Abstract
The prevalence of aromatic residues in the ligand binding site of the GABAA receptor, as with other cys-loop ligand-gated ion channels, is undoubtedly important for the ability of neurotransmitters to bind and trigger channel opening. Here we have examined three conserved tyrosine residues at the GABA binding pocket (β2Tyr97, β2Tyr157, and β2Tyr205), making mutations to alanine and phenylalanine. We fully characterized the effects each mutation had on receptor function using heterologous expression in HEK-293 cells, which included examining surface expression, kinetics of macroscopic currents, microscopic binding and unbinding rates for an antagonist, and microscopic binding rates for an agonist. The assembly or trafficking of GABAA receptors was disrupted when tyrosine mutants were expressed as αβ receptors, but interestingly not when expressed as αβγ receptors. Mutation of each tyrosine accelerated deactivation and slowed GABA binding. This provides strong evidence that these residues influence the binding of GABA. Qualitatively, mutation of each tyrosine has a very similar effect on receptor function; however, mutations at β2Tyr157 and β2Tyr205 are more detrimental than β2Tyr97 mutations, particularly to the GABA binding rate. Overall the results suggest that interactions involving multiple tyrosine residues are likely during the binding process.
Keywords: GABAA receptor, GABA binding, surface expression, kinetics
Introduction
At the heart of synaptic transmission is the binding of presynaptically released neurotransmitters with postsynaptic ionotropic receptors. The interaction of neurotransmitter and receptor sets in motion a cascade of structural rearrangements that culminates with receptor activation and channel opening. For the pentameric cys-loop receptors, which include GABAA, nACh, glycine, and 5-HT3 receptors, the site of neurotransmitter binding has been pinpointed to the conserved extracellular regions at inter-subunit interfaces (Connolly and Wafford, 2004). The five subunits of each receptor are arranged around a central ion-conducting pore, and each subunit consists of a large extracellular N-terminal domain, followed by a transmembrane domain with four α-helices. Binding sites exist at multiple interfaces where several discontinuous loops from both subunits of an interface form a binding pocket. A concentration of aromatic residues exists at the binding pocket and these residues are believed to, at least partially, mediate neurotransmitter binding (Figure 1A). Termed the “aromatic box”, this structural motif has been postulated to provide a source of hydrophobicity that creates a water-free environment suitable for the docking of a ligand, and to bind positively charged ligands via cation-π interactions (Dougherty and Stauffer, 1990). Indeed a cation-π interaction involving an aromatic residue that stabilizes binding of the ligand has been identified in each cys-loop receptor by using a sophisticated strategy involving the incorporation of fluorinated aromatic residues with decreasing cation-π binding affinity at prospective cation-π sites (Zhong et al., 1998; Beene et al., 2002; Celie et al., 2004; Padgett et al., 2007; Pless et al., 2008).
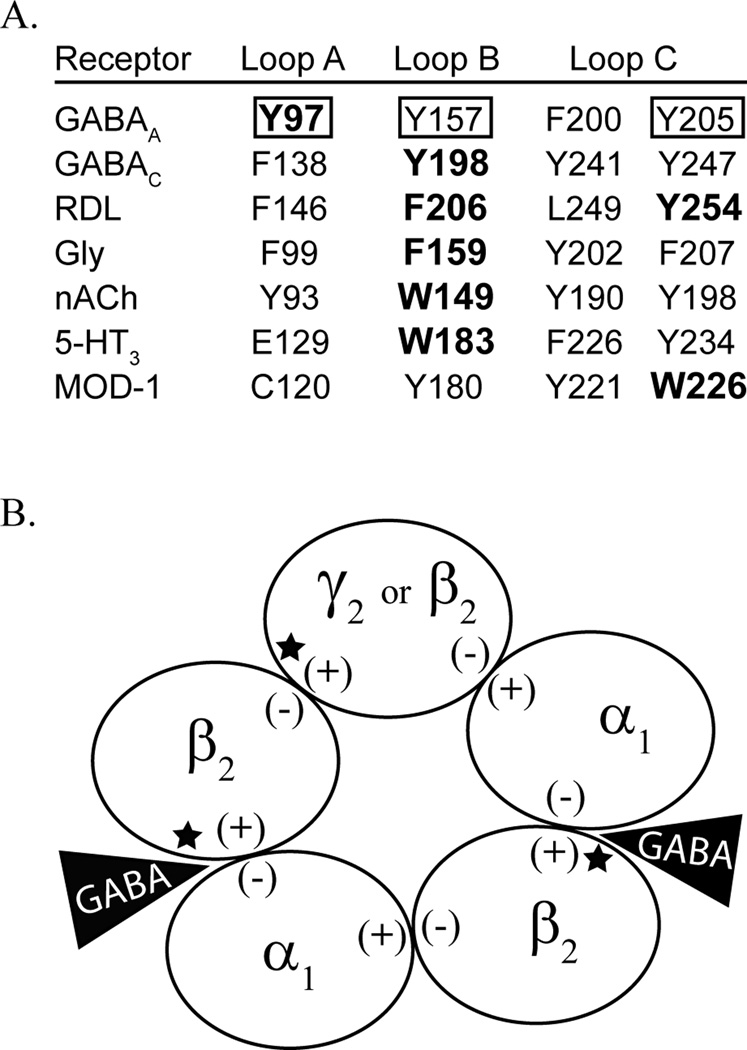
Figure 1.
The present study investigates tyrosine mutations of the β2 subunit. A) Alignment of the tyrosine residues in the binding pocket of the GABAA receptor, denoted by a box, with aromatic residues across the cys-loop family of receptors. Residues known to contribute to cation-π interactions are in bold. B) Depicted is a GABAA receptor that is comprised of 2α1, 2β2, and a fifth subunit that can be β2 or γ2. The (+) and (−) face of each subunit is labeled. The GABA binding sites, at the β/α interfaces, are indicated by a black triangle, and the locations of the mutations studied here are denoted by a star.
The GABAA receptor, which mediates inhibitory signaling in the mammalian brain, has three aromatic tyrosine residues in the binding pocket found on different loops of the β subunit (β2Tyr97-Loop A, β2Tyr157-Loop B, β2Tyr205-Loop C). Site-directed mutagenesis studies have found that mutation of any of these residues causes a decrease in GABA potency and may prevent expression of functional receptors (Amin and Weiss, 1993; Boileau et al., 2002; Padgett et al., 2007). Potential cation-π interactions involving Tyr97, Tyr157, or Tyr205 have been examined, and of these only Tyr97 was found to participate in a cation-π interaction, presumably with the amine group of GABA (Padgett et al., 2007). The other two tyrosines were found to influence GABA potency in a manner uncorrelated with cation-π binding affinity, but attributed to the hydroxyl group of the tyrosine.
We currently have a general picture of neurotransmitter binding, but we are continuing to work towards a detailed understanding of how the aromatic box mediates ligand binding and influences receptor activation. Previous studies of the aromatic box residues have relied on comparisons of EC50 values. Although a large increase in the EC50 value can be indicative of a general disruption in GABA binding or activation of the receptor, it fails to distinguish the myriad of effects on receptor function caused by a given mutation. In this study we aimed to explore, in depth, the role of the three tyrosine residues at the GABA binding pocket, and break down the contributions of each residue to a variety of specific receptor characteristics. We characterized how mutations of each tyrosine residue influenced surface expression of receptors, used sub-millisecond ligand application to observe changes to the kinetics of macroscopic desensitization and deactivation, and experimentally measured the binding rates of an agonist and competitive antagonist for each mutation.
