AG/AA) group (Table-1). Frequencies of alleles in the rs3818361 polymorphism were G/G in 40 subjects (70.2%), A/G in 15 subjects (26.3%), and A/A in 2 subjects (3.5%). Thus 29.8% of our participants carried the risk A-allele. The frequency of the minor allele (A) in our sample was 0.16 and that of the major allele (A) was 0.84. There were no significant differences in the age distribution of APOE ε4 alleles between the CR1 risk and non-risk groups (Table-2).
Table-1. Characteristics of participants from the BLSA in the 11C-PiB PET study.
| N | Age | Sex | Race | Number of APOE ε4 carriers (%) | |
|---|---|---|---|---|---|
| Total | 57 | 78.5 (6.3) | 25 F (44%) | 48 W (84%) | 18 (33%) |
| GG | 40 | 78.8 (6.7) | 16 F (40%) | 37 W (93%) | 13 (35%) |
| AG(15)/AA(2) | 17 | 77.8 (5.1) | 9 F (53%) | 11 W (65%) | 5 (29%) |
| Difference (p-value) | 0.59 | 0.37 | 0.0154 | 0.68 |
Data are presented as mean (SD); F=Female, W=Caucasian
Table-2.
Age distribution of APOE ε4 alleles among CR1 risk carriers (AG/AA) and non-risk (GG) groups.
| APOE ε4 non-carriers (years) (SD) | APOE ε4 carriers (years) (SD) | Difference (p-value) | |
|---|---|---|---|
| GG (N=40) | 79.9 (7.5) | 76.5 (4.2) | 0.13 |
| AG/AA (N=17) | 77.4 (6.1) | 78.7 (0.8) | 0.64 |
| Difference (p-value) | 0.31 | 0.25 | Overall p = 0.38 |
CR1 and brain amyloid burden
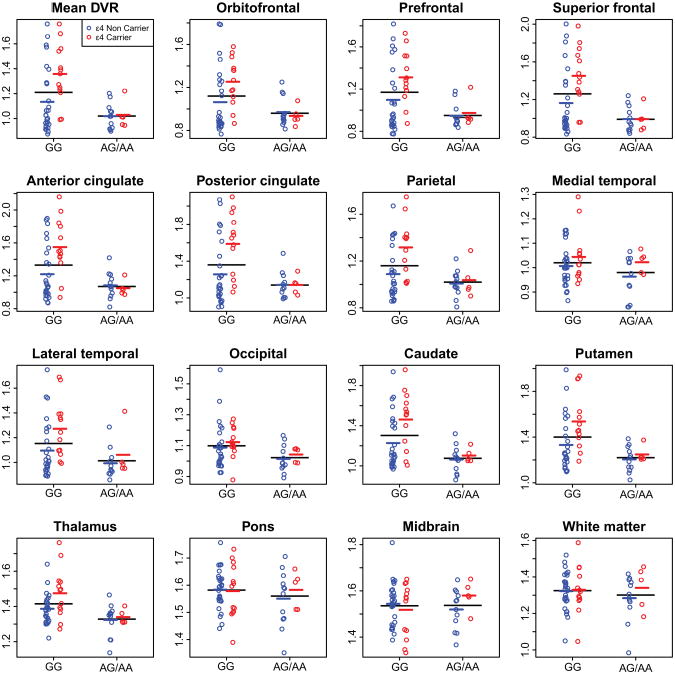
We observed widespread and statistically significant decreases in brain amyloid burden among carriers of the risk allele (AA/AG) of rs3818361 relative to non-carriers (GG). These differences were observed in mean cortical DVR (t(52) = 3.61, padj = 0.0016) orbitofrontal (t(52) = 2.78, padj =0.013), prefrontal (t(52) = 3.76, padj = 0.0011), superior frontal cortex (t(52) = 4.07, padj = 0.0011), anterior (t(52) = 3.85, padj = 0.0011) and posterior cingulate cortex (t(52) = 3.05, padj = 0.0072) and in the parietal (t(52) = 2.76, padj = 0.013), lateral temporal (t(52) = 2.70, padj = 0.014) as well as occipital cortices (t(52) = 2.61, padj = 0.016) (Fig. 1). Significant differences were also observed in the caudate (t(52) = 4.43, padj = 0.0008), putamen (t(52) = 3.91, padj =0.0011) and thalamus (t(52) = 3.80, padj = 0.0011). No significant differences were found in the pons (t(52) = 1.38, padj =0.19), mid brain (t(52) = 0.36, padj = 0.72) and white matter (t(52) = 1.11, padj=0.29), regions associated with non-specific PiB binding (22) and medial temporal (t(52) = 2.03, padj = 0.059). We did not observe significant interactions between age and CR1 group in any of the brain regions examined, indicating similar effects of age on brain amyloid for the risk (AG/AA) and non-risk (GG) groups.
