Abstract
After thymic emigration CD4-T-cells continue to differentiate into multiple effector and suppressor sub-lineages in peripheral lymphoid organs. In-vivo analysis of peripheral CD4-T-cell differentiation has relied on animal models with targeted gene mutations. These are expressed either constitutively, or conditionally after Cre mediated recombination. Available Cre transgenic strains to specifically target T-cells act at stages of thymocyte development that precede thymic selection. Tracing gene functions in CD4-T-cell development after thymic exit becomes complicated when the targeted gene is essential during thymic development. Other approaches to conditionally modify gene functions in peripheral T-cells involve infection of in-vitro activated cells with Cre expressing lenti-, retro-, or adenoviruses, which precludes in-vivo analyses. To study molecular mechanisms of peripheral CD4-T-cell differentiation in-vivo and in-vitro we generated transgenic mice expressing a tamoxifen inducible Cre recombinase (CreERT2) under the control of the CD4 gene promoter. We show here that in CD4CreERT2 mice Cre is inducibly and selectively activated in CD4-T-cells. Tamoxifen treatment both in-vivo and in-vitro results in efficient recombination of loci marked by LoxP sites. Moreover, this strain shows no abnormalities related to transgene insertion. Therefore it provides a valuable tool for studying gene function during differentiation of naïve peripheral CD4-T-cells into effector or suppressor sub-lineages.
Keywords: CD4 T-cell differentiation, CreERT2 recombinase
After successful maturation in the thymus, CD4 T-cells emigrate to peripheral lymphoid organs where they perform their immune functions. In these organs, CD4-T-cells undergo further differentiation into effector cell lineages including the T-helper 1 (Th1), Th2 and Th17 or into T-regulatory-cells (Treg) (Murphy and Reiner 2002). Delineating molecular mechanisms that guide maturation to these sub-lineages requires specific and conditional targeting of developmental regulators in CD4-T-cells after thymic selection and migration to the periphery.
Current approaches to enable conditional loss- or gain-of-function make use of the Cre/LoxP system. Cre is a DNA recombinase from the bacteriophage P1 that recognizes a 34-bp sequence motif (LoxP) and catalyzes excision of the intervening sequences (Sauer 1993). Conditional gene modification in peripheral CD4−T-cells has relied on Cre transgenic strains (Hennet et al. 1995; Lee et al. 2001), as well as Cre expressing lentiviruses (Pfeifer et al. 2001) and adenoviruses (Lee et al. 2001; Prost et al. 2001). However, these aforementioned techniques have drawbacks. First, available Cre transgenic strains to target T-cells express Cre recombinase either from the Lck, or the CD4 gene promoter and mediate in-vivo conditional gene modification prior to thymic selection. This is problematic for determining the role of genes in peripheral T cell differentiation, in particular when the genes of interest have essential functions during thymic development. Examples of such genes include, but are not limited to, molecules that are essential for TCR signaling and thymic selection. Second, other available mouse strains expressing Cre from the Foxp3 gene promoter target genes specifically in Tregs in a constitutive (Liston et al. 2008), or tamoxifen inducible manner (Josefowicz et al. 2012). While this is optimal for studies of gene function in the Treg lineage, it does not address gene functions in CD4 effector sub-lineages. Third, gene modification by Cre expressing lenti-, and retro-viruses can only target ex-vivo CD4 T-cells. Moreover, activation of the T-cells is required for efficient infection (Dardalhon et al. 2001). Fourth, adenovirus based Cre expression requires peripheral T-cells to express the receptor for the coxsackie virus which also results in T-cell activation (Hurez et al. 2002).
To enable conditional in-vivo genetic manipulation and analysis of developmental processes in naïve peripheral CD4-T-cells, we generated transgenic mice expressing a tamoxifen inducible Cre from the CD4 gene promoter. The inducible CreER recombinase has been previously generated through the fusion of Cre to the ligand-binding domain of the estrogen receptor (ER). Subsequent mutagenesis led to the CreERT2 recombinase. This contains three mutations in the human ER moiety and is efficiently activated by the synthetic estrogen-like agonist tamoxifen, but not by endogenous estrogens (Feil et al. 1997; Indra et al. 1999; Kellendonk et al. 1999; Metzger et al. 2003). A number of studies have demonstrated that inducible tissue-specific CreERT2 based recombination systems are a powerful tool for gene targeting (Feil et al. 1997; Hayashi and Mcmahon 2002; Leone et al. 2003). We cloned sequences encoding the CreERT2 fusion downstream of the mouse CD4 gene promoter/enhancer/silencer (Fig. 1A) that has been previously used for transgenic expression of genes specifically in T-cells (Lee et al. 2001).