Methods
cDNA constructs, mutagenesis, and HaloTag® fusion constructs
Human α1, β2, and γ2S subunits inserted into the pcDNA 3.1 vector were used. Mutant β2 and γ2S subunits were created using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA), and double-stranded sequencing of the entire coding region was conducted in order to verify fidelity.
The sequence for the HaloTag® (Promega, Madison, WI) was inserted between the codons for the fourth and fifth amino acid of the mature β2 subunit and of the mature γ2S subunit. These subunits were referred to as β2-Halo and γ2-Halo.
Cell Culture and Transfection
Human embryonic kidney (HEK-293) cells were cultured in Minimum Essential Medium Eagle with Earle’s salts (Mediatech, Manassas, VA) supplemented with 10% newborn calf serum (Thermo Scientific, Waltham, MA) and Penicillin-Streptomycin-Glutamine (Mediatech) in a 37° C incubator under a 5% CO2 atmosphere. For electrophysiology experiments, cells were plated onto 35 mm dishes coated with poly-L-lysine and were transfected 18–24 hours later using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Receptors consisting of α and β subunits, or α, β, and γ subunits were used throughout this study. For αβ receptors, the following amounts of cDNA were transfected: 250 ng enhanced green fluorescent protein, 1.5 µg of α1, and 1.5 µg of β2 (wild-type or mutant). For αβγ receptors the following amounts of cDNA were transfected: 250 ng enhanced green fluorescent protein, 1 µg of α1, 1 µg of β2 (wild-type or mutant), and 3 µg of γ2S (wild-type or mutant). During electrophysiology experiments, enhanced green fluorescent protein served as a marker for cells that were transfected. Cells were recorded from 48–96 hours post-transfection.
For fluorescent imaging experiments, HEK-293 cells were plated onto poly-d-lysine coated glass coverslips that had been placed in the bottom of 24-well plates. The next day transfection with Lipofectamine 2000 was carried out. The media was changed 36 hours post transfection and after an additional 24 hours two-step labeling experiments were performed.
Labeling with HaloTag® technology
Sequential labeling of cells expressing HaloTag® GABAA receptor subunits was performed first with the cell-impermeant HaloTag® AlexaFluor® 488 ligand (Promega) (1µM in media, 20 min), and then washed with warm media for 25 minutes (37° C, 5% CO2). The cells were labeled second with the cell-permeant HaloTag® tetra-methyl-rhodamine (TMR) ligand (Promega) (5 µM in media, 15 min), and then washed in warm media for 25 minutes (37° C, 5% CO2). The cells were fixed immediately after labeling using 4% paraformaldehyde, 0.2% sucrose in 1X PBS (pH 7.5) for 10 minutes room temperature, followed by treatment with 0.2% Triton X-100 in 1X PBS for 10 minutes room temperature. The coverslips were placed onto glass slides with Vectashield mounting medium (Vector Laboratories, Inc., Burlingame, CA).
Fluorescent microscopy and analysis
Imaging was performed using a Nikon A1 confocal microscope (Tokyo, Japan) with a 40x objective. HaloTag® Alexafluor 488 (494 nm excitation, 517 nm emission) and HaloTag® TMR (555 nm excitation, 585 nm emission) images were collected with a Galvano scanner and analyzed using NIS-Elements Advanced Research software (Nikon).
The mean intensity of the HaloTag® AlexaFluor® 488 label and the HaloTag® TMR label was measured for individual cells after subtracting background fluorescence. Using a Bezier hollow tool, a region of interest was established around every cell that appeared to have intracellular HaloTag® TMR labeling, and hence expression of GABAA receptor subunits. The percent surface expression was calculated as the mean intensity of HaloTag® AlexaFluor® 488 divided by the sum of HaloTag® AlexaFluor® 488 and HaloTag® TMR label mean intensities.
Electrophysiology
All recordings for this study were collected by voltage-clamp electrophysiology using outside-out patches excised from HEK-293 cells and held at −60 mV. Recordings were made using borosilicate glass pipettes filled with (in mM): 140 KCl, 10 EGTA, 2 MgATP, 20 phosphocreatine and 10 HEPES, pH 7.3. Rapid solution exchange was accomplished using a multibarreled flowpipe array (Vitrodynamics, Rockaway, NJ) mounted on a piezoelectric bimorph (Vernitron, Bedford, OH), which was driven by a computer-controlled current source. GABAA receptor agonists and antagonists were dissolved in the perfusion solution, which contains (in mM) 145 NaCl, 2.5 KCl, 2 CaCl2, 1 MgCl2, 10 HEPES and 4 Glucose, pH 7.4. For extracellular solutions that contained >30 mM GABA, the concentration of NaCl was reduced to 95 mM, and a combination of sucrose and GABA was added to compensate for the reduced osmolarity. The pipette solution was adjusted in conjunction, reducing the KCl concentration to 90 mM, and adding 50 mM K-gluconate to maintain a constant Cl− driving force. GABA and SR-95531 were obtained from Sigma-Aldrich Chemicals, St Louis, MO. Currents were low-pass filtered at 2–5 kHz with a four-pole Bessel filter, and data were collected at 20 kHz using an Axopatch 200B amplifier (Axon Instruments, Foster City, CA) and an ITC-1600 digitizer (InstruTech, Port Washington, NY), controlled by Axograph X software (Axograph Scientific, Sydney, AUS). Macroscopic current ensembles were collected with 15-second intervals between consecutive solution applications.
Antagonist unbinding experiments
Outside-out patches were pre-equilibrated in SR-95531 for 750 ms, and then rapidly switched to a solution containing saturating GABA. Home-written Matlab (Mathworks, Natick, MA) routines were used for the deconvolution of the time course of antagonist unbinding from this evoked current (Jones et al., 2001). The time course of antagonist unbinding was fit with an exponential function, yielding koff-SR and the percentage of receptors occupied by antagonist at equilibrium in the absence of GABA (Jones et al., 1998). The experiment was repeated several times, pre-equilibrating in different test concentrations of SR-95531. KD-SR was determined by plotting the antagonist occupancy versus concentration. The binding rate of SR-95531 (kon-SR) was calculated from the measured values of koff-SR and KD-SR.
Measuring the microscopic binding rate of GABA
The microscopic binding rate of GABA (kon-GABA) was measured using "race" experiments (Jones et al., 1998). Agonist (having an unknown binding rate) was co-applied with an antagonist, which had a previously determined binding rate (kon-ant, see "Antagonist Unbinding Experiments" above). The amplitude of the current evoked by co-application of agonist and antagonist was compared to the amplitude of current evoked by agonist alone, and this ratio (Iag-ant/Iag-only) was called Irace. The agonist binding rate, kon-agonist, was calculated using the following equation: kon-agonist = ([antagonist]·kon-ant)/([agonist]·(1/Irace − 1)) (Jones et al., 1998).
Statistical methods
Graphpad Prism 4 (GraphPad Software, Inc., San Diego, CA) was employed for performing statistical significance tests, as well as fitting concentration-response curves. For statistical analysis of fluorescent microscopy data an arcsine transform was performed on the percent surface expression, providing a normal distribution of the data. These values were used with a Student's t-test in order to compare mutant and wild-type receptors. Significant differences between macroscopic currents or microscopic kinetic rates of control and mutant receptors were tested using ANOVA with Dunnett’s post-test. All values are presented as the mean ± SEM.
