Abstract
Active transport along the microtubule lattice is a complex process that involves both the Kinesin and Dynein superfamily of motors. Transportation requires sophisticated regulation much of which occurs through the motor’s tail domain. However, a significant portion of this regulation also occurs through structural changes that arise in the motor and the microtubule upon binding. The most obvious structural change being the manifestation of asymmetry. To a first approximation in solution, kinesin dimers exhibit two-fold symmetry, and microtubules, helical symmetry. The higher symmetries of both the kinesin dimers and microtubule lattice are lost on formation of the kinesin-microtubule complex. Loss of symmetry has functional consequences such as an asymmetric hand-over-hand mechanism in plus-end directed kinesins, asymmetric microtubule binding in the Kinesin-14 family, spatially biased stepping in dynein, and cooperative binding of additional motors to the microtubule. This review focuses on how the consequences of asymmetry affect regulation of motor heads within a dimer, dimers within an ensemble of motors, and suggests how these asymmetries may affect regulation of active transport within the cell.
Keywords: Kinesin, Microtubules, Molecular Motors, Dynein, Asymmetric macromolecular assemblies
Introduction
Within the intricate network of microtubules, two families of molecular motors, the kinesin superfamily and dynein, work together and against each other to transport material in both directions along the microtubule and to crosslink, slide, and organize microtubules during spindle formation. This is a highly regulated process where a significant portion of motor targeting and regulation occurs through the cargo domain. However, the motor domain itself plays a large role in determining the direction of travel and cooperativity between motors. The high complexity arising from multiple types of motors transporting many types of cargos in both directions demands a sophisticated and adaptable level of communication between motors in order to ensure energy efficient and timely transportation. This communication can be broken into three main categories: communication between motor heads when they exist as functional dimers, communication between dimers in a working ensemble of motors, and long-range communication between motors on the same microtubule. Much of this communication appears to begin upon binding to the microtubule and in some cases occurs through the microtubule lattice itself. Therefore, a knowledge of the structural changes that occur when a motor interacts with a microtubule is critical to understanding how this communication occurs. The most profound structural change that occurs upon interaction of a motor and microtubule is a decrease in the intrinsic symmetry of the macromolecular assembly compared to that of the isolated motors and microtubules in solution. Hence, this review focuses first on the structural and functional asymmetry within the kinesin superfamily that is believed to be an essential component of their motor activity, and then briefly extends these considerations to the dynein superfamily of motors. The kinesin superfamily includes plus-end directed motors, minus-end directed motors, and kinesins that regulate microtubule dynamics and can be divided into 14 subfamilies (Lawrence et al., 2004). Despite these differences, all kinesins contain a structurally conserved motor domain that includes a microtubule and ATP binding site, and they most often operate as dimers. In general, the position of the motor domain within the polypeptide sequence is a predictor of motor direction. Motors with N-terminal motor domains typically exhibit processive, plus-end directed motility, while C-terminal motor domains exhibit nonprocessive minus-end directed motility (Figure 1). Mechanistically, the direction of travel is related to the structure of the neck linker where this consists of approximately 15 class-specific amino acid residues that are located directly C-terminal to the motor domain in plus-end directed kinesins or lie immediately before the motor domain in minus-end directed motors (Sablin et al., 1998). In addition to opposite directions of travel, subfamilies of kinesin exhibit different characteristic run lengths, speeds, stalling forces, and cooperativity reflecting the diverse function of these motors. These features are due in part to differing communication between heads in the molecular dimers, however, the structural and mechanistic details in many instances are still unclear.
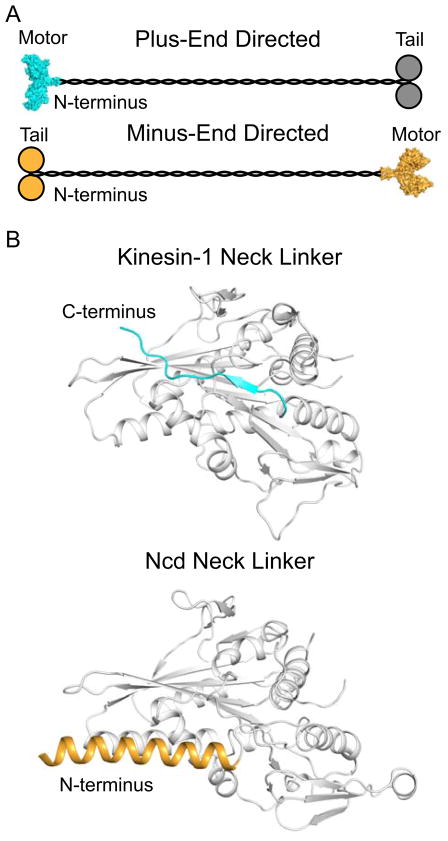
Figure 1. Topological Organization of Plus- and Minus-End Directed Kinesins.
A. Overall topology of plus- and minus-end directed motors (cyan and orange, respectively) where plus-end directed motors contain N-terminal motor domains and C-terminal cargo domains, and minus-end directed motors contain N-terminal cargo domains and C-terminal motor domains. A surface representation of a dimeric Kinesin-1 motor domain (pdb accession 2Y65) is shown as a representation structure of a plus-end directed motor, and a surface representation of a dimeric Ncd motor domain (pdb accession 2NCD) is shown as a representative structure of a minus-end directed motor (Sablin et al., 1998; Kaan et al., 2011). The cargo domains of the plus-end directed motors are depicted in grey as some plus-end kinesins, such as Kinesin-1, utilize a second polypeptide chain for their cargo domains. B. Structure of the neck linker in Kinesin-1 and Ncd (pdb accession 2Y65 and 2NCD) show two radically different conformations. In plus-end directed motors, the neck linker is located C-terminal of the motor domain (grey) and exhibits a random coil and β-strand secondary structure. The depicted orientation represents a parallel docked position. In minus-end directed motors, the neck linker is located N-terminal to the motor domain (grey) and exists as an α-helix which forms a coiled-coil with the second head in the dimer. The depicted position represents a pre-powerstroke orientation. The neck linker region in minus-end directed motors is not as well defined as in plus-end directed motors; the region highlighted here represents the section of coiled-coil that makes direct contacts with the motor domain and is required for wild-type motility (Sablin et al., 1998; Rank et al., 2012).
Processive, plus-end directed kinesins undergo an asymmetric hand-over-hand mechanism (Svoboda et al., 1993; Hackney, 1994; Hoenger et al., 1998; Hancock and Howard, 1999; Asbury et al., 2003; Kaseda et al., 2003; Yildiz et al., 2004). This requires the heads to operate as a functional dimer where communication between the heads is necessary to synchronize stepping and prevent premature release from the microtubule. This is thought to occur through strain in the neck linker, neck linker orientation, and chemical gating (Rice et al., 1999; Yildiz et al., 2008; Clancy et al., 2011). In contrast, the Kinesin-14 family which undergo minus-end directed motility are nonprocessive (Vale and Fletterick, 1997; deCastro et al., 2000; Foster and Gilbert, 2000; Fink et al., 2009). These motors use a powerstroke mechanism where only one head is directly involved in each motile cycle (Wendt et al., 2002; Endres et al., 2005); however, recent studies suggest functional importance for the second head of the dimer (Foster et al., 2001; Sproul et al., 2005; Kocik et al., 2009; Rank et al., 2012).