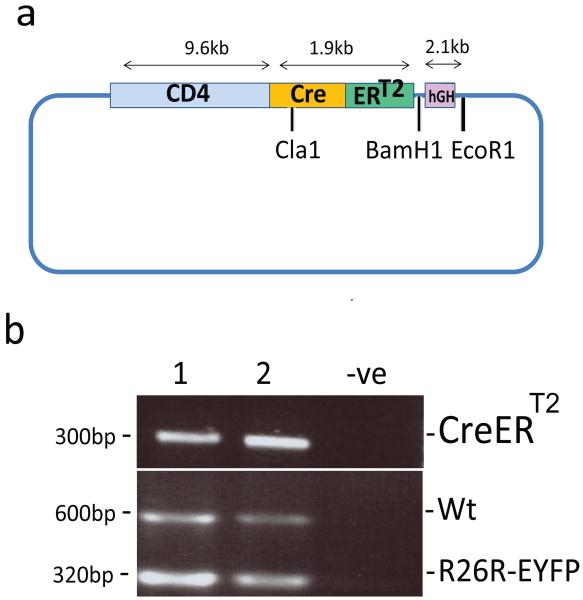
Figure 1. Generation of CD4-CreERT2 mice.
(a) Schematic diagram of the construct used for the generation of the CD4CreERT2 transgenic mouse strain. Colored boxes show the Cd4 gene promoter/enhancer/silencer the Cre the ERT2 moiety and the HGH polyadenylation as indicated. Restriction sites and sizes of the fragments used for the cloning are shown (see Methods). (b) Representative genotyping PCR analysis of 4 weeks old CD4CreERT2/R26R-EYFP double transgenic mice. A negative control (-ve) is also shown.
Transgenic mice were subsequently generated by microinjection of the CD4CreERT2 cassette into the pronuclei of C57BL/6 fertilized oocytes. Two founder lines were obtained carrying the transgene. Neither of the founder lines showed aberrant phenotype(s) associated with transgene homozygosity. To measure the efficiency and inducibility of Cre recombinase activity in T cells, the two CD4CreERT2 founder lines (11 and 19) were independently crossed with the R26R-EYFP reporter strain of Cre activity (Soriano 1999; Srinivas et al. 2001) (Fig. 1B). In these mice, the gene encoding the enhanced yellow fluorescent protein (EYFP) preceded at the 5′ end by a Cre excisable “Stop” cassette is inserted into the Rosa 26 locus by homologous recombination. Cre activity mediates excision of the “Stop” cassette leading to EYFP expression that can be detected by fluorescence-activated cell sorting (FACS) analysis. The fraction of EYFP positive cells after tamoxifen treatment identifies cells that have experienced Cre activity (Srinivas et al. 2001).
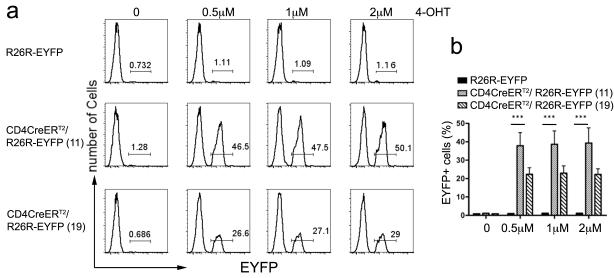
We measured tamoxifen mediated Cre activation in CD4CreERT2/ R26R-EYFPdouble transgenic mice both in-vitro and in-vivo. For in-vitro analyses, we harvested white blood cells from the spleen and lymph nodes of CD4CreERT2 / R26R-EYFP (founder 19 and founder 11) mice as well R26R-EYFP controls. These cells were cultured in medium containing increasing concentrations of 4-hydroxy tamoxifen (4-OHT) ranging from 0.5 μM - 2 μM for 3 days. FACS analysis showed that 4-OHT treatment, induced EYFP expression in ~50% and ~30% of the CD4-T-cells in the CD4CreERT2/ R26R-EYFP founder lines 11 and 19 respectively (Fig. 2A) as compared to no treatment. Moreover, control R26R-EYFP cells did not show EYFP expression. Thus, founder line 11 showed more effective induction of CreERT2 compared to founder line 19 and was used for further analyses.