Results
Electrophysiological responses were altered by mutation of binding pocket tyrosines
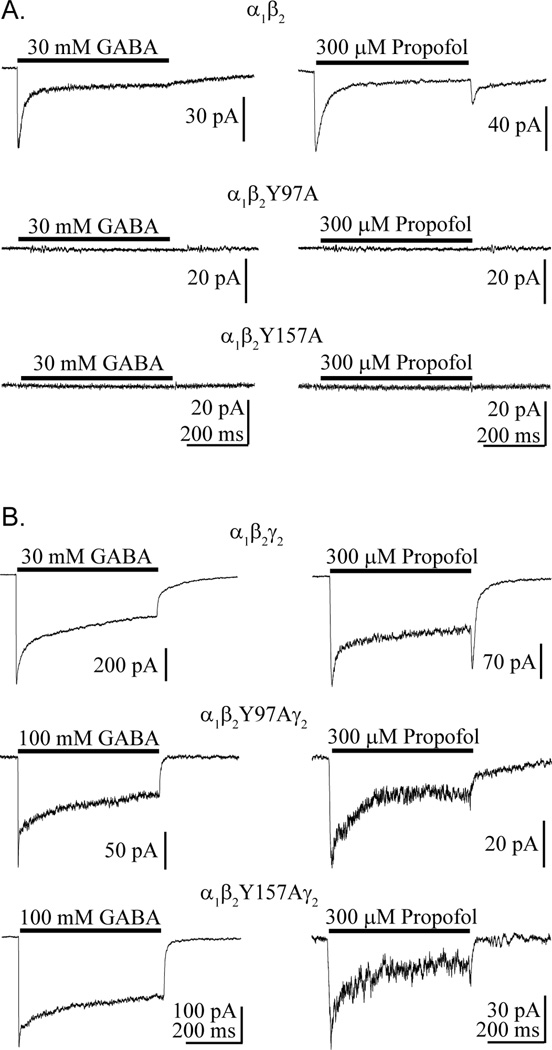
The residues Tyr97, Tyr157, and Tyr205, which lie on the (+) face of the β2 subunit (Figure 1B), were individually mutated to alanine, and the mutant constructs were separately transfected with the α1 construct in HEK-293 cells. GABA application (30–100 mM) to outside-out patches pulled from these cells never evoked substantial currents (Figure 2A). Propofol (100–300 µM), which has a distinct binding site from the GABA binding site, also failed to evoke current with patches pulled from these cells. Interestingly, when these β2 mutant subunits were expressed with α1 and γ2, GABA application consistently evoked a current response for Y97A and Y157A (Figure 2B). Y205A, which is not shown, failed to yield currents in response to 100 mM GABA or 300 µM propofol when expressed with α1 and γ2.
Figure 2.
GABA and propofol responses from receptors containing mutant β2 subunits. Currents were evoked by a 500 ms application of GABA or propofol (black bar) to patches pulled from HEK-293 cells expressing control and mutant receptor types comprised of α and β subunits (A) or α, β, and γ subunits (B).
β2Y97A and β2Y157A prevented surface expression of GABAA receptors when expressed only with α1
Previously, several mutations on the β2 subunit at the GABA binding site were found to abolish GABAA receptor function (Amin and Weiss, 1993; Newell et al., 2004), suggesting a role for such residues in assembly of the β(+)/α(−) inter-subunit interface; however it has not been verified whether a) non-functional receptors reach the cell surface, b) transport of fully assembled receptors to the cell surface is disrupted, or c) the subunits fail to completely oligomerize. In order to clarify the impact each mutation in this study has on GABAA receptor biogenesis and function, the cell surface expression of receptors was assessed. This was accomplished using a HaloTag® (Promega), which has been commercially designed with various fluorescent ligands that are highly specific, covalently bind, and are either cell-permeable or cell-impermeable. Responses of outside-out patches with α1β2-Haloγ2 receptors to GABA application were indistinguishable from α1β2γ2 receptors when comparing current amplitude and EC50-GABA. The HaloTag® did not alter the electrophysiological expression profile of β2-HaloY97A or β2-HaloY157A, which only exhibited GABA-evoked currents when expressed with both α1 and γ2 subunits.
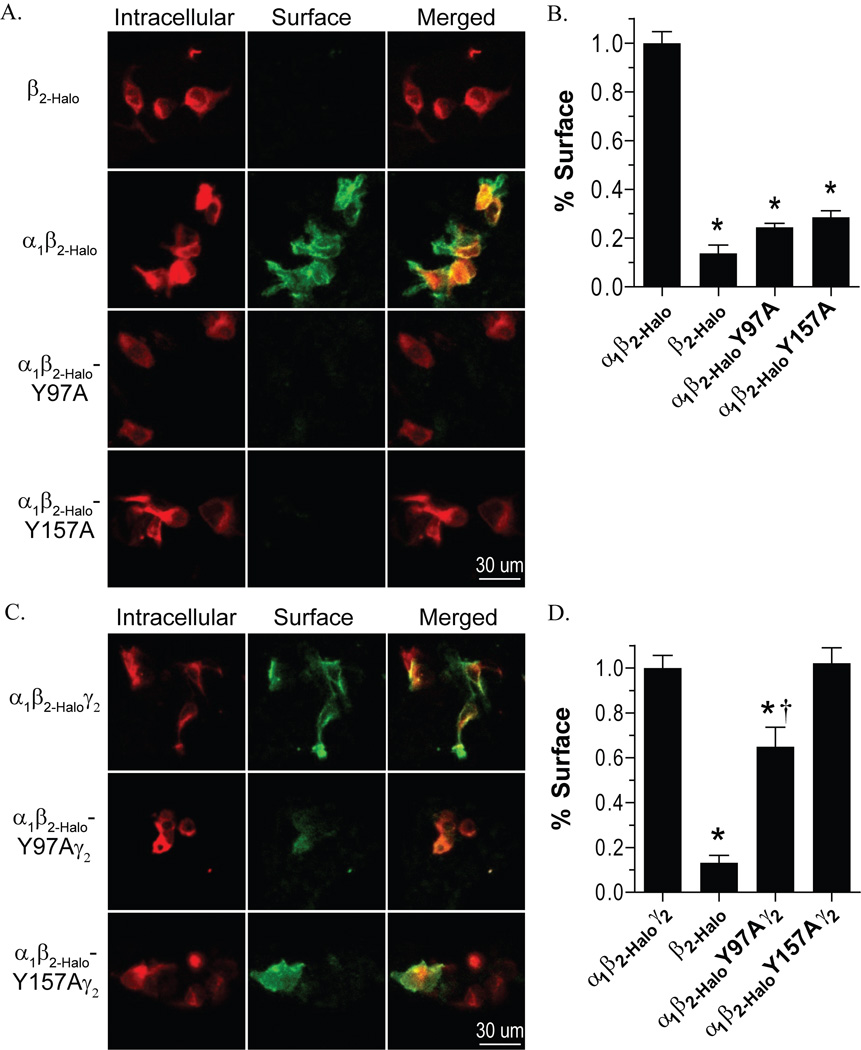
Sequential labeling of the cell surface with the cell-impermeant HaloTag® AlexaFluor® 488 ligand, followed by intracellular labeling with the cell-permeant HaloTag® TMR ligand revealed that α1β2-Halo and α1β2-Haloγ2 receptors exhibited pronounced surface labeling (Figure 3). Expression of β2-Halo alone exhibited no surface labeling (Figure 3A). This was in agreement with previous reports of the β2 subunit being unable to reach the cell surface without being assembled into a pentamer with α subunits or other isoforms (Connolly et al., 1996). Therefore, β2-Halo alone was used as a negative control and α1β2-Halo and α1β2-Haloγ2 receptors were used as positive controls for surface expression.