Figure 1.
Scatter plots showing the inter-group (AA/AG; risk carriers versus GG; risk non-carriers) differences in mean cortical and regional PiB DVRs. Individual values are shown in open circles. Red and blue circles denote APOE ε4 carriers and non-carriers respectively. Black lines indicate mean values in risk and non-risk groups; colored lines indicate mean values for APOE ε4 carriers and non-carriers.
In addition to differences in mean level of amyloid burden in association with CR1 genotype, we also observed a statistically significant increase in variability in brain amyloid burden in risk non-carriers (GG) of rs3818361 relative to the risk group (AG/AA). These differences were found in mean cortical DVR (χ2 (1) = 19.3, padj <.0001), orbitofrontal (χ2 (1) = 16.4, padj = 0.0001), prefrontal (χ2 (1) = 21.8, padj<.0001), superior frontal cortex (χ2 (1) = 23.0, padj <.0001), anterior (χ2 (1) = 18.3, padj<.0001) and posterior cingulate cortices (χ2 (1) = 19.9, padj<.0001) and in the parietal (χ2 (1) = 10.3, padj = 0.0023), lateral temporal (χ2 (1) = 7.1, padj = 0.012) as well as occipital cortices (χ2 (1) = 5.0, padj = 0.036). Significant increases in variability in PiB DVR in the non-risk group were also observed in the caudate (χ2 (1) = 19.1, padj < .0001), putamen (χ2 (1) = 19.1, padj<.0001) and thalamus (χ2 (1) = 4.5, padj = 0.045). No significant differences in variability of PiB DVR were found in medial temporal cortex (χ2 (1) = 1.4, padj = 0.30), pons (χ2 (1) = 0.2, padj=0.65), mid brain (χ2 (1) = 1.0, padj = 0.37) and white matter (χ2 (1) = 0.2, padj=0.65).
In order to confirm that our observations of statistically significant differences in both mean levels and variability of brain amyloid burden between the CR1 risk (AA/AG) and non-risk (GG) groups were not driven by differences in racial distribution, we repeated the above analyses after excluding all African American participants and obtained identical results (supplemental tables S1 and S2).
To investigate factors responsible for the increased variability in brain amyloid deposition in non-carriers of the CR1 risk allele, we investigated whether APOE genotype modified the effect of CR1 on brain amyloid. We found significant interactions between APOE genotype and CR1 group in several brain regions, indicating differential effects of APOE genotype on amyloid burden for risk versus non-risk groups in these regions. Among individuals who did not carry the risk allele of rs3818361 (GG), APOE ε4 carriers showed greater brain amyloid burden than APOE ε4 non-carriers. In contrast, amyloid burden was similar for APOE ε4 carriers and non-carriers in the risk (AG/AA) group. Significant interactions between CR1 and APOE genotype were observed for mean cortical DVR (t(50) = −2.76, padj = 0.029), orbitofrontal (t(50) = −2.79, padj=0.029), superior frontal (t(50) = −3.02, padj = 0.029), anterior (t(50) = −3.71, padj = 0.0080) and posterior cingulate (t(50) = −2.71, padj= 0.029). Similar effects were observed in the caudate (t(50) = −2.57, padj = 0.035) .
Finally, in order to replicate our main finding of reduced brain amyloid burden among CR1 risk allele carriers in an independent sample, we analyzed 11C-PiB PET data available in 22 cognitively normal older individuals in the ADNI sample. The mean age of this sample (77.1±6.2 years) was similar to participants in our own 11C-PiB PET study. The CR1 risk (N=4) and non-risk groups (N=18) were well-matched in age and sex as well as the number of APOE ε4 carriers (table-3). Similar to our findings, there were no significant differences in MMSE scores and domain-specific (memory, language, executive function, visuospatial function and attention) cognitive performance between the CR1 risk and non-risk groups. Identical to our findings in the BLSA sample, we found that CR1 risk allele carriers showed a significantly lower global brain amyloid burden than non-risk allele carriers in the ADNI dataset (1.308±0.308 and 1.619±0.328 respectively; p=0.049).
