Abstract
Heterozygous mutations in the EFTUD2 were identified in 12 individuals with a rare sporadic craniofacial condition termed Mandibulofacial dysostosis with microcephaly (MIM 610536). We present clinical and radiographic features of three additional patients with de novo heterozygous mutations in EFTUD2.. Although clinical features overlap with findings of the original report (choanal atresia, cleft palate, maxillary and mandibular hypoplasia, and microtia), microcephaly was present in two of three patients and cognitive impairment was milder in those with head circumference proportional to height. Our cases expand the phenotypic spectrum to include epibulbar dermoids and zygomatic arch clefting. We suggest that craniofacial computed tomography studies to assess cleft of zygomatic arch may assist in making this diagnosis. We recommend consideration of EFTUD2 testing in individuals with features of oculo-auriculo-vertebral spectrum and bilateral microtia, or individuals with atypical CHARGE syndrome who do not have a CHD7 mutation, particularly those with a zygomatic arch cleft. The absence of microcephaly in one patient indicates that it is a highly variable phenotypic feature.
Keywords: craniofacial development, EFTUD2, epibulbar dermoid, craniofacial microsomia, oculo-auriculo-vertebral spectrum (OAVS), choanal atresia
Introduction
Mandibulofacial dysostosis is a clinically and etiologically heterogeneous group of conditions characterized by significant malar and mandibular hypoplasia. Although many distinct mandibulofacial and acrofacial dysostoses syndromes have been described clinically, causative mutations have only been indentified for Treacher Collins (OMIM154500), Miller (OMIM 263750), and Nager (OMIM 154400) syndrome. Recently, heterozygous mutations in EFTUD2 (elongation factor Tu GTP binding domain containing 2) gene were identified in 14 individuals with a rare sporadic craniofacial condition termed Mandibulofacial dysostosis with microcephaly (OMIM 610536) [Bernier et al., 2012; Lines et al., 2012; Need et al., 2012]. The phenotype reported in this condition included microcephaly, choanal atresia, cleft palate, maxillary and mandibular hypoplasia, microtia, and developmental delay. Microcephaly was described as both progressive and severe [Guion-Almeida et al., 2006; Lines et al., 2012; Need et al., 2012; Wieczorek et al., 2009]. Distinguishing Mandibulofacial dysostosis caused by EFTUD2 from the other known craniofacial dysostosis can be challenging, and there are numerous reports of individuals with craniofacial dysostosis that have not yet been distinctly classified. Given the phenotypic overlap between the mandibulofacial dysostosis syndromes and other craniofacial conditions, it is important to have a detailed delineation of this phenotype.
We present clinical and radiographic features of three additional unrelated patients whose facial gestalt and dysmorphic features lead us to the identification of de novo heterozygous mutations in EFTUD2 via exome sequencing, prior to publication of the initial 12 cases. Our report expands the clinical features in patients with EFTUD2 mutations and demonstrates an overlap with oculo-auriculo-vertebral spectrum (OAVS) and CHARGE syndrome.
Materials and Methods
Clinical and Radiological Assessment
Patients were ascertained from the Craniofacial Center at Seattle Children's Hospital following routine referral for clinical treatment. Phenotypic data was obtained by direct physical exam, questioning, review of medical records and 3-dimensional (3D) computed tomography (CT) scans. An experienced clinical geneticist and craniofacial pediatrician were responsible for each diagnosis. Each participant provided informed consent and the study was approved by the institutional review board of Seattle Children’s Hospital.
Molecular Genetics Analysis
We performed exome capture and sequencing to determine the molecular etiology of the three unrelated patients that we identified as having a strikingly similar phenotype. We considered autosomal-dominant and recessive models because the mode of inheritance was unclear. We also sequenced the unaffected parents to perform de novo variant analysis.
Briefly, exome enrichment was performed on an Illumina-compatible shotgun library generated from 1ug of genomic DNA from each sample. Each shotgun library was hybridized to DNA probes targeting ~300,000 coding regions (Roche Nimblegen SeqCap EZ Exome V2.0), then sequenced on an Illumina HiSeq 2000 with paired-end 50bp reads. For each sample, we required 8× coverage over at least 90% of the exome target, and 20× coverage over at least 80% of the target. Sequence data was mapped using BWA to a custom human genome build (hg19). Variations (SNVs and indels) were called with the GATK UnifiedGenotyper run in “multisample” mode to provide a complete data set for each trio.