Figure 3.
Surface expression of receptors containing mutant β2 subunits. A and B) Expression of Halo-tagged subunits was determined by labeling with a cell-impermeable Alexafluor 488 ligand (green), followed by labeling with a cell-permeable TMR ligand (red), and images were collected by confocal microscopy. C and D) The percent of surface expression was quantified by measuring the mean intensity of the AlexaFluor 488 label relative to the total of AlexaFluor 488 label plus TMR label mean intensity of individual cells. The percent surface expression is normalized to the wild-type α1β2-Halo receptor (C) or α1β2-Haloγ2 receptor (D). * Denotes significant difference when compared to the wild-type receptor and † denotes a significant difference when compared to the negative control (β2-Halo only), Student’s t-test (p < 0.05).
Comparison of the cell surface label and the intracellular label revealed only intracellular labeling for both β2-HaloY97A and β2-HaloY157A when expressed with α1 (Figure 3A). These results were quantitatively assessed by comparing the percent surface label for individual cells. Both α1β2-HaloY97A and α1β2-HaloY157A demonstrated significantly reduced surface expression compared to wild-type α1β2-Halo, and were indistinguishable from the negative control (Figure 3C). Also, there was no difference in the average intracellular label intensity for cells expressing the wild-type or mutant receptors (data not shown), indicating overall subunit expression was not compromised by these mutations.
When expressed with α1 and γ2, β2-HaloY157A surface expression was visible (Figure 3B). We presumed the β2-HaloY157A subunit was associated with α1 and γ2 when it reached the cell surface because previous studies showed β2 and γ2 subunits did not reach the cell surface without the α1 subunit (Bollan et al., 2003). Quantitatively, the percent surface expression was not different from the positive control (Figure 3D). β2-HaloY97A, however, exhibited reduced surface labeling when expressed with α1 and γ2 (Figure 3B). The percent surface expression for α1β2-HaloY97Aγ2 was significantly reduced compared to wild-type receptors, but was still greater than the negative control (Figure 3D).
The ability of γ2 to rescue the functional expression of the β2 mutants may be the result of a reduction in the number of subunit interfaces that contain a mutation. In order to evaluate if this was a simple dosage effect and if the accumulation of mutations at subunit interfaces inhibited functional expression, we tested whether the γ2 subunits with the conserved aromatic residue mutated to alanine (β2Tyr97 aligns with γ2Phe112, and β2Tyr157 aligns with γ2Tyr172) could still rescue the functional expression of the corresponding β2 mutants. HEK-293 cells transfected with α1β2Y97Aγ2-HaloF112A displayed neither GABA-evoked (100 mM) nor propofol-evoked (300 µM) current. In contrast, patches pulled from cells expressing α1β2Y157Aγ2-HaloY172A repeatedly exhibited a current response to GABA, indicating functional surface expression of these receptors.
To test whether the γ2 mutations reduced the surface expression of receptors, labeling and fluorescence microscopy were performed (Supplemental Figure 1A). α1β2γ2-HaloF112A exhibited only 75% surface expression compared to wild-type receptors, while the percent surface expression for α1β2γ2-HaloY172A was equivalent to the wild-type (Supplemental Figure 1B). Combination of the γ2-HaloF112A subunit with β2Y97A completely abolished surface expression, whereas combination of γ2-HaloY172A and β2Y157A still exhibited the equivalent surface expression as wild-type receptors.
Mutation of binding pocket tyrosines caused qualitatively similar changes in macroscopic kinetics
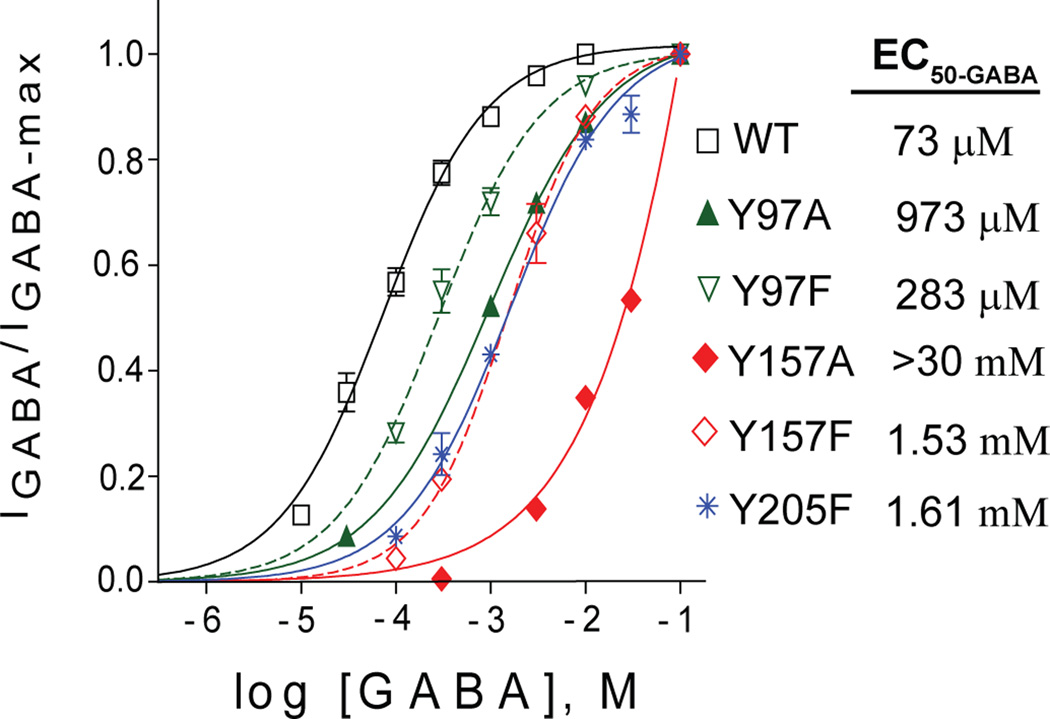
Mutation of these tyrosines undoubtedly alters GABA activation of the receptor, as indicated by previous concentration-response studies (Amin and Weiss, 1993; Padgett et al., 2007). In order to elucidate the side-chain requirements for normal receptor function, we compared mutations to alanine, which is a small hydrophobic side-chain, and mutations to phenylalanine, which maintains the aromatic ring of the wild-type residue but lacks the hydroxyl group at the C4 position. Receptors comprised of α, β, and γ subunits were used, therefore the mutations were present only at the interfaces that form a binding pocket. The alanine mutation of Tyr97 caused a 14-fold increase in EC50-GABA, which was partially rescued by phenylalanine, still increasing the EC50-GABA 4-fold (Figure 4). Tyr157 and Tyr205 mutations showed a similar pattern, although the effects were more severe. Y157A and Y157F increased the EC50-GABA more than 400-fold and 21-fold, respectively. Y205A completely disrupted GABAA receptor function, while Y205F increased EC50-GABA 22-fold.
Figure 4.