Table-3. Characteristics of participants from ADNI in the 11C-PiB PET study.
| N | Age | Sex | Race | Number of APOE ε4 carriers (%) | |
|---|---|---|---|---|---|
| Total | 22 | 77.1 (6.2) | 8 F (36.4%) | 20 W (90.9%) | 6 (27.3%) |
| GG | 18 | 76.9 (6.3) | 5 F (27.8%) | 18 W (100%) | 5 (27.8%) |
| AG/AA | 4 | 78.1 (6.3 | 3 F (75%) | 2 W (50%) | 1 (25%) |
| Difference (p-value) | 0.75 | 0.12 | 0.026 | 1.00 |
Data are presented as mean (SD); F=Female, W=Caucasian
Discussion
Our main aim in this study was to examine the relationship between the AD risk variant SNP rs3818361 in the CR1 gene and in vivo brain amyloid burden measured with 11C-PiB PET in non-demented older individuals. In light of recent GWAS studies that showed a greater risk of AD in carriers of the A-allele of this SNP (2, 27), as well as an interaction between the CR1 and APOE genes in conferring risk for AD, our primary goal was to examine whether the carriers of the risk allele of CR1 had significant differences in brain amyloid burden relative to non-carriers and whether the CR1 × APOE interaction might influence brain amyloid deposition in cognitively normal older individuals. We found widespread and statistically significant decreases in brain amyloid burden in individuals carrying one or two copies of the risk allele (AG/AA) relative to risk non-carriers (GG) in the BLSA sample and also confirmed this finding in an independent sample from the ADNI dataset.
In addition, we observed significantly greater variance in brain amyloid deposition in the non-risk group (GG), an effect that appears to be influenced in part by APOE genotype. Thus, among GG but not AG/AA individuals, APOE ε4 carriers exhibited greater amyloid deposition in several brain regions relative to APOE ε4 non-carriers (Figure-1).
To the best of our knowledge, this is the first report of an association between genetic variation in the CR1 gene and brain amyloid deposition quantified by in vivo PET imaging in non-demented older individuals. Our findings run counter to the direction of effect on brain amyloid deposition observed in non-demented carriers of the APOE ε4 allele, the most robust genetic risk factor for sporadic AD. While the CR1 risk allele was associated with decreased fibrillar amyloid in non-demented individuals in the current study, we and others have demonstrated increased brain amyloid levels in carriers of the APOE ε4 allele relative to non-carriers in cognitively normal older individuals (21, 22). Our present findings also suggest that CR1 risk allele may modify the relationship between APOE genotype and brain amyloid deposition. This finding is especially relevant against the background of the index GWAS study by Lambert and colleagues which demonstrated a differential effect of the CR1 rs3818361 SNP on AD risk between APOE ε4 carriers and non-carriers (2). Our current findings further suggest that the CR1 × APOE interaction also influences an alternative phenotype relevant to early changes in AD pathogenesis by showing that this interaction modulates brain amyloid deposition even in cognitively normal older individuals.
Our findings merit examination in the light of a recent study by Brouwers and colleagues (28) which showed that four CR1 SNPs in two haplotype blocks were associated with elevated CSF Aβ1-42 levels in AD patients; a finding that is similarly counter-intuitive in suggesting that CR1-associated risk for AD may not be associated with increased brain Aβ accumulation. A recent study however did not find an association between other CR1 SNPs associated with AD risk and CSF levels of Aβ (29) suggesting that these findings indicate a complex relationship between polymorphic variations in CR1 and regulation of brain Aβ clearance. Brouwers et al. (28) also showed that the common AD risk association with CR1 may be explained by a low copy number repeat (LCR) in high LD with the risk variant that encodes a longer isoform (CR1-S) of the CR1 protein. This longer isoform has an increased number of C3b/C4b cofactor activity sites, which may have a positive effect on Aβ clearance through a C3b-mediated mechanism. However, similar to our present findings, this mechanism suggests that CR1-associated risk for AD in older individuals may not be mediated through increased accumulation of Aβ in the brain.
Alternative mechanisms that may mediate the association between CR1 and brain amyloid levels include its role as an inhibitor of complement activity. However, the net effects of CR1-mediated complement modulation on AD pathogenesis are unclear. Such effects may include, for instance, both a deleterious reduction in C3b-mediated clearance of neurotoxic Aβ species from the brain as well as a potentially protective effect through limiting immune-mediated damage of healthy neurons (30).