Figure 2. In-vitro activation in CD4CreERT2 in CD4+ T cells.
(a) Lymphocytes isolated from spleen and lymph nodes of the indicated mice were cultured for 3 days in medium containing the indicated concentrations of 4-OHT. Treated cells were stained for surface expression of CD4 and analyzed by FACS. Line histograms show representative EYFP expression in gated CD4 T-cells for the indicated treatment and mouse strain. (b) Histogram bars show the average frequency of EYFP+ CD4 T-cells after 4-OHT treatment of lymphocytes isolated from 4 independent mice for each indicated strain and 4-OHT concentration p< 0.0001. Results are representative of two independent experiments.
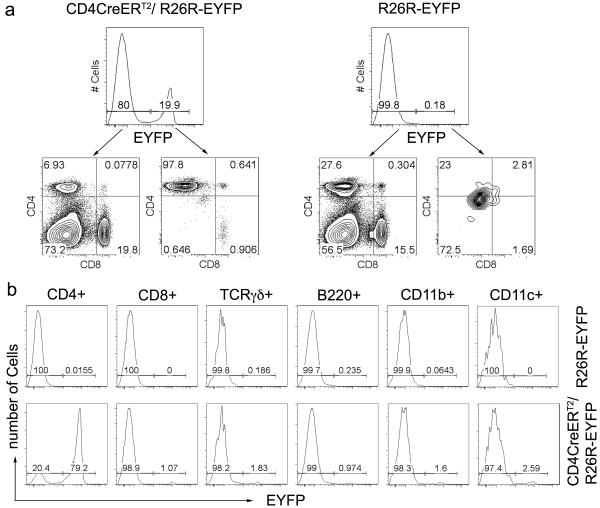
Inducible activation of CD4CreERT2 in-vivo was measured, by tamoxifen administration to 6-8 weeks old CD4CreERT2/ R26R-EYFP mice. Treatment was carried out by daily intragastric gavage of tamoxifen resuspended in peanut oil (5mg daily) for 3-5 days. After treatment, white blood cells from spleen and lymph nodes of the treated mice were harvested and analyzed. R26R-EYFP mice treated with tamoxifen showed little or no expression of the EYFP reporter in blood overall (0.18), (Fig. 3A). Tamoxifen treatment of CD4CreERT2/ R26R-EYFP mice induced Cre activity, leading to EYFP expression specifically in CD4-T-cells, which accounted for ~98% of all EYFP+ lymphocytes (Fig. 3A). Importantly Cre activity was induced in the majority of peripheral CD4-T-cells (~79%). By contrast other subsets including CD11b+ cells, reported to express CD4, showed little or no EYFP expression (1%-2.5%) (Fig. 3B).
Figure 3. In-vivo Cre activation.
The indicated mice were treated in-vivo with tamoxifen (See Methods). EYFP expression was analyzed as a measure of Cre activation and representative FACS plots of the indicated parameters are shown. In (a) histogram plots show EYFP expression in un-gated live cells from the spleen of the indicated mice. Tow parameter contour plots show CD4 versus CD8 expression in gated YFP+ and YFP− cells as shown by the arrows. (b) Line histograms show EYFP expression in cells harvested from the spleen of the indicated mice and gated for surface expression of the indicated markers. Data are representative of three independent experiments.