Mutation of β2 subunit tyrosine residues in the binding pocket shifts the GABA concentration-response curve rightward. Responses of αβγ receptors to a series of GABA concentrations were normalized for the maximum evoked current. The data was fit with a sigmoidal curve: Y= Ymin + (Ymax − Ymin) / (1+10^(LogEC50 − X)*HillSlope)).
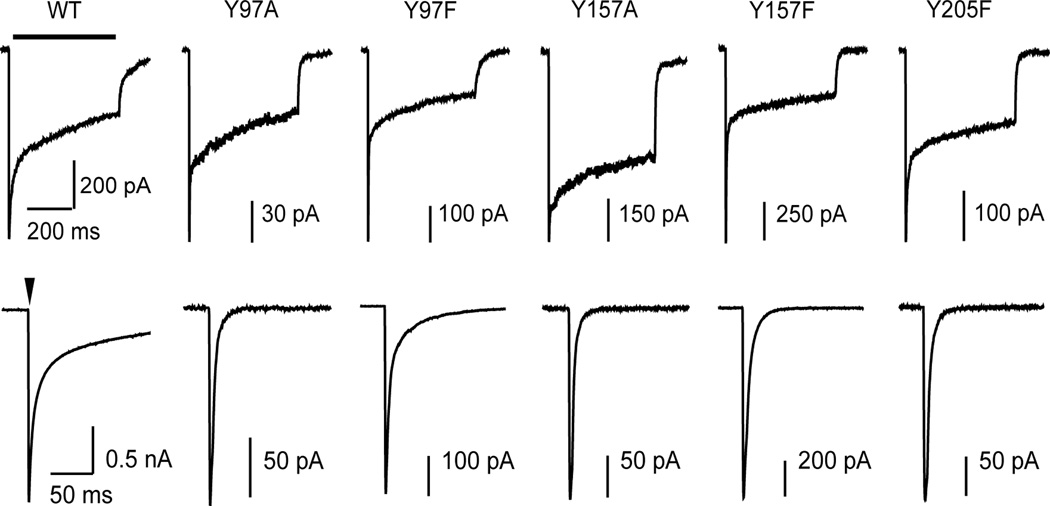
Macroscopic deactivation and desensitization were examined to further explore the role of each tyrosine in receptor function. It was found that mutations made to the three tyrosine residues all resulted in an increased rate of deactivation (Figure 5). Bi-exponential fits of the deactivation phase revealed an acceleration of both the fast and slow components, leading to a significant change in the weighted time constant of decay for each mutation (Table 1). For Tyr97 and Tyr157 the effects of phenylalanine mutations were less severe than alanine mutations, but either mutation significantly increased the rate of deactivation. For Y205A, GABA-evoked response could not be achieved, therefore only the macrokinetic effects of Y205F were assessed. Desensitization was unchanged for Y97A, Y157F, and Y205F (Table 1). For Y97F desensitization was deeper and more rapid than wild-type. For Y157A desensitization did not reach the same extent as wild-type; however, interpretation is limited because the maximal concentration of GABA (100 mM) could not be confirmed as saturating.
Figure 5.
Mutating Tyr97, TyrY157, and TyrY205 to either alanine or phenylalanine results in more rapid macroscopic deactivation. In the upper row are representative currents evoked by long (500 ms, bar) pulses and in the lower row are representative currents evoked by short (4 ms, arrow head) pulses of GABA (10 mM for wild-type; 100 mM for all mutants). Macroscopic desensitization and deactivation are quantified by fitting each phase with an exponential decay function (see Table 1).
Table 1.
Summary of exponential fits to deactivation and desensitization
| Deactivation (4 ms pulse) |
% τfast | τfast (ms) | % τslow | τslow (ms) | τw (ms) | n | |
|---|---|---|---|---|---|---|---|
| α1β2γ2 | 76±2 | 7.3±0.8 | 24±2 | 124.5±12.7 | 37.4 ± 5.4 | 14 | |
| α1β2Y97Aγ2 | 84±2 | 5.2±0.8 | 16±2 | 51.3±20.0** | 11.8 ± 2.7 ** | 6 | |
| α1β2Y97Fγ2 | 74±2 | 3.9±0.3* | 26±2 | 45.4±4.7** | 14.7 ± 1.6 ** | 5 | |
| α1β2Y157Aγ2 | 84±8 | 1.8±0.5** | 16±2 | 17.1±5.2** | 3.1 ± 1.1 ** | 4 | |
| α1β2Y157Fγ2 | 73±9 | 4.1±0.7* | 27±2 | 17.5±2.9** | 8.5 ± 2.1 ** | 5 | |
| α1β2Y205Fγ2 | 75±4 | 2.6±0.1** | 25±2 | 15.0±2.8** | 5.2 ± 0.4 ** | 8 | |
| Desensitization (500 ms pulse) |
% τfast | τfast (ms) | % τslow | τslow (ms) | % remaining |
τw (ms) | n |
| α1β2γ2 | 32±3 | 7.4±1.0 | 29±2 | 247.8±12.6 | 39±2 | 130 ± 11 | 39 |
| α1β2Y97Aγ2 | 35±4 | 3.6±1.2 | 32±3 | 272.3±22.4 | 33±8 | 136 ± 20 | 12 |
| α1β2Y97Fγ2 | 56±3** | 4.0±0.5 | 25±2 | 170.0±12.0** | 19±2** | 58 ± 9 ** | 10 |
| α1β2Y157Aγ2 | 10±6* | 2.2±1.1 | 29±2 | 190.1±34.9 | 61±4** | 139 ± 14 | 4 |
| α1β2Y157Fγ2 | 43±2* | 6.0±0.6 | 24±1 | 244.9±10.6 | 33±1 | 96 ± 10 | 17 |
| α1β2Y205Fγ2 | 28±3 | 10.±1.3 | 31±2 | 280.0±22.8 | 41±2 | 160 ± 18 | 11 |
Deactivation and desensitization phases were fit with bi-exponential equations, Y= A1 · e−t/τfast + A2 · e−t/τslow and Y= A1 · e−t/τfast + A2 · e−t/τslow + C, performed with Axograph. The weighted time constant (τw) was computed (A1/(A1 + A2)) · τfast + (A2/(A1+A2)) · τslow. Significant differences between wild-type and mutant receptors were calculated using ANOVA with a Dunnett’s posttest
p < 0.05;
p < 0.01
Antagonist unbinding was accelerated by mutation to binding pocket tyrosines
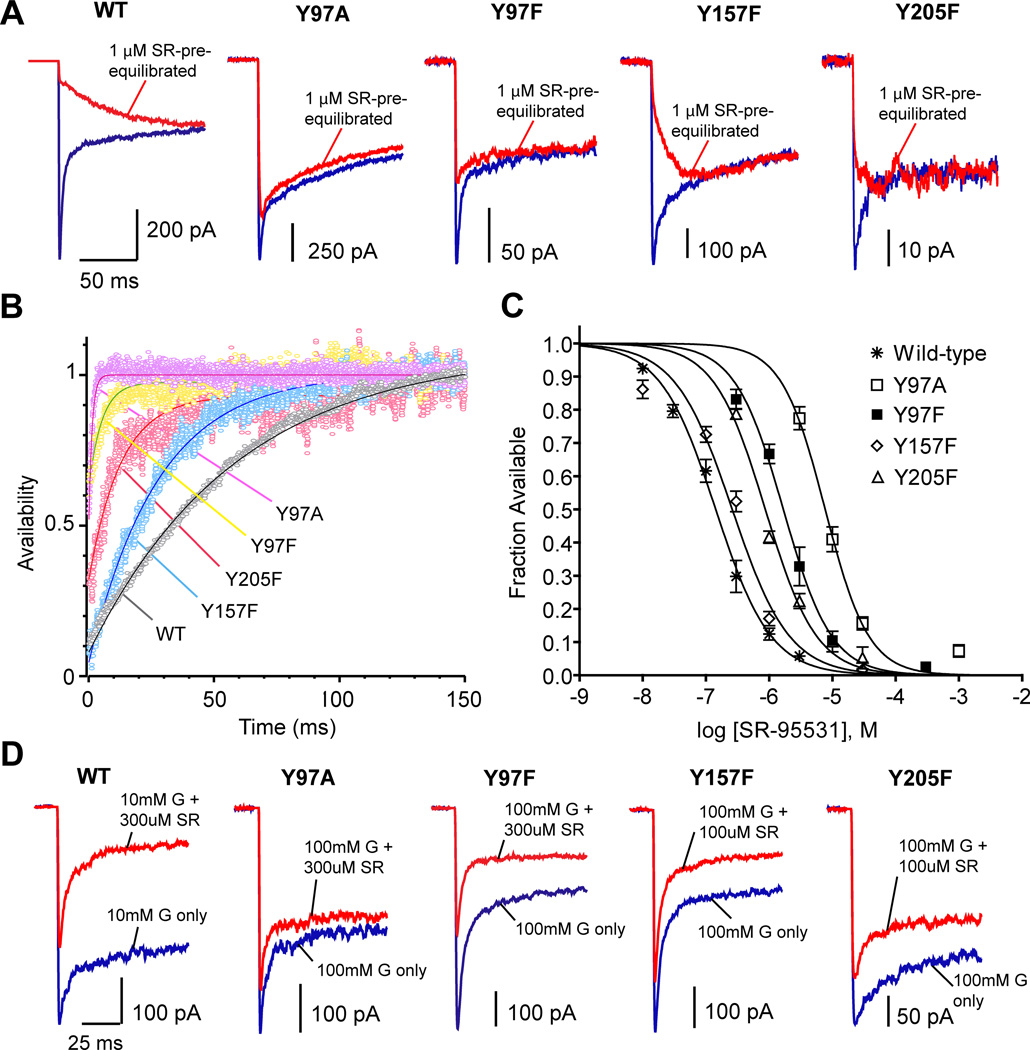
Competitive antagonists such as SR-95531 prevent GABA binding because they bind at the same location. Also, GABA and SR-95531 share a structural motif with a carboxy group and an amino group separated by a three-carbon chain. In order to examine if any of the tyrosines mediate the interaction of such a ligand with the GABAA receptor, we examined the effect that mutating each tyrosine has on the binding and unbinding rates of SR-95531. Using a deconvolution method described in detail byJones et al. (2001), we first obtained the unbinding rate from macroscopic currents. Figure 6A illustrates the response to GABA following pre-equilibration in SR-95531 for wild-type and mutant receptors. As