A complete model of the mechanism of both plus-end and minus-end directed motors requires a detailed knowledge of the structural changes in each head during their interaction with microtubules, coupled with an understanding of how the state of one head influences the other since most of the motile cycle occurs when kinesins are bound to microtubules. Likewise an understanding of the changes induced in the microtubule lattice by kinesins is needed to account for the cooperative binding exhibited by kinesins that operate as ensembles. All of these biological processes introduce asymmetry into an otherwise symmetric system. The relationship between symmetry and asymmetry is the focus of this review which is directed towards recent developments in our knowledge of kinesins, but touches briefly upon analogies to Dynein.
Asymmetry between Kinesin Heads
Although there is some evidence that even in solution kinesin dimers can be asymmetric (Stone et al., 1999; Kozielski et al., 2001), to a first approximation most kinesins can be considered to be chemically symmetric dimers in solution, whereas microtubules consist of helically symmetric arrangements of tubulin heterodimers. The symmetry of the individual components is lost the moment the macromolecular assemblies interact because kinesins cannot exhibit helical symmetry and the microtubule lattice does not embody a two-fold axis of symmetry. In solution, two heads tethered by a coiled-coil can be structurally equivalent by being related by a two-fold rotational axis parallel to the coiled-coil, and this state has been observed in crystal structures for both the Kinesin-14 Ncd and Kinesin-1 (Figure 2) (Sablin et al., 1996; Kaan et al., 2011). Any symmetry between heads in solution likely represents an average state, but this is lost the instant one head binds to the microtubule lattice. Each head then becomes structurally unique which has kinetic and motile consequences. In plus-end directed processive kinesins, this is most obvious in the kinetic cycle where the nucleotide state of each head is controlled out of phase by the state of the other (Rosenfeld et al., 2003; Klumpp et al., 2004). For Kinesin-1 and the Kinesin-7 CENP-E, asymmetry between the nucleotide-state of the motor heads is established the instant the first head binds to the microtubule (Hackney, 1988; Ma and Taylor, 1997; Gilbert et al., 1998; Sardar and Gilbert, 2012). Asymmetry also manifests itself in a more subtle manner in the limping of homodimeric motor constructs.
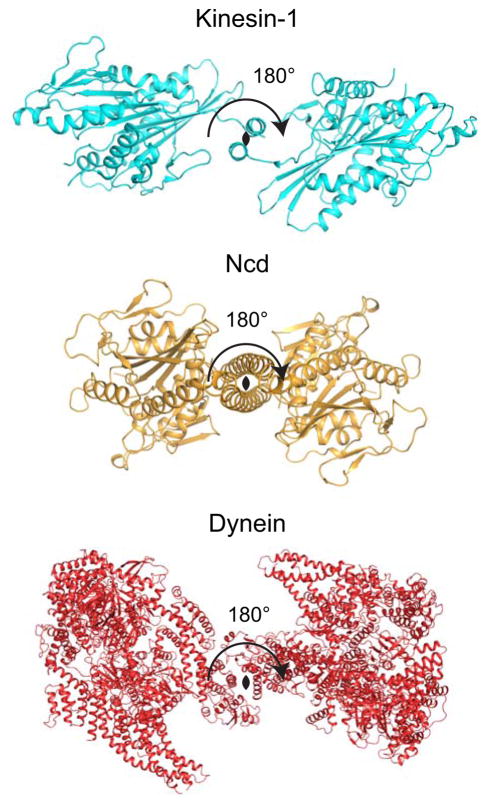
Figure 2. Symmetry between Motor Domains in a Dimer.
Cartoon representations of dimeric structures of Kinesin-1 (pdb accession 2Y5W, cyan), Ncd (2NCD, orange), and dynein (4AKG, red) viewed looking down the coiled-coil with the 2-fold axis depicted as a black oval (Sablin et al., 1998; Kaan et al., 2011; Schmidt et al., 2012). Both Kinesin-1 and dynein show close to a 2-fold symmetry between heads. Variation from 2-fold symmetry likely reflects flexibility within the neck linker/GST linker and effects of crystal packing. The Ncd structure shows crystallographic 2-fold symmetry where only one polypeptide is found within the asymmetric unit of the unit cell.
At its most basic level, limping of homodimeric motor constructs is a result (and proof) of an asymmetric hand-over-hand walk (Asbury et al., 2003; Higuchi et al., 2004). Limping is defined as alternate and distinct stepping rates as measured by single molecule studies, and strict alternation in rates implies that alternate steps are mechanistically unique. This excludes symmetric model of motions such as the symmetric hand-over-hand or inchworm mechanism. Further proof of the asymmetric hand over hand mechanism comes from two key experiments. The Gelles group demonstrated that microtubules do not rotate in response to stepping by a Kinesin-1 attached to a coverslip through a torsionally stiff coiled-coil (Hua et al., 2002). This result is inconsistent with a symmetric hand-over-hand mechanism since the latter would require a 180° rotation with each Kinesin-1 step. Yildiz et al. further showed by labeling each head in a Kinesin-1 dimer with a different fluorescent probe that each Kinesin-1 head takes a 16 nm step excluding an inchworm model (Yildiz et al., 2004). Further studies of an asymmetric hand-overhand mechanism have proven (Fehr et al., 2009) that limping is due to each head experiencing different vertical load during the motile cycle as predicted (Xie et al., 2007). This requires the same motor domain to undergo two intrinsically different trajectories during the motile cycle (Figure 3). Two trajectories are required otherwise the stepping motion would generate rotation of the cargo which is not observed.
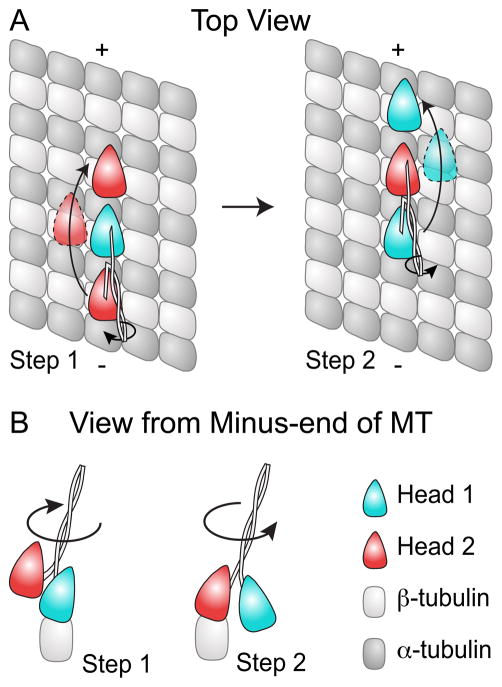
Figure 3. Motor Domain Trajectories during Asymmetric Hand-over-Hand Mechanism.
(A) A schematic representation of the different trajectories experienced by kinesin dimer heads during an asymmetric hand-over-hand stepping motion. In this model, head 2 (red) travels to the left of the protofilament, when viewed from the minus-end of the microtubule, and a greater distance above the protofilament while stepping compared to head 1 (cyan), which swings to the right of the protofilament during its step. (B) Stepping of the heads also introduces twist within the coiled-coil; however as each step introduces twist in the opposite direction, the net rotation within the cycle is zero. This is more clearly illustrated in the view from the minus-end of the microtubule.