It is interesting to note that recent studies examining the effect of APOE genotype on AD risk associated with genetic variation in CR1 have been inconsistent. While a recent meta-analysis showed no evidence for an interaction between APOE genotype and CR1 in mediating risk for AD (31), other reports suggest that the increased risk of AD in carriers of the risk variant of rs3818361 is strongest in APOE ε4 carriers (2). Our current results suggest a complex interaction between CR1 and APOE that influences brain amyloid levels in non-demented older individuals.
Our findings showing widespread decreases in brain amyloid burden in non-demented carriers of an AD risk variant gene may also be relevant to recent efforts aimed at lowering Aβ production or enhancing its clearance in asymptomatic individuals at increased genetic risk for AD (32, 33). It is worth noting in this context that some previous studies suggest that Aβ deposition in the brain may be a protective adaptive response to neuronal stress and therapeutic strategies against it may exacerbate the disease process (34). Our results showing lower brain amyloid burden in non-demented carriers of the AD risk variant CR1 suggest that at least in this group of older individuals, further lowering brain amyloid levels may be of doubtful clinical benefit. Furthermore, by showing a robust interaction between the CR1 and APOE genes, our findings also suggest that clinical outcomes of such therapeutic approaches in presymptomatic individuals may be determined by complex gene-gene interactions.
It must be noted that the participants included in the 11C-PiB PET study described herein are derived from the neuroimaging substudy of the BLSA and represent a highly educated and healthy sample of non-demented older individuals. The non-demented individuals in our study remained so over intensive follow-up of more than 10 years since their entry into the neuroimaging substudy of the BLSA. Our findings may thus suggest robust compensatory mechanisms in at-risk participants in this cohort that serve to maintain cognitive health. We must also acknowledge that although we were able to independently replicate our main finding of lower brain amyloid in CR1 risk carriers in the ADNI sample, the small number of subjects in the replication analyses did not allow us to test the presence of a CR1 × APOE interaction on brain amyloid in this sample.
Our findings merit consideration in the light of a recent study on the effect of the CR1 rs6656401 SNP on neuritic plaque burden in AD. In their study reporting an association between the rs6656401A SNP in CR1 and higher neuritic plaque burden in the brain, Chibnik and colleagues studied 553 older individuals who came to autopsy, of whom 220 carried a pathological diagnosis of AD (6). It is worth noting that the minor allele frequency (MAF) in our current report (0.16) is comparable to that in their autopsy sample (0.17). However, there are a number of methodological differences between our study and that of Chibnik and colleagues. The latter study was based on an autopsy sample of individuals consisting of both pathologically confirmed AD cases as well as non-AD control subjects. It is not clear whether the association of the rs6656401A SNP with neuritic plaque burden in their study remained significant when the analysis was restricted to healthy controls and whether there was a statistical interaction between the CR1 and APOE risk alleles. Another methodological distinction between our current report and theirs is our use of in vivo amyloid imaging to quantify brain fibrillar amyloid burden in a variety of brain regions that are not typically examined in post-mortem brain tissue using CERAD criteria (35). It also is notable that the observed MAF in the rs3818361 SNP in our study is also comparable to the index GWAS study by Lambert and colleagues in a European population where the MAF for this SNP among more than 8000 control subjects was reported to be 0.19 (2). Similarly, a recent meta analysis of studies describing the association of the CR1 rs3818361 SNP with AD risk included six separate cohorts with a range of the MAF among more than 19,000 control subjects being 0.17 to 0.23 (36). Nevertheless, replication of our present findings using in vivo amyloid imaging in a larger sample that is more representative of community-dwelling elderly and inclusive of individuals with cognitive impairment may be informative.
Conclusions
In summary, our findings suggest a complex effect of the common AD risk variant CR1 on brain amyloid deposition and its modulation by APOE genotype. These findings are relevant to emerging disease-modifying treatments targeting brain Aβ deposition in pre-symptomatic individuals at risk for AD.
Supplementary Material
Acknowledgments
We are grateful to the Baltimore Longitudinal Study of Aging participants and neuroimaging staff for their dedication to these studies and the staff of the Johns Hopkins University PET facility for their assistance. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD. (http://biowulf.nih.gov).This work was supported in part by research and development contract N01-AG-3-2124 from the Intramural Research Program, National Institute on Aging, National Institutes of Health. The replication analysis in this report was based on data from the ADNI study (National Institutes of Health (NIH) Grant U01 AG024904; RC2AG036535). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC; Johnson & Johnson Pharmaceutical Research & Development LLC; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30AG010129 and K01AG030514.