the waveform of the pre-equilibrated current is the convolution of the GABA current and the antagonist unbinding time-course (Jones et al., 2001), the antagonist unbinding time-course was extracted from this data by deconvolving the GABA-only current from the pre-equilibrated current (Figure 6B). The time course of unbinding (τunbinding) was accelerated by mutation of each tyrosine. The microscopic unbinding rate for each receptor type, which is equal to 1/τunbinding, is presented in Table 2. The data concerning the microscopic kinetics for Y97A was also presented in a previous study of the binding pocket (Tran et al., 2011), while data for all other mutants are presented for the first time here. Mutation of Tyr97, either to alanine or phenylalanine, caused the most dramatic increase in the unbinding rate. Mutation of Tyr157 or Tyr205 to phenylalanine also caused a significant increase in the unbinding rate. Alanine mutations of Tyr157 and Tyr205 were too disruptive to complete analysis of microscopic kinetics.
Figure 6.
Antagonist unbinding and race experiments. A) Currents evoked by a control pulse of a saturating concentration of GABA (blue) are overlayed with currents evoked by the same GABA solution following pre-equilibration in 1 µM SR-95531 (red) for each receptor type. B) Plots show the deconvolution of GABA-evoked currents with SR-95531 pre-equilibration from those without pre-equilibration. Curve fits (solid lines) reveal the time course of SR-95531 unbinding. C) Concentration-response curves for the equilibrium antagonist occupancy in the absence of GABA were fit to the normalized hill equation I/Imax = 1 − 1/[(KD-SR/[SR-95531])N + 1]. D) Sample raw traces recorded from race experiments. Currents evoked by simultaneous application of GABA and SR-95531 (red traces) are overlayed with the currents evoked by GABA alone (blue traces) for each receptor type.
Table 2.
Summary of microscopic rates determined for SR-95531 and GABA
| KD-SR (µM) | koff-SR (s−1) | kon-SR (M−1s−1) | kon-GABA (M−1s−1) | |
|---|---|---|---|---|
| Wild-type | 0.14 | 15.9 ± 0.8 | (1.14 ± 0.10) × 108 | (7.36 ± 0.42) × 106 |
| Y97A | 7.68 | 832.1 ± 91.5 * | (1.08 ± 0.12) × 108 | (2.17 ± 0.13) × 106 * |
| Y97F | 1.67 | 188.9 ± 9.7* | (1.13 ± 0.06) × 108 | (5.66 ± 0.62) × 106 † |
| Y157F | 0.26 | 44.4 ± 1.6 * | (1.71 ± 0.06) × 108 | (1.60 ± 0.31) × 106 * |
| Y205F | 0.87 | 121.0 ± 7.9 * | (1.39 ± 0.09) × 108 | (6.03 ± 0.44) × 105 * |
Significant differences between wild-type and mutant receptors were calculated using ANOVA with a Dunnett’s post-test
p < 0.01;
p < 0.05
The antagonist unbinding experiment was repeated with pre-equilibration at different SR-95531 concentrations, and the microscopic affinity constant, KD-SR, was obtained by plotting the fraction of receptors that were unbound, and therefore available, at time zero versus antagonist concentration (Figure 6C). This plot was fit with a normalized Hill equation for antagonist. Each mutation caused an increase in KD-SR. The microscopic binding rate was then calculated using the equation kon = koff/KD. None of the mutations examined here caused a significant change in kon-SR.
GABA binding rates were reduced by mutation to binding pocket tyrosines
One microkinetic parameter we can readily measure for GABA is the binding rate (kon-GABA), which can serve as evidence for a residue’s participation in GABA binding. kon-GABA was measured using race experiments, as described previously byJones et al. (1998). This process first involves determining the binding rate for a competitive antagonist, in this case SR-95531, which was shown in the previous section (Figure 6A–C). Then the binding rate of GABA can be determined by performing a race experiment in which GABA and SR-95531 are co-applied (Figure 6D). The peak current resulting during co-application is compared to the current evoked by application of GABA alone. The extent to which the peak current is reduced by the presence of antagonist depends on the relative binding rates of the two compounds and the relative concentrations available. Since kon-SR has been determined by the antagonist unbinding experiment, kon-GABA can be calculated as kon-GABA = [SR-95531] kon-SR/([GABA](1/Irace −1)) (Jones et al., 1998), where Irace is the ratio of the peak response of co-application to the peak response of GABA alone.