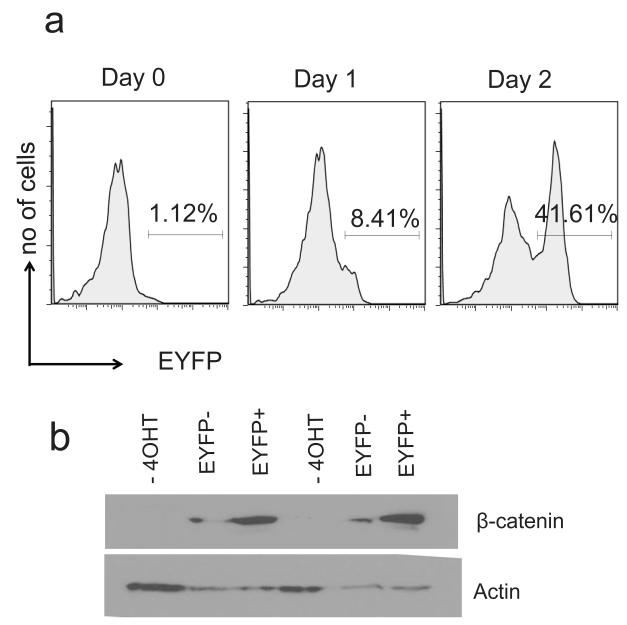
To provide proof of principle that the CD4CreERT2 mice could be used to study gene function in peripheral T-cell development, we crossed CD4CreERT2/ R26R-EYFP mice to mice in which the third exon of β-catenin is marked by LoxP sites (Ctnnb1ex3). We have previously shown that stabilization of β-catenin, through deletion of the 3rd exon of the gene using CD4 or Lck Cre transgenes, interferes with thymocyte development, alters the outcome of thymic T-cell selection, and induces T-cell lymphomas (Gounari et al. 2002; Guo et al. 2007; Kovalovsky et al. 2009). These functions of β-catenin during thymic T-cell development complicate analysis of its roles in peripheral CD4-T-cell differentiation. The targeted 3rd exon of β-catenin encodes phosphorylation sites that are responsible for the degradation of the protein (Harada et al. 1999). Excision of this exon renders β-catenin stable and leads to its accumulation. We treated T cells isolated from spleen and lymph nodes of CD4CreERT2/ R26R-EYFP/ Ctnnb1ex3 compound mutant mice with tamoxifen or vehicle and measured the course of EYFP upregulation by FACS (Fig 4a). Maximum induction of EYFP was detected after 2-3 days of treatment. To provide evidence for Cre mediated stabilization of β-catenin we independently sorted EYFP+ and EYFP− cells and measured the levels of β-catenin by western blot in lysates prepared from the sorted cells. Untreated cells showed low levels of β-catenin as previously documented for T-cells (Guo et al. 2007; Kovalovsky et al. 2009). EYFP+ cells showed elevated levels of β-catenin compared to EYFP− cells, indicating Cre mediated stabilization of the protein. This finding confirmed that EYFP expression correlates with Cre activity (Fig. 4b), which can be specifically and inducibly targeted to peripheral CD4 T-cells in this novel mouse model.
Figure 4. Stabilization of β-catenin in CD4 T-cells correlates with YFP expression.
CD4+ T cells from the spleen and lymph nodes of CD4CreERT2/R26R-EYFP/ Ctnnb1ex3 compound mutant mice were treated with 0.5 μM 4-OHT. (a) Histogram plots show EYFP expression for the indicated length of treatment. EYFP+ and EYFP− cells were sorted on day 3 of treatment (b) Shows Western Blot analysis for β-catenin in lysates from the indicated sorted populations of CD4-T-cells. Data show cells isolated from two independent mice. Actin is shown as a loading control. One of 2 independent experiments is shown.
In conclusion, we report here the generation of a CD4CreERT2 transgenic strain and demonstrate that it can be used effectively to determine gene function during peripheral CD4-T-cell differentiation. We show that Cre expression in these mice is tightly regulated for inducibility as well as for specific expression in CD4 T-cells. We also show that Cre activity can be induced in these mice both in-vitro and in-vivo. Furthermore, by targeting two independent loci, we show that, following activation by tamoxifen, Cre efficiently mediate deletion of LoxP marked sequences in our new mouse model. We anticipate that the CD4CreERT2 mouse model will be a valuable tool to specifically determine the physiological and pathological functions of genes during differentiation of naïve peripheral CD4-T-cells into effector (TH1, TH2, TH17) and suppressor (Treg) sub-lineages.
Methods
R26R-EYFP reporter mice(Srinivas et al. 2001) were a kind gift from Dr. Frank Constantini (Columbia University). All animal studies were conducted according to the guidelines of IACUC of University of Chicago.