Footnotes
Potential conflicts of interest: Dr. Wong discloses the following: Consulting for Amgen, Funded Research in addition to NIH: Avid, Biotie, GE, Intracellular, Johnson and Johnson, Lilly, Lundbeck, Merck, Orexigen, Otsuka, Roche, and Sanofi-Aventis. Dr. Thambisetty is a named inventor on a patent application related to blood biomarkers for Alzheimer's disease filed by his previous employer, Kings College, London. Dr. Saykin has served as a consultant to Baxter International Inc., Bristol-Myers Squibb, and Pfizer Inc.; and has received research support from Pfizer Inc., Eli Lilly and Company, Siemens AG, Welch Allyn Inc., the NIH (R01 CA101318 [PI], R01 AG19771 [PI], RC2AG036535 [Genetics Core Leader], P30 AG10133-18S1 [Imaging Core Leader], and U01 AG032984 [Site PI and Chair, Genetics Working Group]), the Indiana Economic Development Corporation (IEDC #87884), and the Foundation for the NIH. All other authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 3.Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, et al. Replication of CLU, CR1, and PICALM associations with alzheimer disease. Arch Neurol. 2010;67:961–964. doi: 10.1001/archneurol.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corneveaux JJ, Myers AJ, Allen AN, Pruzin JJ, Ramirez M, Engel A, et al. Association of CR1, CLU and PICALM with Alzheimer's disease in a cohort of clinically characterized and neuropathologically verified individuals. Hum Mol Genet. 2010;19:3295–3301. doi: 10.1093/hmg/ddq221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chibnik LB, Shulman JM, Leurgans SE, Schneider JA, Wilson RS, Tran D, et al. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann Neurol. 2010 doi: 10.1002/ana.22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGeer EG, Klegeris A, McGeer PL. Inflammation, the complement system and the diseases of aging. Neurobiol Aging. 2005;26(1):94–97. doi: 10.1016/j.neurobiolaging.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Resnick SM, Goldszal AF, Davatzikos C, Golski S, Kraut MA, Metter EJ, et al. One-year age changes in MRI brain volumes in older adults. Cereb Cortex. 2000;10:464–472. doi: 10.1093/cercor/10.5.464. [DOI] [PubMed] [Google Scholar]
- 9.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 10.Petersen RC. Mild cognitive impairment as a diagnostic entity. J Intern Med. 2004;256:183–194. doi: 10.1111/j.1365-2796.2004.01388.x. [DOI] [PubMed] [Google Scholar]
- 11.Resnick SM, Sojkova J, Zhou Y, An Y, Ye W, Holt DP, et al. Longitudinal cognitive decline is associated with fibrillar amyloid-beta measured by [11C]PiB. Neurology. 2010;74:807–815. doi: 10.1212/WNL.0b013e3181d3e3e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Driscoll I, Zhou Y, An Y, Sojkova J, Davatzikos C, Kraut MA, et al. Lack of association between 11C-PiB and longitudinal brain atrophy in non-demented older individuals. Neurobiol Aging. 2011;32:2123–2130. doi: 10.1016/j.neurobiolaging.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sojkova J, Beason-Held L, Zhou Y, An Y, Kraut MA, Ye W, et al. Longitudinal cerebral blood flow and amyloid deposition: an emerging pattern? J Nucl Med. 2008;49:1465–1471. doi: 10.2967/jnumed.108.051946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terracciano A, Balaci L, Thayer J, Scally M, Kokinos S, Ferrucci L, et al. Variants of the serotonin transporter gene and NEO-PI-R Neuroticism: No association in the BLSA and SardiNIA samples. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:1070–1077. doi: 10.1002/ajmg.b.30932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, et al. Alzheimer's Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement. 2010;6:265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res. 1990;31:545–548. [PubMed] [Google Scholar]