Although studies of limping have focused on the Kinesin-1 family, all motors that utilize a processive hand-over-hand mechanism experience asymmetry both in the chemical state of each head and in the trajectory if the cargo does not rotate. Asymmetry is more pronounced in members of the Kinesin-2 family which exist as heterodimers. Kif3a (Aizawa et al., 1992; Kondo et al., 1994) forms a heterodimer with either the motor Kif3b (Yamazaki et al., 1995) or Kif3c (Muresan et al., 1998; Yang and Goldstein, 1998), and both Kif3a heterodimers are thought to undergo a processive hand-over-hand motion (Muthukrishnan et al., 2009). The evolutionary development of a heterodimeric kinesin raises the question if the heads have been optimized to perform maximally at different vertical loads. Artificially formed homodimeric Kif3a and Kif3b motors display similar motility rates and run lengths to Kif3a/b heterodimers (Muthukrishnan et al., 2009). However, Kif3b and Kif3c, but not Kif3a, contain a conserved proline in the neck region, which has been implied as the region sensitive to changes in vertical loads (Fehr et al., 2009). There are also sequence conserved differences between Kif3a, Kif3b, and Kif3c in the microtubule binding region particularly in loop 11 which may affect microtubule binding affinity (Figure 4). More detailed single molecule studies, kinetics and microtubule binding studies are needed to elucidate the role of these conserved differences in the mechanism and biological benefit of heterodimerization in the Kinesin-2 family.
Figure 4. Sequence Conserved Differences within heterodimeric Kinesin-2 Motors.
Kif3a, Kif3b, and Kif3c show conserved sequence differences between themselves. Conserved differences within the microtubule binding region, particularly loop 11, and the neck linker suggests that these motors have evolved to have different microtubule binding interactions and to respond differently to tension within the neck linker. It is possible that these differences within the neck linker optimize the motors to work at the different vertical loads, and that differences within the microtubule binding region bias one motor to bind first to the microtubule. Sequences aligned using T-coffee server (Notredame et al., 2000), and further analyzed utilizing Jalview (Clamp et al., 2004; Waterhouse et al., 2009).
Like the Kinesin-1 and -2 subfamilies, motors of the Kinesin-5 subfamily exhibit a processive, plus-end directed, asymmetric hand-over-hand motion (Valentine et al., 2006). However, the homodimeric Kinesin-5 member Eg5 displays apparent asymmetric binding of ADP in solution which biases the weakly ADP bound head to bind to microtubules first (Krzysiak et al., 2008). This suggests that in solution Eg5 dimer heads must be structurally unique resulting in asymmetric affinities for ADP. This is in strong contrast to homodimeric Kinesin-1 heads which are indistinguishable in solution. Electron paramagnetic resonance (EPR) studies of Eg5 dimers and monomers show two distinct populations of motor heads, presumed to be the tight and weak ADP bound heads, supporting the idea of a structurally asymmetric dimer (Waitzman et al., 2011). The ratio between these two populations as seen by EPR is constant for both monomeric and dimeric motors and the heads appear to switch rapidly between the two states (Waitzman et al., 2011). The EPR data together with the MANT-ADP titrations from the Krzysiak study suggest that within the dimer the Eg5 heads switch freely between the tight and weak ADP binding state; however, it is energetically unfavorable for both heads of the dimer to be in either the weak or tight ADP binding state resulting in asymmetric heads within the dimer. EPR studies also suggest that this dynamic equilibrium between heads is lost upon binding of the first motor head to the microtubule as the asymmetry is imposed (Waitzman et al., 2011).
Structural studies of Eg5 show a novel position of the neck linker which is docked perpendicular to the motor domain, and residues necessary to stabilize this conformation are conserved only within the Kinesin-5 subfamily (Turner et al., 2001). The asymmetric ADP binding in solution may be a result of this novel docked neck linker position as this conformation structurally prevents formation of the symmetrically related solution phase structure seen in Kinesin-1 (Kaan et al., 2011). Assuming one head within the dimer displays perpendicular docking of the neck linker as seen in the ADP crystal structure (Turner et al., 2001), this raises two questions: what is the structure of the neck linker in the second head, and which conformation represents tight ADP binding? Answers to these questions awaits further study.
The Kinesin-14 family of C-terminal motors also exhibit functional and structural asymmetry. Some of these consist of homodimers whereas others consist of a heterodimer where one polypeptide chain includes a motor domain and the other carries a motor homology domain that exhibits the kinesin fold but lacks a nucleotide binding site. In both homo and heterodimers, the two heads appear to play radically different roles in the motile cycle where at first sight only one head appears necessary to generate the powerstroke. Indeed, engineered heterodimers of the well studied homodimeric Kinesin-14 Ncd can promote motility with only a single motor domain, and cryo-EM studies suggest binding of only one head during the nucleotide-free and AMPPNP states (Wendt et al., 2002; Endres et al., 2005; Cope et al., 2010). Yet, sequence conservation of the dimer and kinetic studies suggest functional importance of the second head. Solution studies indicate cooperative interactions between the heads of Ncd (Foster et al., 1998; Mackey and Gilbert, 2000), where each head exhibits different affinities for ADP in solution. In principle, this biases the weakly ADP bound head to bind the microtubule first similar to Eg5 (Foster et al., 2001). Additionally, studies of heterodimeric Ncd motors where one head has been inactivated show clear interactions between motor domains that affect ATPase activity and microtubule binding (Kocik et al., 2009).
The most profound example of asymmetry is seen in the unique Kinesin-14 motor Kar3 which operates as a heterodimer with either the non motor protein Cik1 or Vik1 (Page and Snyder, 1992; Page et al., 1994; Manning et al., 1999; Barrett et al., 2000). Surprisingly, Vik1, and presumably Cik1, contains a C-terminal motor homology domain which retains the canonical kinesin motor domain fold but lacks an ATP binding site. At first sight this might be seen as support for a second head whose only task is to stabilize the coiled-coil required for the function of the lever arm, but remarkably both Vik1 and Cik1 bind to microtubules more tightly than Kar3 (Allingham et al., 2007; Chen et al., 2011). This demands a role for the second head as has recently been demonstrated for both Cik1 and Vik1 (Chen et al., 2011; Rank et al., 2012).
During the motile cycle, Vik1 binds first to the microtubule on an adjacent protofilament to Kar3 utilizing a novel microtubule binding site that is not well understood (Rank et al., 2012). Kar3 then binds to the microtubule and releases ADP. During the nucleotide-free state, Vik1 dissociates from the microtubule allowing Kar3 to uptake ATP and undergo a powerstroke very similar to Ncd’s proposed mechanism (Rank et al., 2012). Cik1, like Vik1, binds to the microtubule independently of Kar3 (Chen et al., 2011). However unlike Vik1, Cik1 targets Kar3 to the microtubule plus-ends, where this localization is critical for Kar3Cik1’s microtubule depolymerase activity (Sproul et al., 2005; Allingham et al., 2007). These studies suggest that both heads in the Kinesin-14 dimer bind to the microtubule during the ADP state utilizing two unique binding sites. The role of this second and novel microtubule binding site is unclear, but appears to affect cooperative binding to the microtubule as discussed below. Evolutionary loss of the ATP binding site from Vik1 and robust motility from single headed Ncd dimers suggests that the second head is not directly necessary for force production, but clearly it also demonstrates that the second head can play a role that extends beyond simply providing structural support of the adjoining coiled-coil.