Generation of the CD4CreERT2 construct
The pGEM plasmid containing 9.6kb of the mouse Cd4 gene promoter/enhancer/silencer (Lee et al. 2001) was a kind gift from Dr. Chris Wilson (University of Washington, Seattle). cDNA encoding CreERT2 was PCR amplified from an NLS CreERT2 retroviral construct (a gift form Dr. Klaus Rajewsky, Harvard Medical School) and a ClaI-BamH1 fragment (1.9kb) was prepared from the PCR amplified product by restriction digestion. The pGEM-CD4Cre construct was restriction digested with ClaI and EcoRI. The released fragment, containing part of Cre and the human growth hormone (hGH) polyadenylation sequences, was further digested with BamH1 and EcoR1 to isolate a 2.1 kb BamH1-EcoRI fragment containing only the hGH polyadenylation signal. The CD4CreERT2 construct was assembled using a three fragment ligation; (a) the ClaI-EcoRI fragment containing pGEM sequences the Cd4 promoter and part of the Cre cDNA, (b) the ClaI-BamHI PCR amplified fragment containing the remaining Cre cDNA sequences and the ERT2 moiety, (c) the small BamHI-EcoRI fragment containing the hGH polyadenylation signal.
Transgenic Mouse Production
The CD4CreERT2 transgenic mice were generated by pronuclear injection of the CD4CreERT2 targeting construct into fertilized eggs of pseudo-pregnant C57BL/6 females. After establishing germ-line transmission, two founder lines (11 and 19) were first backcrossed to produce homozygotes. Then they were bred independently onto the R26R-EYFP and Ctnnb1ex3 strains to generate CD4CreERT2/R26R-EYFP/Ctnnb1ex3 compound mutant mice.
Breeding and Genotyping of Mice
Two independent founder lines of CD4CreERT2 mice (11 and 19) were crossed to the R26R-EYFP strain (Soriano 1999; Srinivas et al. 2001) bearing a EYFP allele the can only be expressed after Cre mediated excision of a STOP cassette. Progeny was genotyped for the presence of the CreERT2 allele using Cre specific PCR primers (Cre-Forward: 5′ TCA AGG CCA GAC TAG GCT GCC TAT 3′ and Cre-Reverse: 5′ TCT CTG TGG CTG GCA GTT TCT CCA 3′). To generate CD4CreERT2/R26R-EYFP/Ctnnb1ex3 compound mutant mice; CD4CreERT2/R26R-EYFP mice were crossed onto the Ctnnb1ex3 mutant mice and genotyped as previously described (Scheller et al. 2006). Our transgenic mice will be available to the research community upon acceptance of the manuscript.
Tamoxifen treatment
In-vitro Tamoxifen treatment
4-Hydroxytamoxifen (4-OHT, Sigma H7904) was dissolved in ethanol (20 mg/ml) and stored at −20°C for further experiments. Cells harvested from the mice were cultured in IMDM (life technologies) containing 10% fetal bovine serum (Sigma), 103 U/ml penicillin–streptomycin (life technologies) and 0.5 μM-2 μM 4-OHT for 3 days before FACS analysis.
In-vivo Tamoxifen treatment
Tamoxifen (TM, tamoxifen-free base, Sigma T5648) was dissolved in peanut oil as described before (Metzger and Chambon, 2001). Briefly, 100 mg of tamoxifen was suspended in 100 μl ethanol and dissolved in 1 ml peanut oil (Sigma, P2144). This 100 mg/ml tamoxifen solution was sonicated for 1-2 minutes and then stored at −20°C. For in-vivo administration the solution was thawed at 55°C. 50 μl (5 mg) were administrated daily to six-weeks old mice by intragastric gavage for 3-5 days. A day after final treatment, the animals were euthanized, and cells were collected from the spleen and peripheral lymph nodes for analysis.
Fluorescent Assisted Cell Sorting (FACS) Analysis
Cells were harvested and stained with fluorochrome conjugated antibodies against CD4, CD8, B220, Mac-1 and CD11c surface markers (eBioscience) and the expression of EYFP in different cell types was determined by an LSR II (BD Biosciences) FACS analyzer.
Western blot
CD4 T cells were purified from spleen and lymph nodes of CD4CreERT2/R26R-EYFP/Ctnnb1ex3 compound mutant mice using negative MACS bead separation according to the manufacturer’s instructions (Miltenyi Biotec, Sunnyvale, CA). Isolated cells were treated with 4-OHT (0.5 μM) for 3 days. Treated cells were independently sorted for CD4+EYFP− and CD4+EYFP+ cells. Lysates were prepared using from the sorted cells using RIPA buffer (20 mM TrisHCl pH 7.5, 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% deoxycholic acid, 0.1% SDS) supplemented with a cocktail of protease inhibitors (Roche Diagnostics GmbH, Germany). Western blot was probed with a monoclonal antibody to β-catenin (BD Biosciences) or β-actin (Thermo Fisher).