- 17.Jagust WJ, Bandy D, Chen K, Foster NL, Landau SM, Mathis CA, et al. The Alzheimer's Disease Neuroimaging Initiative positron emission tomography core. Alzheimers Dement. 2010;6:221–229. doi: 10.1016/j.jalz.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price JC, Klunk WE, Lopresti BJ, Lu X, Hoge JA, Ziolko SK, et al. Kinetic modeling of amyloid binding in humans using PET imaging and Pittsburgh Compound-B. J Cereb Blood Flow Metab. 2005;25:1528–1547. doi: 10.1038/sj.jcbfm.9600146. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y, Resnick SM, Ye W, Fan H, Holt DP, Klunk WE, et al. Using a reference tissue model with spatial constraint to quantify [11C]Pittsburgh compound B PET for early diagnosis of Alzheimer's disease. Neuroimage. 2007;36:298–312. doi: 10.1016/j.neuroimage.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aisen PS, Petersen RC, Donohue MC, Gamst A, Raman R, Thomas RG, et al. Clinical Core of the Alzheimer's Disease Neuroimaging Initiative: progress and plans. Alzheimers Dement. 2010;6:239–246. doi: 10.1016/j.jalz.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reiman EM, Chen K, Liu X, Bandy D, Yu M, Lee W, et al. Fibrillar amyloid-beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer's disease. Proc Natl Acad Sci U S A. 2009;106:6820–6825. doi: 10.1073/pnas.0900345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thambisetty M, Tripaldi R, Riddoch-Contreras J, Hye A, An Y, Campbell J, et al. Proteome-based plasma markers of brain amyloid-beta deposition in non-demented older individuals. J Alzheimers Dis. 2010;22:1099–1109. doi: 10.3233/JAD-2010-101350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villemagne VL, Pike KE, Chetelat G, Ellis KA, Mulligan RS, Bourgeat P, et al. Longitudinal assessment of Abeta and cognition in aging and Alzheimer disease. Ann Neurol. 2011;69:181–192. doi: 10.1002/ana.22248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. controlling the false Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B-Methodol. 1995;57:289–300. [Google Scholar]
- 25.Swaminathan S, Shen L, Risacher SL, Yoder KK, West JD, Kim S, et al. Amyloid pathway-based candidate gene analysis of [(11)C]PiB-PET in the Alzheimer's Disease Neuroimaging Initiative (ADNI) cohort. Brain Imaging Behav. 2012;6:1–15. doi: 10.1007/s11682-011-9136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP, et al. Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound-B. Ann Neurol. 2004;55:306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- 27.Jun G, Naj AC, Beecham GW, Wang LS, Buros J, Gallins PJ, et al. Meta-analysis confirms CR1, CLU, and PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol. 2010;67:1473–1484. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brouwers N, Van Cauwenberghe C, Engelborghs S, Lambert JC, Bettens K, Le Bastard N, et al. Alzheimer risk associated with a copy number variation in the complement receptor 1 increasing C3b/C4b binding sites. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kauwe JS, Cruchaga C, Karch CM, Sadler B, Lee M, Mayo K, et al. Fine mapping of genetic variants in BIN1, CLU, CR1 and PICALM for association with cerebrospinal fluid biomarkers for Alzheimer's disease. PLoS One. 2011;6:e15918. doi: 10.1371/journal.pone.0015918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aiyaz M, Lupton MK, Proitsi P, Powell JF, Lovestone S. Complement activation as a biomarker for Alzheimer's disease. Immunobiology. 2011 doi: 10.1016/j.imbio.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 31.Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, et al. Replication of BIN1 Association with Alzheimer's Disease and Evaluation of Genetic Interactions. J Alzheimers Dis. 2011 doi: 10.3233/JAD-2011-101932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golde TE, Schneider LS, Koo EH. Anti-abeta therapeutics in Alzheimer's disease: the need for a paradigm shift. Neuron. 2011;69:203–213. doi: 10.1016/j.neuron.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reiman EM, Langbaum JB, Tariot PN. Alzheimer's prevention initiative: a proposal to evaluate presymptomatic treatments as quickly as possible. Biomark Med. 2010;4:3–14. doi: 10.2217/bmm.09.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HG, Casadesus G, Zhu X, Takeda A, Perry G, Smith MA. Challenging the amyloid cascade hypothesis: senile plaques and amyloid-beta as protective adaptations to Alzheimer disease. Ann N Y Acad Sci. 2004;1019:1–4. doi: 10.1196/annals.1297.001. [DOI] [PubMed] [Google Scholar]
- 35.Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, Brownlee LM, et al. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41:479–486. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- 36.Antunez C, Boada M, Lopez-Arrieta J, Moreno-Rey C, Hernandez I, Marin J, et al. Genetic association of complement receptor 1 polymorphism rs3818361 in Alzheimer's disease. Alzheimers Dement. 2011;7:e124–129. doi: 10.1016/j.jalz.2011.05.2412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.