Cooperative Binding of Kinesins to Microtubules
In vivo microtubules exist in a B-lattice form where protofilaments are arranged laterally such that adjacent α-tubulin show an axial stagger of 0.9 nm on adjacent protofilaments (McIntosh et al., 2009). There is a single break in this helical symmetry, called the seam, where two protofilament exhibit A-lattice contacts and the α-tubulin are staggered by 4.9 nm. Symmetry within the microtubule lattice is further lost upon binding of a single kinesin motor since strictly speaking the αβ tubulin heterodimers surrounding that motor are no longer symmetry equivalent. This event profoundly affects the microenvironment of the kinesin bound tubulin dimer and has consequences towards binding of subsequent kinesins which are a result of either perturbations in the helical symmetry of the underlying microtubule and/or head-to-head contacts with the bound kinesin. Electron microscopy has shown that microtubules undergo structural changes in response to saturation binding of kinesin motor domains where helical symmetry is preserved (Krebs et al., 2004) so that it can be assumed that localized structural changes will occur when a single or small group heads binds. Both Kinesin-1 and Kinesin-14 motors exhibit cooperative binding to the microtubule, and this cooperativity appears to not only affect the binding affinity of subsequent motors but also their spatial organization. This spatial coordination likely ensures that all motors within an ensemble are positioned such that each can contribute work.
Kinesin-14 dimers operate in vivo as ensembles to generate minus-end directed motility, even though individual motors are nonprocessive (deCastro et al., 1999). Efficient recruitment of motors is therefore essential to their biological function. Ncd dimers exhibit strong cooperative binding in the nucleotide-free and AMPPNP states where one head is bound to the microtubule and the second head is removed but tethered. When microtubules are decorated with Ncd dimers at under-saturating conditions, most microtubules are either completely decorated with Ncd heads, partially decorated along a few protofilaments, or undecorated as shown by electron microscopy (EM) (Figure 5) (Wendt et al., 2002). The presence of microtubules with a combination of fully decorated protofilaments and bare protofilaments suggests that this cooperative binding proceeds in the axial direction (Wendt et al., 2002).
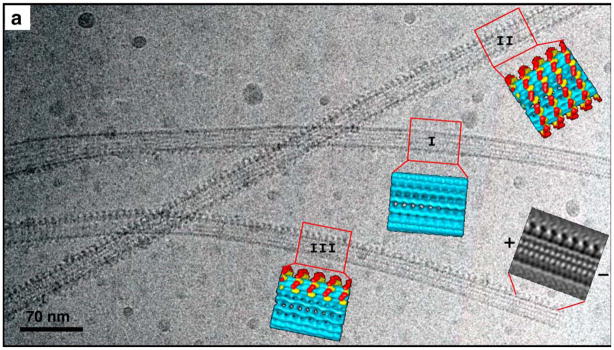
Figure 5. A cryo-EM image of microtubules decorated with dimeric Ncd at subsaturating ratios reveals a cooperative binding mechanism in the axial direction, along individual protofilaments.
While one of the microtubules is mostly decorated (I), partial decoration (III) and complete decoration (II) co-exist in the same area. A Fourier-filtered image of the microtubule and the motors in a side projection allows determination of the polarity of the microtubule according to the shape of the dimeric motor construct (Adapted from Figure 1 of (Wendt et al., 2002)).
Very similar cooperative decoration has also been observed for the Kinesin-14 heterodimer Kar3Vik1 in the nucleotide-free and AMPPNP states. During these states Kar3 and not Vik1 is assumed to be bound to the microtubule (Cope et al., 2010; Rank et al., 2012). In the ADP state, Vik1 is thought to be tightly bound to the microtubule with Kar3 either bound or occluding a tubulin dimer on an adjacent protofilament, and under these conditions no cooperative binding behavior is observed (Rank et al., 2012). This suggests that cooperativity is directly related to Kar3 binding. Interestingly, immunofluorescent studies of the Kar3 motor domain by itself complexed with AMPPNP demonstrate stochastic binding to the microtubule which implies that Vik1 is needed for cooperative binding of Kar3. In contrast, Kar3Cik1 · AMPPNP exhibits targeted binding to the microtubule plus ends and Kar3Vik1 · AMPPNP cooperatively binds along the length of the microtubule (Sproul et al., 2005; Allingham et al., 2007). This then suggests that cooperative binding and microtubule targeting is not related solely to the Kar3 motor domain and instead relies on the Vik1 and Cik1 motor homology domain and/or the adjoining coiled-coil.
Binding of a plus-end directed processive kinesin motor also affects binding of additional motors to the microtubule in terms of binding affinity and spatial arrangements. However, this cooperative behavior is only seen at close to or above stoichiometric ratios of motors to microtubule binding sites, which is in contrast to the Kinesin-14 motors. Furthermore, the degree of cooperativity varies between families. For example, Eg5 dimers, which in vivo operate as ensembles to slide antiparallel microtubules relative to each other, exhibit cooperative binding to the microtubule as detected by sigmoidal binding profiles during cosedimentation studies with microtubules in the AMPPNP and nucleotide-free states (Krzysiak et al., 2006). In contrast, Kinesin-1 dimers likely work individually or in small groups when transporting large cargos such as vesicles and organelles (Shubeita et al., 2008), and consistent with their biological behavior, cooperative binding with Kinesin-1 is only observed through EM studies suggesting this class of motor exhibits lower cooperativity than Eg5.
Kinesin-1 dimers exhibit cooperative binding to microtubules in the AMPPNP state as shown by EM studies similar to Kinesin-14 motors (Vilfan et al., 2001). However unlike Kinesin-14’s which only bind to the microtubule through one head under cooperative conditions, both Kinesin-1 heads bind under cooperative conditions, and Kinesin-1 dimers occasionally display an additional 16 nm periodicity upon binding indicating that when both dimer heads are bound to the same protofilament they exhibit lateral alignment along the microtubule. This leads to the periodic presence of the neck linker every 16 nm (Hoenger et al., 1998; Thormahlen et al., 1998; Vilfan et al., 2001; Skiniotis et al., 2003). The lateral alignment seen for Kinesin-1 is in stark contrast to the longitudinal (along a protofilament) cooperative binding seen in the Kinesin-14’s (Figure 6). Mechanistically this is consistent with the need for the individual heads of a Kinesin-1 dimer to bind on the same protofilament, whereas the Kinesin-14 heads appear to bind adjacent protofilaments (Rank et al., 2012). This suggests that the Kinesin-14’s and the Kinesin-1’s have different mechanisms to promote cooperative binding, where the Kinesin-14’s exhibit strong cooperative binding that proceeds first along the axial direction of the microtubule and Kinesin-1’s exhibit very weak cooperative binding that can include lateral alignment along the protofilament.