We performed de novo variant analysis on the three trios. Briefly, variants in the affected proband were screened against the variants in the unaffected parents to discover alleles that could not be derived via parental transmission. A genotype call for each parent was required, and the read depth for each parent was required to be 8 or greater. Selected variations meeting these criteria were submitted to the EVS server (http://evs.gs.washington.edu/EVS) to screen against 1537 NHLBI Exome Sequencing Project (ESP) individuals whose exomes were sequenced at the University of Washington with the same exome target and sequencing platform. Additional filtering was applied to the trios to remove variants seen in dnSNP build 134 and only missense, stop gain, stop loss, canonical splice and indel variants were kept for further analysis.
Results
Clinical and Radiological Features of the Patients
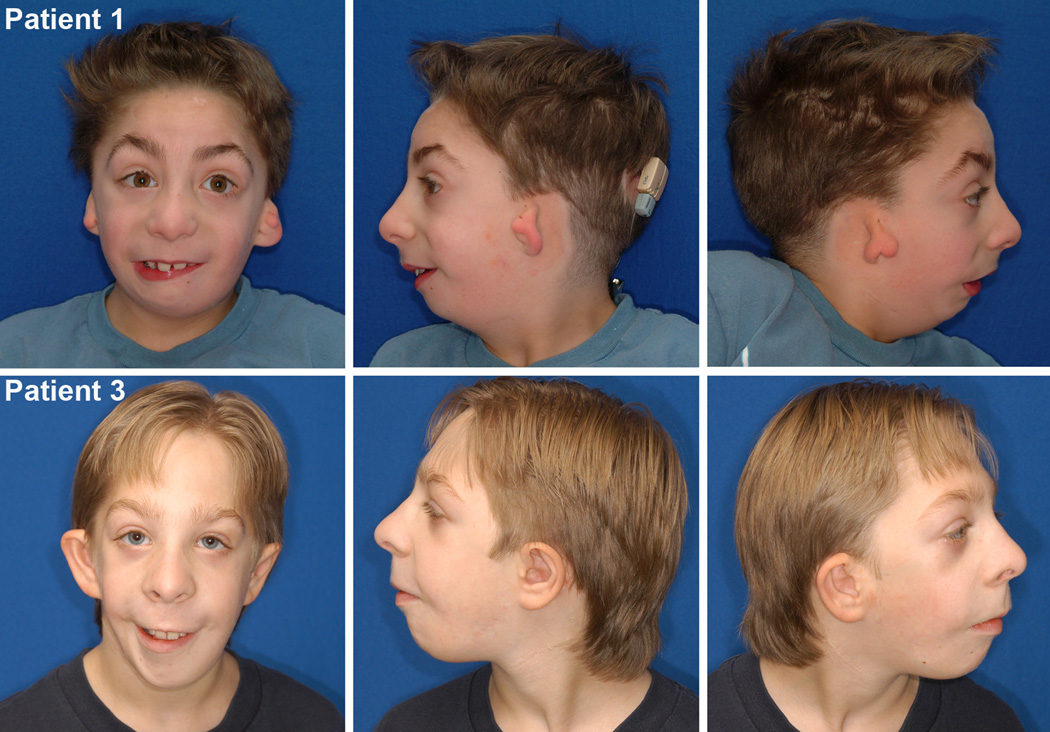
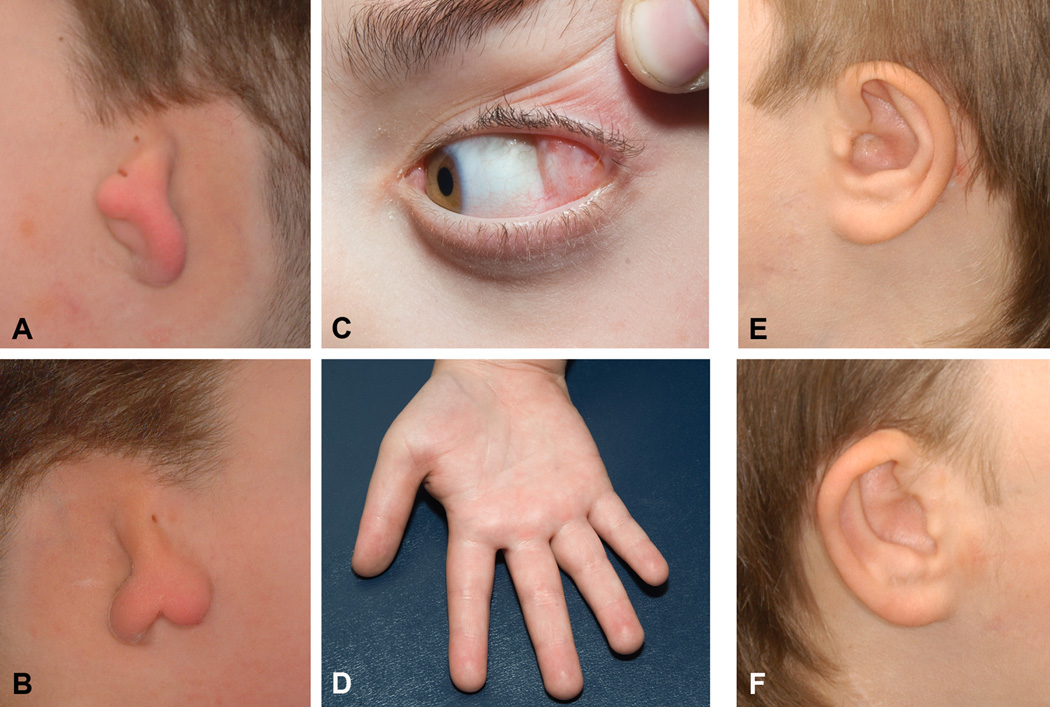
Prior to our investigation each patient had been given the tentative diagnoses of atypical OAVS or CHARGE syndrome. Their phenotype was characterized by choanal atresia, maxillary and mandibular hypoplasia, zygomatic arch clefting, bilateral symmetric microtia with preauricular skin tags and hearing loss, and developmental delay (Figs 1 and 2). Microcephaly, as defined by OFC smaller than 2 SD below mean, was present in two patients and one presented an epibulbar dermoid on the left eye. We did not observe any lower eyelid coloboma or metopic ridge as described in the previous patients with mutations on this gene (Table I) [Lines et al; Need at al].
Figure 1.