Acknowledgments
We wish to thank members of the Gounari Lab for helpful discussions. This work was supported by National Institutes of Health Grants R21AI076720 (F.G.), American Cancer Society Grant ACS/RSG, LIB-113428 (F.G), Chicago Biomedical Consortium (F.G.). The authors declare no conflict of interest.
References
- Dardalhon V, Jaleco S, Kinet S, Herpers B, Steinberg M, et al. IL-7 differentially regulates cell cycle progression and HIV-1-based vector infection in neonatal and adult CD4+ T cells. Proc Natl Acad Sci U S A. 2001;98:9277–9282. doi: 10.1073/pnas.161272698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- Gounari F, Aifantis I, Martin C, Fehling HJ, Hoeflinger S, et al. Tracing lymphopoiesis with the aid of a pTalpha-controlled reporter gene. Nat Immunol. 2002;3:489–496. doi: 10.1038/ni778. [DOI] [PubMed] [Google Scholar]
- Guo Z, Dose M, Kovalovsky D, Chang R, O’neil J, et al. Beta-catenin stabilization stalls the transition from double-positive to single-positive stage and predisposes thymocytes to malignant transformation. Blood. 2007;109:5463–5472. doi: 10.1182/blood-2006-11-059071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. Embo J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Mcmahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Hennet T, Hagen FK, Tabak LA, Marth JD. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc Natl Acad Sci U S A. 1995;92:12070–12074. doi: 10.1073/pnas.92.26.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurez V, Hautton RD, Oliver J, Matthews RJ, Weaver CK. Gene delivery into primary T cells: overview and characterization of a transgenic model for efficient adenoviral transduction. Immunol Res. 2002;26:131–141. doi: 10.1385/ir:26:1-3:131. [DOI] [PubMed] [Google Scholar]
- Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, et al. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellendonk C, Tronche F, Casanova E, Anlag K, Opherk C, et al. Inducible site-specific recombination in the brain. J Mol Biol. 1999;285:175–182. doi: 10.1006/jmbi.1998.2307. [DOI] [PubMed] [Google Scholar]
- Kovalovsky D, Yu Y, Dose M, Emmanouilidou A, Konstantinou T, et al. Beta-catenin/Tcf determines the outcome of thymic selection in response to alphabetaTCR signaling. J Immunol. 2009;183:3873–3884. doi: 10.4049/jimmunol.0901369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Leone DP, Genoud S, Atanasoski S, Grausenburger R, Berger P, et al. Tamoxifen-inducible glia-specific Cre mice for somatic mutagenesis in oligodendrocytes and Schwann cells. Mol Cell Neurosci. 2003;22:430–440. doi: 10.1016/s1044-7431(03)00029-0. [DOI] [PubMed] [Google Scholar]
- Liston A, Lu LF, O’carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008;205:1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger D, Indra AK, Li M, Chapellier B, Calleja C, et al. Targeted conditional somatic mutagenesis in the mouse: temporally-controlled knock out of retinoid receptors in epidermal keratinocytes. Methods Enzymol. 2003;364:379–408. doi: 10.1016/s0076-6879(03)64022-x. [DOI] [PubMed] [Google Scholar]
- Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- Pfeifer A, Brandon EP, Kootstra N, Gage FH, Verma IM. Delivery of the Cre recombinase by a self-deleting lentiviral vector: efficient gene targeting in vivo. Proc Natl Acad Sci U S A. 2001;98:11450–11455. doi: 10.1073/pnas.201415498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost S, Sheahan S, Rannie D, Harrison DJ. Adenovirus-mediated Cre deletion of floxed sequences in primary mouse cells is an efficient alternative for studies of gene deletion. Nucleic Acids Res. 2001;29:E80. doi: 10.1093/nar/29.16.e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B. Manipulation of transgenes by site-specific recombination: use of Cre recombinase. Methods Enzymol. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- Scheller M, Huelsken J, Rosenbauer F, Taketo MM, Birchmeier W, et al. Hematopoietic stem cell and multilineage defects generated by constitutive beta-catenin activation. Nat Immunol. 2006;7:1037–1047. doi: 10.1038/ni1387. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]