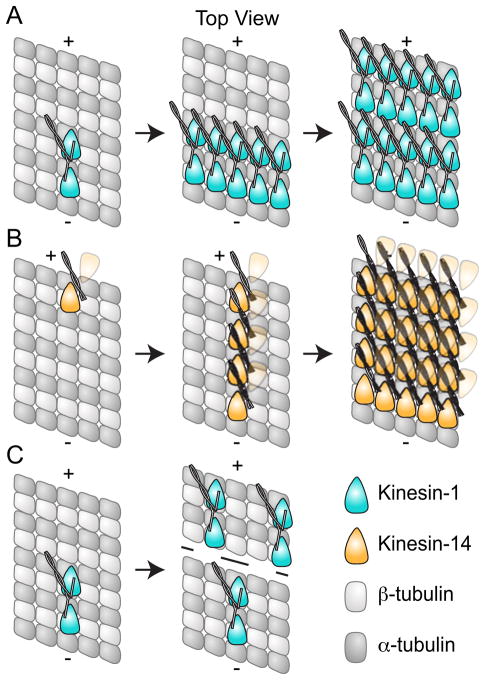
Figure 6. Cooperative binding to the Microtubule.
A. Kinesin-1 (cyan) dimers exhibit cooperative binding to the microtubule, and the presence of 16 nm layer lines in cryo-EM suggests this proceeds in an lateral manner (Vilfan et al., 2001). Kinesin-14 dimers (orange) also display cooperative binding; however in contrast to Kinesin-1, this appears to proceed along the protofilament (Wendt et al., 2002; Cope et al., 2010). The second head in the Kinesin-14 dimer is depicted as partially transparent since cooperative binding is only observed when one head of the dimer is bound; for Kar3Vik1, binding of both heads in the ADP state appeared to abolish cooperative binding (Rank et al., 2012). C. Kinesin-1 dimers also exhibit long range cooperative binding where binding of one dimer biases additional dimers to bind further towards the plus-end of the microtubule (Muto et al., 2005).
Cooperative binding of kinesin motor domains to the microtubule lattice requires that binding of one dimer to the microtubule increases the chance of a second dimer binding either through head-to-head contacts and/or through changes to the microtubule lattice. Although further studies are needed to distinguish between these two mechanisms, there are several lines of evidence that support changes in the microtubule lattice upon binding of a kinesin motor. Electron microscopy has shown that microtubules undergo structural changes in response to binding of kinesin motor domains (Krebs et al., 2004). Additionally, binding of a Kinesin-1 dimer to the microtubule also appears to bias additional Kinesin-1 dimers to bind further towards the plus-end of the microtubule (Muto et al., 2005) (Figure 6). This is different from the cooperative binding observed by EM where each motor head was bound directly next to another Kinesin-1 head. The cooperativity observed by Muto et al. must occur through changes in the microtubule lattice as there is no contact between motors.
Dynein
The dynein family is the second major class of microtubule molecular motors, and these motors utilize ATP to produce minus-end directed motility. Dynein motors have no structural relationship to the kinesin motors and instead belong to the ATPase associated with diverse cellular activities (AAA+) superfamily. Each motor head is composed of six AAA domains, a linker, and stalk which contains the microtubule-binding domain at its tip. Similar to most kinesins, dynein motors operate as functional dimers and exhibit a processive stepping mechanism with an average step size of 8 nm towards the minus-end of the microtubule (Wang et al., 1995; King and Schroer, 2000; Mallik et al., 2004; Reck-Peterson et al., 2006). However, dynein steps show great variability in size and direction, and the two motor heads appear to straddle adjacent protofilaments (Wang et al., 1995; Mallik et al., 2004; Reck-Peterson et al., 2006; Qiu et al., 2012). Additionally Dynein motors appear to display very little coordination between motor heads within the dimer, especially when the motor domains are close together on the microtubule lattice, and instead rely on a high duty ratio for processive motion (Shima et al., 2006; DeWitt et al., 2012; Qiu et al., 2012).
Like Kinesin motors, Dynein also exhibits an inherent structural and functional asymmetry between motor domains where the left, as determined by looking down the microtubule towards the minus end, motor domain is more likely to be the lagging head and the right motor domain more likely to be the leading head (DeWitt et al., 2012; Qiu et al., 2012). Yet, crystal structures of the dynein motor in solution also show a pseudo two-fold symmetry between motor domains (Figure 2) suggesting that dynein, like Kinesins, transitions from a structurally symmetric solution conformation to an asymmetric conformation once bound to microtubules (Carter et al., 2011; Schmidt et al., 2012). Recent X-ray crystal structures of the microtubule binding domain (Carter et al., 2008) and low (Carter et al., 2011; Kon et al., 2011) and high (Kon et al., 2012; Schmidt et al., 2012) resolution structures of the motor domain are beginning to allow detailed structural based analysis of dynein’s mechanism as reviewed in (Cho and Vale, 2012).
Binding of dynein to the microtubule occurs through a small globular domain attached to the six AAA domains by a long stalk (Carter et al., 2008) and occludes the same tubulin binding site as kinesin (Mizuno et al., 2004). Dynein exhibits a lower binding affinity to microtubules compared to Kinesin-1 and this combined with its ability to step laterally may allow it to bypass kinesin motors on the microtubule (Mizuno et al., 2004). Future work is needed to elucidate whether dynein exhibits cooperative binding to microtubules similar to kinesin.
Perspectives/Conclusions
The kinesin superfamily demonstrates again the importance of symmetry and asymmetry in biology. Asymmetry between motor domains is an inherent consequence of a two-fold symmetric motor binding to a helically symmetric microtubule lattice. This structural asymmetry results in different nucleotide binding affinities for each head when two heads are bound to the lattice and provides the directionality of movement. Asymmetry is also present in the hand-over-hand mechanism in processive plus-end directed motors where each motor head undergoes a unique trajectory during the motile cycle and the nucleotide states of the heads are kept out of phase. Within the Kinesin-14’s, asymmetry leads to a motile cycle where only one head directly participates in the powerstroke mechanism and the other head appears to function in microtubule binding, cooperativity, and ensemble organization. Asymmetry between Dynein heads is less well understood but appears to bias the spatial orientation of the heads on the microtubule during the motile cycle. These examples clearly show that asymmetry is universally required in order to distinguish and coordinate motor heads during the motile cycle.
Within plus-end processive motors, an asymmetric hand-over-hand mechanism most obviously functions to eliminate increasing torque within the coiled-coil and cargo rotation as the motor travels. However, this mechanism may also aid in maintaining the nucleotide states of the two heads out of phase, which is crucial to successful processive motility, by structurally ensuring that each head is unique during the motile cycle. Within the Kinesin-14 motors, asymmetry between heads appears to allow the two heads within the dimer to perform two completely different functions. Here one head appears to drive the ATP-promoted powerstroke while the other head functions to target the motor to the microtubule lattice.
One of the most striking consequences of asymmetry within motors and the microtubule lattice is cooperative binding of motors. Cooperative binding to the microtubule leads to a fascinating potential for regulation between motors as this cooperativity appears to not only affect binding affinity but also how motors spatially align. This spatial alignment appears to be motor class specific and likely ensures that each motor is position such that they perform maximally as a group. Kinesin-14 motors exhibit strong cooperative binding and arrange themselves along protofilaments consistent with their need to work as ensembles and binding to adjacent protofilaments. In contrast, processive plus-end directed Kinesin-1 dimers align laterally along a microtubule allowing binding of the second head to a tubulin dimer along the same protofilament, and also display long range cooperative binding of motors further towards the plus-end of the microtubule.