Photographs of Patients 1 and 3 showing frontal and lateral views.
Figure 2.
Photographs of patients showing detailed craniofacial phenotype. A–B: External ears of Patient 1 presenting microtia type 3. C–D: Epibulbar dermoid and proximally placed thumb in Patient 1. E–F: Patient 3 external ears presenting microtia type 1.
Table I.
Clinical and Radiological features of patients with EFTUD2 mutations and comparison with 14 patients previously described in the literature.
| Patient 1 | Patient 2 | Patient 3 | Lines et al, 2012 | Need et al, 2012 | |
|---|---|---|---|---|---|
| Sex | M | M | M | 7M/ 5F | n.r. |
| Birth weight (g) | 2693 (−2SD) | n.r. | 3890 (+1SD) | −2.5SD to +0.5SD | n.r. |
| OFC at birth (cm) | n.r. | n.r. | 32 (+1SD) | −3.5SD to −1SD | n.r. |
| OFC (cm) | 51.8 (−1SD) | 49 (−4SD) | 52.5 (−3SD) | −6SD to −3SD | n.r. |
| Height (cm) | 137.6 (−1SD) | 147.3 (−4SD) | 159.8 (−3SD) | −2SD to 0SD | n.r. |
| Microcephaly | No | OFC proportionate to height | OFC proportionate to height | 12/12 | 2/2 |
| Metopic ridge | No | No | No | 7/11 | n.r. |
| Facial asymmetry | Yes | Yes | Yes | n.r. | 1/2 |
| Choanal atresia | Yes | Yes | Yes | 6/10 | n.r. |
| Epibulbar dermoid | Left | No | No | n.r. | n.r. |
| Lower eyelid cleft | No | No | No | 3/12 | n.r. |
| Cleft of zygomatic arch | Unilateral (left side) | Bilateral | Bilateral | n.r. | n.r. |
| Microtia | Bilateral (grade 3) | Bilateral (grade 2) | Bilateral (grade 1) | 11/11 | 2/2 |
| Preauricular skin tags | Bilateral | Bilateral | Unilateral –left | 10/12 | n.r. |
| External auditory canal | Atretic | Small | Stenotic | 7/10 | n.r. |
| Hearing loss | Conductive | Sensorineural | Conductive | 10/11 | 2/2 |