It was observed that Y97A reduced kon-GABA 3.4-fold and Y97F only reduced kon-GABA 1.3-fold (Table 2). The conservative phenylalanine mutations of Tyr157 and Tyr205 were more detrimental, decreasing kon-GABA by 4.6-fold and 12-fold, respectively.
Discussion
The importance of several tyrosine residues in the GABA binding pocket has been evident since the first mutagenesis studies in this region. Yet, until now the precise contributions of each residue to the function of the receptor have not been described. Here, we have for the first time characterized the effect that mutation of each residue has on surface expression, macroscopic kinetics, and binding rates for GABA. The tyrosines of the binding pocket serve a dual function, required for receptor biogenesis and stability of the receptor-ligand complex.
Tyr97 and Tyr157 have distinct influences over surface expression of αβ and αβγ receptors
Substituted cysteine accessibility method experiments have demonstrated that Tyr97, Tyr157, and Tyr205 face the aqueous environment at the GABA binding pocket (Wagner and Czajkowski, 2001; Boileau et al., 2002; Newell et al., 2004). Since residues that are positioned on a subunit such that they face an inter-subunit interface are in an ideal location to mediate protein-protein interactions, we suspect that the tyrosines may influence receptor assembly. The absence of surface labeling when β2Y97A or β2Y157A is expressed with α1 supports the role of each residue in assembly or trafficking of the receptor. The interpretation of results with Halotag® fusion proteins assumes that labeling of the Halotag® represents labeling of an intact subunit. Although it remains to be investigated, our electrophysiology data corroborates the presence or absence of receptors at the cell surface in each case. The possibility exists that these mutations do not only interfere with the oligomerization of subunits, but may also alter subunit stability during the biogenesis process; however, the ability of the mutated β2 subunits to form functional receptors when expressed with α1 and γ2 makes it less likely that the mutations cause severe misfolding or substantial degradation of the subunit. Further biochemical analysis would be needed to confirm at what point between translation and surface expression the biogenesis of the receptors is disrupted.
The putative disruption of assembly by these mutations could manifest at the β(+)/α(−) interface (the GABA binding site) or the β(+)/β(−) interface, where they are present. The β(+)/β(−) interface only exists when expressing α and β, not when expressing α, β, and γ (Figure 1B). For both β2Y97A and β2Y157A, when expressed with α1 and γ2, functional expression of receptors resulted, demonstrating that the mutations do not absolutely prohibit the oligomerization of the β(+)/α(−) interface. However, based on surface labeling results, β2Y97A caused a reduction in receptors at the cell membrane when expressed with α1 and γ2. The sensitivity of surface expression to mutation at Tyr97 suggests that the residue plays a general role in assembly or trafficking of receptors, and the complete failure of assembly for αβ receptors may arise due to an accumulation of disturbances at three interfaces.
The normal expression of α1β2-HaloY157Aγ2 and α1β2Y157Aγ2-HaloY172A indicates that Tyr157 is specifically important for the assembly of a β(+)/β(−) interface, but not a γ(+)/β(−) interface or β(+)/α(−) interfaces. Although Tyr157 is present at the β(+)/α(−) interface, the results here suggest that it is not a major assembly determinant of this interface.
Tyrosine residues are critical to GABA binding
In the course of this study we consistently found common effects associated with the mutation of each tyrosine residue believed to be in or near the GABA binding pocket. This included a rightward shift of the GABA concentration response, an increased rate of deactivation, and a decreased GABA binding rate. Considering that desensitization was only minimally affected by the mutations, these results support the participation of these tyrosine residues primarily in the formation and stabilization of the receptor-ligand complex.
Our analysis of the macroscopic desensitization phase and macroscopic deactivation phase provides an indication of the kinetic transitions that may be altered by a given mutation, but specific kinetic transitions cannot simply be assigned to the observed phases in the macroscopic currents. The macroscopic desensitization phase is governed by transitions into and out of an unknown number of receptor states, and includes desensitization and resensitization transitions, as well as opening and closing transitions. Changes in any one of these kinetic transitions may manifest themselves in the observed macroscopic desensitization phase. The macroscopic deactivation phase is governed by all of the same transitions that govern the desensitization phase, but it additionally includes unbinding steps. Accelerated deactivation rates can result from increases in the closing rate, resensitization rate, or unbinding rate. Although we do not exclude all conceivable gating changes, a significant acceleration of the deactivation phase and largely unchanged desensitization phase (as seen with Y97A, Y157F, and Y205F) is consistent with an increase in the GABA unbinding rate (Wagner et al., 2004; Bianchi et al., 2007).
The measured changes in binding rates and plausible changes in unbinding rates would account for the large EC50 shifts. The tyrosine residues may contribute to ligand affinity in a variety of ways: directly interacting with the ligand, hydrophobically shielding the binding pocket to optimize energy of binding, or indirectly maintaining structural integrity of the binding pocket. Although we cannot exclude the possibility that mutation of the tyrosine residues caused long-range disturbances of protein conformations, which contributed to the affinity changes seen, these residues appear critical for both formation and stability of the receptor-ligand complex. Additionally, it is intriguing that mutation of each tyrosine caused a large increase in the SR-95531 unbinding rate without altering the binding rate. Distinct biophysical mechanisms may exist for binding SR-95531 and then stabilizing the bound complex (Jones et al., 2001), and these tyrosine residues only contribute to the stability of the bound state of the receptor with SR-95531. The unique effects of tyrosine mutations on SR-95531 kinetics compared to GABA kinetics also support postulations that agonists and antagonists have different interactions with the binding pocket (Jones et al., 2001).
The effects of mutating these tyrosine residues on GABA kinetics are similar to those seen with the mutation of β2R207 or α1R67 (Goldschen-Ohm et al., 2011). Both arginines were found to be critical for the rapid and stable binding of GABA. Also, both arginines were considered candidates to interact directly with GABA. It may very well be the case that multiple arginines and tyrosines mutually interact with GABA to stabilize its carboxyl group and amino group, respectively.
Attempting to pinpoint direct interactions between specific residues and GABA is a difficult venture.Padgett et al. (2007) proposed a ligand-receptor model in which the amino group of GABA forms a cation-π bond only with the aromatic face of Tyr97. Removing the aromatic at position 97 of β2 (i.e. Y97A) would be expected to cause severe impairment in the receptor’s ability to bind GABA; however, compared to mutations of numerous other residues in the binding pocket, including Y157F, Y205F, F200I (Tran et al., 2011), and R67A (Goldschen-Ohm et al., 2011), Y97A caused only a subtle change in kon-GABA. The story must be more complex than this one residue, Tyr97, stabilizing the amino group of GABA. Considering the significant influence on GABA binding and unbinding kinetics of several tyrosine residues presented here, it appears that all three of the tyrosines are reasonable candidates for interacting directly with GABA. As a similar example,Lummis et al. (2011) demonstrated that two aromatic residues of the binding pocket (on Loop B and C) contribute to cation-π interactions in a single cys-loop receptor. The identification of only one cation-π interaction in the GABAA receptor does not preclude participation of multiple tyrosines in the binding of ligand via electrostatic interactions or other forces.