Lateral alignment of processive plus-end directed kinesins likely does not occur under average physiological conditions as this cooperative binding is only seen under high stoichiometric ratios of motors to microtubule binding sites. Additionally, single molecule studies suggest that two Kinesin-1 molecules bound to a microtubule do not work collectively during transport at sub-stalling forces (Leduc et al., 2007; Rogers et al., 2009; Jamison et al., 2010). However, there is evidence of Kinesin-1 motors working cooperatively under super-stall and accelerating cargo forces, and in these cases close spatial arrangement of motors is critical to this cooperative force generation (Jamison et al., 2010; Jamison et al., 2012). It is likely that under these high force loads that stalling of the leading Kinesin-1 motor allows additional motors to ‘catch-up’ on the microtubule lattice leading to a local high concentrations of motors, and under these conditions lateral alignment may occur allowing motors to work collectively to generate force. Recent single molecule studies indicate that local interactions likely affect stepping rates and microtubule affinities at high applied loads for Kinesin-1 (Uppulury et al., 2012).
Clearly the mechanisms by which Kinesin-1 and Kinesin-14 motors cooperatively bind to microtubules are different, and this raises the tantalizing potential for motors to be able to regulate binding of different classes of motors. For example, does binding of a minus-end directed Kinesin-14 motor not only recruit binding of additional Kinesin-14 motors but also reduce the binding affinity for a plus-end directed motor? Additionally, does binding of non-motor microtubule associated proteins affect the binding of different classes of motors? There is evidence that the manner in which microtubules are stabilized (taxol, GMPCPP, 3R tau, or 4R tau) leads to differences in Kinesin-1 velocities and processivity (McVicker et al., 2011; Peck et al., 2011). In vivo, this could provide an elegant way to regulate motility or transport where different classes of motors bind within the complex microtubule network. Additionally this regulation could serve to minimize occasions when motors traveling in different directions would collide on the microtubule leading to dissociation thus ensuring cargo transportation occurs to the correct location in an efficient time scale with minimal energy expenditure.
Acknowledgments
This work was supported by an NIH grant to I.R. (GM086351), and K.C.R. was supported by an NIH training grant (T32-GM07215). We thank Susan Gilbert (Rensselaer Polytechnic Institute), Andreas Hoenger (University of Colorado at Boulder), and Sarah Rice (Northwestern University) for helpful discussions, and Andreas Hoenger for providing Figure 5.
Definitions
- Kinesin-14 family
Subfamily of the kinesin superfamily characterized by C-terminal motor domains and non-processive motility directed towards the minus-end of the microtubule.
- Processive motion
Kinesin dimers that are able to take multiple steps before dissociating from the microtubule lattice are termed processive. Processivity is accomplished by always having one motor head in the dimer tightly bound to the microtubule at all stages of the motile cycle.
- Nonprocessive motion
Nonprocessive motors completely dissociate from the microtubule after each motile cycle and must rebind before beginning a new cycle. To promote efficient motility these motors typically operate as ensembles.
- B-lattice microtubules
Microtubules where protofilaments are arranged laterally such that α-tubulin lie adjacent to an α-tubulin on the neighboring protofilament. There is a single break in this arrangement, called the seam, where an α-tubulin lies adjacent to a β-tubulin.
References
*Articles of special interest
- Aizawa H, Sekine Y, Takemura R, Zhang Z, Nangaku M, Hirokawa N. Kinesin family in murine central nervous system. The Journal of cell biology. 1992;119:1287–1296. doi: 10.1083/jcb.119.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allingham JS, Sproul LR, Rayment I, Gilbert SP. Vik1 modulates microtubule-Kar3 interactions through a motor domain that lacks an active site. Cell. 2007;128:1161–1172. doi: 10.1016/j.cell.2006.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asbury CL, Fehr AN, Block SM. Kinesin moves by an asymmetric hand-overhand mechanism. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JG, Manning BD, Snyder M. The Kar3p kinesin-related protein forms a novel heterodimeric structure with its associated protein Cik1p. Mol Biol Cell. 2000;11:2373–2385. doi: 10.1091/mbc.11.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Garbarino JE, Wilson-Kubalek EM, Shipley WE, Cho C, Milligan RA, Vale RD, Gibbons IR. Structure and functional role of dynein’s microtubule-binding domain. Science. 2008;322:1691–1695. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Rayment I, Gilbert SP. Kinesin Kar3Cik1 ATPase pathway for microtubule crosslinking. J Biol Chem. 2011;286:29261–29272. doi: 10.1074/jbc.M111.255554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C, Vale RD. The mechanism of dynein motility: Insight from crystal structures of the motor domain. Biochimica et biophysica acta. 2012;1823:182–191. doi: 10.1016/j.bbamcr.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Clancy BE, Behnke-Parks WM, Andreasson JO, Rosenfeld SS, Block SM. A universal pathway for kinesin stepping. Nat Struct Mol Biol. 2011;18:1020–1027. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope J, Gilbert S, Rayment I, Mastronarde D, Hoenger A. Cryo-electron tomography of microtubule-kinesin motor complexes. J Struct Biol. 2010;170:257–265. doi: 10.1016/j.jsb.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deCastro MJ, Fondecave RM, Clarke LA, Schmidt CF, Stewart RJ. Working strokes by single molecules of the kinesin-related microtubule motor ncd. Nat Cell Biol. 2000;2:724–729. doi: 10.1038/35036357. [DOI] [PubMed] [Google Scholar]
- deCastro MJ, Ho CH, Stewart RJ. Motility of dimeric ncd on a metal-chelating surfactant: evidence that ncd is not processive. Biochemistry. 1999;38:5076–5081. doi: 10.1021/bi9829175. [DOI] [PubMed] [Google Scholar]
- DeWitt MA, Chang AY, Combs PA, Yildiz A. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science. 2012;335:221–225. doi: 10.1126/science.1215804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres NF, Yoshioka C, Milligan RA, Vale RD. A lever-arm rotation drives motility of the minus-end-directed kinesin Ncd. Nature. 2005;439:875–878. doi: 10.1038/nature04320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr AN, Gutierrez-Medina B, Asbury CL, Block SM. On the origin of kinesin limping. Biophys J. 2009;97:1663–1670. doi: 10.1016/j.bpj.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G, Hajdo L, Skowronek KJ, Reuther C, Kasprzak AA, Diez S. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat Cell Biol. 2009;11:717–723. doi: 10.1038/ncb1877. [DOI] [PubMed] [Google Scholar]
- Foster KA, Correia JJ, Gilbert SP. Equilibrium binding studies of non-claret disjunctional protein (Ncd) reveal cooperative interactions between the motor domains. J Biol Chem. 1998;273:35307–35318. doi: 10.1074/jbc.273.52.35307. [DOI] [PubMed] [Google Scholar]