| Inner ear | Normal | Incompletely formed lateral semicircular canal and dilated vestibule (unilateral) |
Abnormal enlarged connection between the cochlea and the vestibule (bilateral) |
1/12 | n.r. |
| Middle ear | Hypoplastic, dysplastic ossicles | -- | Malformed ossicles | n.r. | n.r. |
| Mandibular hypoplasia | Yes | Yes | Mandibular asymmetry | n.r. | n.r. |
| Malar hypoplasia | Yes | Yes | Yes | 11/12 | n.r. |
| Micrognathia | Yes ( asymmetric) | Yes | Yes | 12/12 | n.r. |
| Cleft palate | Bifid uvula | No | No | 7/12 | 1/2 |
| CHD | No | Small VSD | No | 7/12 | 1/2 |
| Kidney | No | Non-specified anomalies (normal function) |
No | n.r. | n.r. |
| Thumb abnormalities | Proximally placed | No | Proximally placed | 5/11 | 1/2 |
| Cervical spine | Normal | --- | Stable rotatory sub-luxation of C2 with respect to C1 |
n.r. | 1/2 (C2– C5 fusion |
| Personality | Cooperative, answer questions appropriately, no unusual behaviors |
Shy and distractible | Obsessive-compulsive behaviors Happy and interactive |
n.r. | n.r. |
| Developmental delay | Mild | Moderate-Severe | Mild | 12/12 | 2/2 |
| Seizures | No | Yes | Yes** | 5/10 | n.r. |
OFC: Occipitofrontal circumference / CHD: Congenital heart defect / n.r.: not reported
Post-operative catatonia lasting several days. Unknown etiology
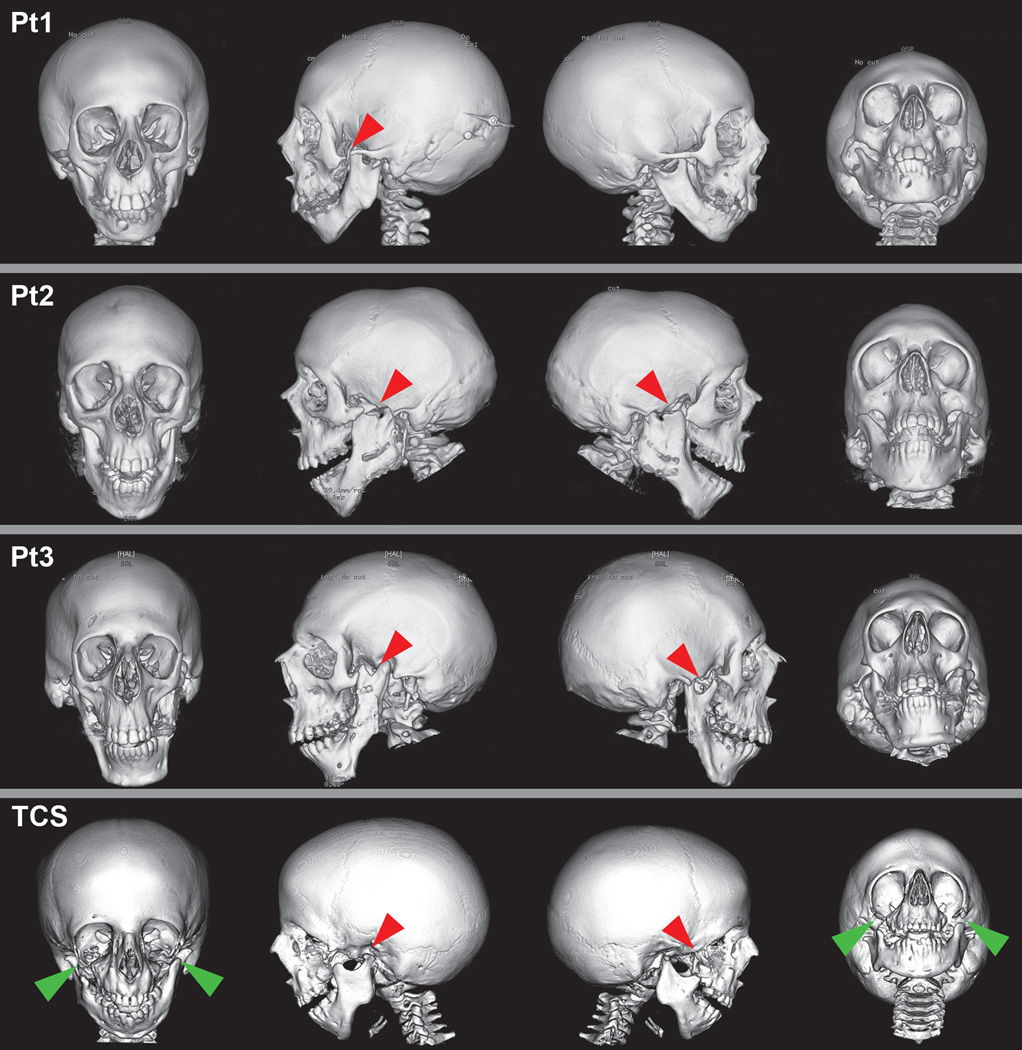
The radiographic features included zygomatic arch clefting described as complete absence of the zygomatic arches in cases 2 and 3 and unilateral cleft of zygomatic arch in case 1, and severe bilateral maxillary hypoplasia (Fig 3, Table I). Abnormalities of the middle and inner ear included malformed ossicles and dilated vestibule in two patients.