Tyr97, Tyr157, and Tyr205 have different side-chain requirements
This study also aimed to assess the relative significance of the hydroxyl group from each of the three tyrosines at the GABA binding pocket by mutating each tyrosine to phenylalanine and measuring the direct effects on kon-GABA. The results show that the hydroxyl group on Tyr97 makes little contribution to GABA binding, while the hydroxyl groups on Tyr157 and Tyr205 greatly influence the binding rate of GABA. The relative indifference of GABA binding to the absence of a hydroxyl group at position 97 is consistent with the finding that the benzene ring of Tyr97, not the hydroxyl group, participates in an interaction critical for ligand affinity (Padgett et al., 2007). A possible hydrogen bond with Tyr157 or Tyr205 has been suggested by substitution of 4-F-Phe at either position, or 4-MeO-Phe at Tyr157, which resulted in receptors with wild-type EC50 values (Padgett et al., 2007). Presumably, an electronegative atom at C4 is favored for normal receptor function. Whether a hydroxyl group mediates an interaction with another residue, the protein backbone, or with GABA remains unclear.
Conclusion
We have homed in on the residues critical to GABA binding using detailed assessment of mutations of tyrosine residues believed to surround the GABA binding pocket. Along the way we confirmed the importance of these residues in assembly. It is of significance that our measurement of microscopic kinetics establishes a specific role in binding for each tyrosine. These studies provided detailed kinetic characterizations for binding pocket mutations that will aid in the evolving description of how neurotransmitter binding leads to receptor activation and channel opening.
Acknowledgement
This research was supported by the National Institutes of Health [Grant NS055793], awarded to David A. Wagner.
Abbreviations
- HEK
human embryonic kidney
- LGIC
ligand-gated ion channel
- TMR
tetra-methyl-rhodamine
- SR-95531
6-imino-3-(4-methoxyphenyl)-1(6H)-pyridazinebutanoic acid
Footnotes
There is no conflict of interest.
Authorship Contributions
Participated in research design: Tran and Laha
Conducted experiments: Tran and Laha
Performed data analysis: Tran and Laha
Wrote or contributed to the writing of the manuscript: Tran and Laha
References
- Amin J, Weiss D. GABAA receptor needs two homologous domains of the β subunit for activation by GABA, but not by pentobarbital. Nature. 1993;366:565–569. doi: 10.1038/366565a0. [DOI] [PubMed] [Google Scholar]
- Beene DL, Brandt GS, Zhong W, Zacharias NM, Lester HA, Dougherty DA. Cation-pi interactions in ligand recognition by serotonergic (5-HT3A) and nicotinic acetylcholine receptors: the anomalous binding properties of nicotine. Biochemistry. 2002;41:10262–10269. doi: 10.1021/bi020266d. [DOI] [PubMed] [Google Scholar]
- Bianchi MT, Botzolakis EJ, Haas KF, Fisher JL, Macdonald RL. Microscopic kinetic determinants of macroscopic currents: insights from coupling and uncoupling of GABAA receptor desensitization and deactivation. J. Physiol. 2007;584:769–787. doi: 10.1113/jphysiol.2007.142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boileau AJ, Newell JG, Czajkowski C. GABAA receptor β2 Tyr97 and Leu99 line the GABA-binding site: insights into mechanisms of agonist and antagonist actions. J. Biol. Chem. 2002;277:2931–2937. doi: 10.1074/jbc.M109334200. [DOI] [PubMed] [Google Scholar]
- Bollan K, Robertson LA, Tang H, Connolly CN. Multiple assembly signals in gamma-aminobutyric acid (type A) receptor subunits combine to drive receptor construction and composition. Biochem. Soc. Trans. 2003;31:875–879. doi: 10.1042/bst0310875. [DOI] [PubMed] [Google Scholar]
- Celie PH, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, Sixma TK. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- Connolly CN, Krishek BJ, McDonald BJ, Smart TG, Moss SJ. Assembly and cell surface expression of heteromeric and homomeric g-aminobutyric acid type A receptors. J. Biol. Chem. 1996;271:89–96. doi: 10.1074/jbc.271.1.89. [DOI] [PubMed] [Google Scholar]
- Connolly CN, Wafford KA. The cys-loop superfamily of ligand-gated ion channels: The impact of receptor structure on function. Biochem. Soc. Trans. 2004;32:529–534. doi: 10.1042/BST0320529. [DOI] [PubMed] [Google Scholar]
- Dougherty DA, Stauffer DA. Acetylcholine binding by a synthetic receptor: implications for biological recognition. Science. 1990;250:1558–1560. doi: 10.1126/science.2274786. [DOI] [PubMed] [Google Scholar]
- Goldschen-Ohm MP, Wagner DA, Jones MV. Three arginines in the GABAA receptor binding pocket have distinct roles in the formation and stability of agonist- versus antagonist-bound complexes. Mol. Pharmacol. 2011;80:647–656. doi: 10.1124/mol.111.072033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MV, Sahara Y, Dzubay JA, Westbrook GL. Defining affinity with the GABAA receptor. J. Neurosci. 1998;18:8590–8604. doi: 10.1523/JNEUROSCI.18-21-08590.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MV, Jonas P, Sahara Y, Westbrook GL. Microscopic kinetics and energetics distinguish GABA(A) receptor agonists from antagonists. Biophys. J. 2001;81:2660–2670. doi: 10.1016/S0006-3495(01)75909-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lummis SCR, McGonigle I, Ashby JA, Dougherty DA. Two amino acid residues contribute to a cation-? binding interaction in the binding site of an insect GABA receptor. J. Neurosci. 2011;31:12371–12376. doi: 10.1523/JNEUROSCI.1610-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell JG, McDevitt RA, Czajkowski C. Mutation of glutamate 155 of the GABAA receptor beta2 subunit produces a spontaneously open channel: a trigger for channel activation. J. Neurosci. 2004;24:11226–11235. doi: 10.1523/JNEUROSCI.3746-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett CL, Hanek AP, Lester HA, Dougherty DA, Lummis SC. Unnatural amino acid mutagenesis of the GABA(A) receptor binding site residues reveals a novel cation-pi interaction between GABA and beta2Tyr97. J. Neurosci. 2007;27:886–892. doi: 10.1523/JNEUROSCI.4791-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless SA, Millen KS, Hanek AP, Lynch JW, Lester HA, Lummis SCR, Dougherty DA. A Cation-? interaction in the binding site of the glycine receptor is mediated by a phenylalanine residue. J. Neurosci. 2008;28:10937–10942. doi: 10.1523/JNEUROSCI.2540-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PN, Laha KT, Wagner DA. A tight coupling between ?2Y97 and ?2F200 of the GABAA receptor mediates GABA binding. J. Neurochem. 2011;119:283–293. doi: 10.1111/j.1471-4159.2011.07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DA, Czajkowski C. Structure and dynamics of the GABA binding pocket: a narrowing cleft that constricts during activation. J. Neurosci. 2001;21:67–74. doi: 10.1523/JNEUROSCI.21-01-00067.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DA, Czajkowski C, Jones MV. An arginine involved in GABA binding and unbinding but not gating of the GABA(A) receptor. J. Neurosci. 2004;24:2733–2741. doi: 10.1523/JNEUROSCI.4316-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Gallivan JP, Zhang Y, Li L, Lester HA, Dougherty DA. From ab initio quantum mechanics to molecular neurobiology: a cation-pi binding site in the nicotinic receptor. Proc. Natl. Acad. Sci. USA. 1998;95:12088–12093. doi: 10.1073/pnas.95.21.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]