- Foster KA, Gilbert SP. Kinetic studies of dimeric Ncd: evidence that Ncd is not processive. Biochemistry. 2000;39:1784–1791. doi: 10.1021/bi991500b. [DOI] [PubMed] [Google Scholar]
- Foster KA, Mackey AT, Gilbert SP. A mechanistic model for Ncd directionality. J Biol Chem. 2001;276:19259–19266. doi: 10.1074/jbc.M008347200. [DOI] [PubMed] [Google Scholar]
- Gilbert SP, Moyer ML, Johnson KA. Alternating site mechanism of the kinesin ATPase. Biochemistry. 1998;37:792–799. doi: 10.1021/bi971117b. [DOI] [PubMed] [Google Scholar]
- Hackney DD. Kinesin ATPase: rate-limiting ADP release. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:6314–6318. doi: 10.1073/pnas.85.17.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD. Evidence for Alternating Head Catalysis by Kinesin During Microtubule-Stimulated ATP Hydrolysis. Proc Natl Acad Sci USA. 1994;91:6856–6869. doi: 10.1073/pnas.91.15.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock WO, Howard J. Kinesin’s processivity results from mechanical and chemical coordination between the ATP hydrolysis cycles of the two motor domains. Proc Natl Acad Sci USA. 1999;96:13147–13152. doi: 10.1073/pnas.96.23.13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi H, Bronner CE, Park HW, Endow SA. Rapid double 8-nm steps by a kinesin mutant. Embo J. 2004;23:2993–2999. doi: 10.1038/sj.emboj.7600306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenger A, Sack S, Thormahlen M, Marx A, Muller J, Gross H, Mandelkow E. Image reconstructions of microtubules decorated with monomeric and dimeric kinesins: comparison with X-ray structure and implications for motility. J Cell Biol. 1998;141:419–430. doi: 10.1083/jcb.141.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua W, Chung J, Gelles J. Distinguishing inchworm and hand-over-hand processive kinesin movement by neck rotation measurements. Science. 2002;295:844–848. doi: 10.1126/science.1063089. [DOI] [PubMed] [Google Scholar]
- Jamison DK, Driver JW, Diehl MR. Cooperative responses of multiple kinesins to variable and constant loads. The Journal of biological chemistry. 2012;287:3357–3365. doi: 10.1074/jbc.M111.296582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamison DK, Driver JW, Rogers AR, Constantinou PE, Diehl MR. Two kinesins transport cargo primarily via the action of one motor: implications for intracellular transport. Biophysical journal. 2010;99:2967–2977. doi: 10.1016/j.bpj.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaan HY, Hackney DD, Kozielski F. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science. 2011;333:883–885. doi: 10.1126/science.1204824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaseda K, Higuchi H, Hirose K. Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat Cell Biol. 2003;5:1079–1082. doi: 10.1038/ncb1067. [DOI] [PubMed] [Google Scholar]
- King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- Klumpp LM, Hoenger A, Gilbert SP. Kinesin’s second step. Proc Natl Acad Sci U S A. 2004;101:3444–3449. doi: 10.1073/pnas.0307691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocik E, Skowronek KJ, Kasprzak AA. Interactions between subunits in heterodimeric Ncd molecules. J Biol Chem. 2009;284:35735–35745. doi: 10.1074/jbc.M109.024240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, Kurisu G. The 2.8 A crystal structure of the dynein motor domain. Nature. 2012 doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- Kon T, Sutoh K, Kurisu G. X-ray structure of a functional full-length dynein motor domain. Nature structural & molecular biology. 2011;18:638–642. doi: 10.1038/nsmb.2074. [DOI] [PubMed] [Google Scholar]
- Kondo S, Sato-Yoshitake R, Noda Y, Aizawa H, Nakata T, Matsuura Y, Hirokawa N. KIF3A is a new microtubule-based anterograde motor in the nerve axon. The Journal of cell biology. 1994;125:1095–1107. doi: 10.1083/jcb.125.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozielski F, Svergun D, Zaccai G, Wade RH, Koch MH. The overall conformation of conventional kinesins studied by small angle X-ray and neutron scattering. The Journal of biological chemistry. 2001;276:1267–1275. doi: 10.1074/jbc.M007169200. [DOI] [PubMed] [Google Scholar]
- Krebs A, Goldie KN, Hoenger A. Complex formation with kinesin motor domains affects the structure of microtubules. J Mol Biol. 2004;335:139–153. doi: 10.1016/j.jmb.2003.10.039. [DOI] [PubMed] [Google Scholar]
- Krzysiak TC, Grabe M, Gilbert SP. Getting in sync with dimeric Eg5. Initiation and regulation of the processive run. The Journal of biological chemistry. 2008;283:2078–2087. doi: 10.1074/jbc.M708354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzysiak TC, Wendt T, Sproul LR, Tittmann P, Gross H, Gilbert SP, Hoenger A. A structural model for monastrol inhibition of dimeric kinesin Eg5. EMBO J. 2006;25:2263–2273. doi: 10.1038/sj.emboj.7601108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, Malmberg RL, McIntosh JR, Miki H, Mitchison TJ, Okada Y, Reddy AS, Saxton WM, Schliwa M, Scholey JM, Vale RD, Walczak CE, Wordeman L. A standardized kinesin nomenclature. The Journal of cell biology. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc C, Ruhnow F, Howard J, Diez S. Detection of fractional steps in cargo movement by the collective operation of kinesin-1 motors. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10847–10852. doi: 10.1073/pnas.0701864104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YZ, Taylor EW. Interacting head mechanism of microtubule-kinesin ATPase. The Journal of biological chemistry. 1997;272:724–730. doi: 10.1074/jbc.272.2.724. [DOI] [PubMed] [Google Scholar]
- Mackey AT, Gilbert SP. Moving a microtubule may require two heads: a kinetic investigation of monomeric Ncd. Biochemistry. 2000;39:1346–1355. doi: 10.1021/bi991918+. [DOI] [PubMed] [Google Scholar]
- Mallik R, Carter BC, Lex SA, King SJ, Gross SP. Cytoplasmic dynein functions as a gear in response to load. Nature. 2004;427:649–652. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]
- Manning BD, Barrett JG, Wallace JA, Granok H, Snyder M. Differential regulation of the Kar3p kinesin-related protein by two associated proteins, Cik1p and Vik1p. J Cell Biol. 1999;144:1219–1233. doi: 10.1083/jcb.144.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Morphew MK, Grissom PM, Gilbert SP, Hoenger A. Lattice structure of cytoplasmic microtubules in a cultured Mammalian cell. Journal of molecular biology. 2009;394:177–182. doi: 10.1016/j.jmb.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker DP, Chrin LR, Berger CL. The nucleotide-binding state of microtubules modulates kinesin processivity and the ability of Tau to inhibit kinesin-mediated transport. The Journal of biological chemistry. 2011;286:42873–42880. doi: 10.1074/jbc.M111.292987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno N, Toba S, Edamatsu M, Watai-Nishii J, Hirokawa N, Toyoshima YY, Kikkawa M. Dynein and kinesin share an overlapping microtubule-binding site. The EMBO journal. 2004;23:2459–2467. doi: 10.1038/sj.emboj.7600240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan V, Abramson T, Lyass A, Winter D, Porro E, Hong F, Chamberlin NL, Schnapp BJ. KIF3C and KIF3A form a novel neuronal heteromeric kinesin that associates with membrane vesicles. Mol Biol Cell. 1998;9:637–652. doi: 10.1091/mbc.9.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukrishnan G, Zhang Y, Shastry S, Hancock WO. The processivity of kinesin-2 motors suggests diminished front-head gating. Current biology: CB. 2009;19:442–447. doi: 10.1016/j.cub.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto E, Sakai H, Kaseda K. Long-range cooperative binding of kinesin to a microtubule in the presence of ATP. J Cell Biol. 2005;168:691–696. doi: 10.1083/jcb.200409035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. Journal of molecular biology. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Page BD, Satterwhite LL, Rose MD, Snyder M. Localization of the Kar3 kinesin heavy chain-related protein requires the Cik1 interacting protein. J Cell Biol. 1994;124:507–519. doi: 10.1083/jcb.124.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page BD, Snyder M. CIK1: a developmentally regulated spindle pole body-associated protein important for microtubule functions in Saccharomyces cerevisiae. Genes Dev. 1992;6:1414–1429. doi: 10.1101/gad.6.8.1414. [DOI] [PubMed] [Google Scholar]