Figure 3.
Three-dimensional (3D) computed tomography (CT) scans of our cohort of patients and a Treacher Collins patient for comparison of craniofacial abnormalities.
Pt (Patient) 1, Pt2, and Pt3: Frontal, lateral, and submental views of individuals with EFTUD2 mutations on 3D-CT. Red arrows indicate that the bilateral aplasia of the zygomatic arch in patients 2 and 3 and unilateral zygomatic arch clefting in Patient 1.
TCS: 3D-CT of individual with TCS (Treacher Collins syndrome). Green arrows show inferiolateral orbital clefts of the zygoma in addition to zygomatic arch clefts in TCS.
All patients had delayed psychomotor milestones. Patient 1 walked with support at 13 months and spoke around 40 words at 3 years, at 4 years of age he was toilet trained. Patient 3 participated in an early intervention program since age 6 months; he sat without support at 10 months and walked at 24 months. His mother reports “significant language delay” during his childhood, at present his speech is sometimes difficult to understand. A psychological evaluation when he was a 15-year-old revealed an overall cognitive ability below average range (2nd centile), however his processing speed is an area of relative strength in the low average range (13th centile). We do not have detailed information on psychomotor milestones for Patient 2. All three patients attend regular school and receive special education services.
No parent had craniofacial abnormalities, cognitive impairment, or apparent behavioral problems.
Exome sequencing results
Under the dominant model, one gene, EFTUD2 was found with one potentially deleterious variant in all three patients. Under the recessive model, no gene was found with two likely deleterious variants per patient. Sanger sequencing subsequently confirmed that all three novel variants were present in EFTUD2 and that the variants were de novo in all. Two patients presented with missense and one with a splice-site mutation (Table II). These mutations were not observed in 1537 ESP individuals and dbSNP build 134. Further evidence supporting the hypothesis that mutations in EFTUD2 cause this condition was provided by the report by Lines et al [2012] on EFTUD2 mutations in 12 individuals with the same phenotype, serving as a validation cohort to our study.
Table II.
Summary of the EFTUD2 mutations in the three patients
| Patient 1 | Patient 2 | Patient 3 | |
|---|---|---|---|
| EFTUD2 mutation | E794K | Splice-3 | Q401E |
| Genomic position | 17:42930740 | 17:42961093 | 17:42941130 |
| Nucleotide substitution | c.2637 G>A | c.504 -2G>T | c.1458 C>G |
| Amino acid substitution | p.Glu794Lys | ----- | p.Gln401Glu |
| GERP | 5.15 | 5.23 | 5.92 |
| Exome variant server frequency | 0/1537 | 0/1537 | 0/1537 |
GERP: Genomic Evolutionary Rate Profiling.
The mutations are highly conserved as indicated by GERP scores and were not found in a control set of 1,537 human exomes from the NHLBI Exome Sequencing Project
Discussion
Our report of three additional patients with EFTUD2 mutations expands the phenotypic spectrum to include epibulbar dermoids and zygomatic arch clefting. In addition, it confirms a well-defined pattern of abnormalities, including choanal atresia, microtia, preauricular tags, mandibular and malar hypoplasia leading to a recognizable syndrome. The microtia is rather distinctive with most patients presenting with more severe, symmetric hypoplasia of the upper portion of the ear and normal lobes. Craniofacial imaging studies of the zygomatic arch may further assist in making the diagnosis. The constellation of malformations, particularly with an epibulbar lipodermoid and unilateral involvement demonstrated that mutations in EFTUD2 overlap with OAVS phenotype.