- Peck A, Sargin ME, LaPointe NE, Rose K, Manjunath BS, Feinstein SC, Wilson L. Tau isoform-specific modulation of kinesin-driven microtubule gliding rates and trajectories as determined with tau-stabilized microtubules. Cytoskeleton (Hoboken) 2011;68:44–55. doi: 10.1002/cm.20494. [DOI] [PubMed] [Google Scholar]
- Qiu W, Derr ND, Goodman BS, Villa E, Wu D, Shih W, Reck-Peterson SL. Dynein achieves processive motion using both stochastic and coordinated stepping. Nature structural & molecular biology. 2012;19:193–200. doi: 10.1038/nsmb.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rank KC, Chen CJ, Cope J, Porche K, Hoenger A, Gilbert SP, Rayment I. Kar3Vik1, a member of the Kinesin-14 Superfamily, shows a Novel Kinesin Microtubule Binding Pattern. The Journal of Cell Biology. 2012;197:957–970. doi: 10.1083/jcb.201201132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, Pate E, Cooke R, Taylor EW, Milligan RA, Vale RD. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- Rogers AR, Driver JW, Constantinou PE, Kenneth Jamison D, Diehl MR. Negative interference dominates collective transport of kinesin motors in the absence of load. Phys Chem Chem Phys. 2009;11:4882–4889. doi: 10.1039/b900964g. [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Fordyce PM, Jefferson GM, King PH, Block SM. Stepping and stretching. How kinesin uses internal strain to walk processively. J Biol Chem. 2003;278:18550–18556. doi: 10.1074/jbc.M300849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablin EP, Case RB, Dai SC, Hart CL, Ruby A, Vale RD, Fletterick RJ. Direction determination in the minus-end-directed kinesin motor ncd. Nature. 1998;395:813–816. doi: 10.1038/27463. [DOI] [PubMed] [Google Scholar]
- Sablin EP, Kull JF, Cooke R, Vale RD, Fletterick RJ. Three-Dimensional Structure of the Motor Domain of NCD, a Kinesin-Related Motor with Reversed Polarity of Movement. Nature. 1996;380:555–559. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- Sardar HS, Gilbert SP. Microtubule Capture by Mitotic Kinesin CENP-E. The Journal of biological chemistry. 2012 doi: 10.1074/jbc.M112.376830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-A crystal structure. Nature structural & molecular biology. 2012 doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima T, Imamula K, Kon T, Ohkura R, Sutoh K. Head-head coordination is required for the processive motion of cytoplasmic dynein, an AAA+ molecular motor. Journal of structural biology. 2006;156:182–189. doi: 10.1016/j.jsb.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Shubeita GT, Tran SL, Xu J, Vershinin M, Cermelli S, Cotton SL, Welte MA, Gross SP. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 2008;135:1098–1107. doi: 10.1016/j.cell.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiniotis G, Surrey T, Altmann S, Gross H, Song YH, Mandelkow E, Hoenger A. Nucleotide-induced conformations in the neck region of dimeric kinesin. EMBO J. 2003;22:1518–1528. doi: 10.1093/emboj/cdg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul LR, Anderson DJ, Mackey AT, Saunders WS, Gilbert SP. Cik1 targets the minus-end kinesin depolymerase Kar3 to microtubule plus ends. Curr Biol. 2005;15:1420–1427. doi: 10.1016/j.cub.2005.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone DB, Hjelm RP, Jr, Mendelson RA. Solution structures of dimeric kinesin and ncd motors. Biochemistry. 1999;38:4938–4947. doi: 10.1021/bi982374z. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct Observation of Kinesin Stepping by Optical Trapping Interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- Thormahlen M, Marx A, Muller SA, Song Y, Mandelkow EM, Aebi U, Mandelkow E. Interaction of monomeric and dimeric kinesin with microtubules. J Mol Biol. 1998;275:795–809. doi: 10.1006/jmbi.1997.1503. [DOI] [PubMed] [Google Scholar]
- Turner J, Anderson R, Guo J, Beraud C, Fletterick R, Sakowicz R. Crystal structure of the mitotic spindle kinesin Eg5 reveals a novel conformation of the neck-linker. J Biol Chem. 2001;276:25496–25502. doi: 10.1074/jbc.M100395200. [DOI] [PubMed] [Google Scholar]
- Uppulury K, Efremov A, Driver J, Jamison K, Diehl MR, Kolomeisky AB. How the Interplay Between Mechanical and Non-Mechanical Interactions Affects Multiple Kinesin Dynamics. J Phys Chem B. 2012;116:8846–8855. doi: 10.1021/jp304018b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Fletterick RJ. The design plan of kinesin motors. Annu Rev Cell Dev Biol. 1997;13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- Valentine MT, Fordyce PM, Krzysiak TC, Gilbert SP, Block SM. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat Cell Biol. 2006 doi: 10.1038/ncb1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilfan A, Frey E, Schwabl F, Thormahlen M, Song YH, Mandelkow E. Dynamics and cooperativity of microtubule decoration by the motor protein kinesin. J Mol Biol. 2001;312:1011–1026. doi: 10.1006/jmbi.2001.5020. [DOI] [PubMed] [Google Scholar]
- Waitzman JS, Larson AG, Cochran JC, Naber N, Cooke R, Jon Kull F, Pate E, Rice SE. The loop 5 element structurally and kinetically coordinates dimers of the human kinesin-5, Eg5. Biophysical journal. 2011;101:2760–2769. doi: 10.1016/j.bpj.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Khan S, Sheetz MP. Single cytoplasmic dynein molecule movements: characterization and comparison with kinesin. Biophysical journal. 1995;69:2011–2023. doi: 10.1016/S0006-3495(95)80071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt TG, Volkmann N, Skiniotis G, Goldie KN, Muller J, Mandelkow E, Hoenger A. Microscopic evidence for a minus-end-directed power stroke in the kinesin motor ncd. EMBO J. 2002;21:5969–5978. doi: 10.1093/emboj/cdf622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie P, Dou SX, Wang PY. Limping of homodimeric kinesin motors. J Mol Biol. 2007;366:976–985. doi: 10.1016/j.jmb.2006.10.081. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Nakata T, Okada Y, Hirokawa N. KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. The Journal of cell biology. 1995;130:1387–1399. doi: 10.1083/jcb.130.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Goldstein LS. Characterization of the KIF3C neural kinesin-like motor from mouse. Mol Biol Cell. 1998;9:249–261. doi: 10.1091/mbc.9.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]