In addition, maxillary and mandibular hypoplasia, ear abnormalities, and hearing impairment are common features for this condition and CHARGE, Treacher Collins, Miller and Nager syndrome showing a clear clinical overlap between them. Choanal atresia and the middle ear anomalies are common in CHARGE and although EFTUD2 patients do not present severe limb deficiencies similar to Nager and Miller syndromes, mild thumb abnormalities were observed in a significant proportion of patients [Lines et al., 2012; Need et al., 2012]. Recently, Bernier et al [2012] reported a mutation in EFTUD2 in a patient previously diagnosed with Nager syndrome. The individual’s phenotype, besides microcephaly, was not described. However, this finding suggests that the proportion of radial abnormalities in MMFD might be underestimated, particularly if we consider that the cases screened for EFTUD2 mutations were selected based on other features and those with more severe radial abnormalities might have been excluded.
The clinical overlap between these conditions had already suggested that abnormal development of the derivatives of the first and second branchial arches may share common broad molecular pathogenetic mechanisms. This suspicion has lately been confirmed by the multiple gene discoveries for these conditions. U5-116kD, the protein encoded by EFTUD2, is a spliceosomal GTPase involved in the assembly and disassembly of the spliceosome to the pre-mRNA transcript[Staley and Woolford, 2009]. SF3B4, the gene identified in Nager syndrome, encodes SAP49, also a critical component of the spliceosome [Bernier et al., 2012]. Genes involved in pre-rRNA transcription are responsible for TCS (TCOF1, POLR1D, and POLR1C) [Dauwerse et al., 2011; 1996] and CHARGE syndrome is caused by mutation of the DNA binding protein CHD7 which associates with rDNA and has a role as a positive regulator of rRNA synthesis, essential for transcription regulation[Vissers et al., 2004; Zentner et al., 2010]. In Miller syndrome the gene (DHODH) encodes an enzyme involved in pyrimidine biosynthesis [Ng et al., 2010]. The study of other individuals with undiagnosed mandibulofacial and other craniofacial dysostoses might reveal mutations in other genes with similar roles. Future studies integrating these molecular pathways will advance our knowledge about the development of the craniofacial skeleton and the role of splicing abnormalities in craniofacial development.
In the clinical setting, we recommend consideration of EFTUD2 testing in individuals affected by craniofacial dysostosis with zygomatic arch clefts, or those with atypical CHARGE syndrome who do not have a CHD7 mutation. Although usually the ear abnormalities in CHARGE affect the lower part, and in MMFD the upper part, we believe that because of the large variability in the ear phenotype in both syndromes, testing is justified, particularly given the benefit of a potential conclusive diagnosis for the management of the patient and genetic counseling of the family. Additionally, consider testing patients with developmental delay, bilateral symmetric microtia and zygomatic arch clefts and epibulbar dermoids or choanal atresia.
The absence of microcephaly in one patient indicates that it is a highly variable phenotypic feature. We also remark that in the two other patients the diagnosis was of “relative microcephaly”, i.e., small head circumference in proportion to the height. In reviewing the data reported by Lines et al. [2012], we observed that one patient also presented with relative microcephaly, which may be associated with a much better neurodevelopmental prognosis than absolute microcephaly.
Whole-exome sequencing has been used successfully to identify the causative genes for many rare disorders in unrelated sporadic cases or in affected families with small pedigrees. The discovery at virtually the same time of this new syndrome by two different research teams validates this method, and, more importantly, reinforces the importance of detailed phenotypic characterization for grouping of similar phenotypes and successful identification of causative genes in syndromes with unknown etiology.
ACKNOWLEDGMENTS
This work was supported by the Jean Renny Endowment for Craniofacial Research and by the Grant Number K99DC011282 from the National Institute on Deafness and Other Communication Disorders, and a grant from National Human Genome Research Institute, HG005608.
REFERENCES
- Bernier FP, Caluseriu O, Ng S, Schwartzentruber J, Buckingham KJ, Innes AM, Jabs EW, Innis JW, Schuette JL, Gorski JL, Byers PH, Andelfinger G, Siu V, Lauzon J, Fernandez BA, McMillin M, Scott RH, Racher H, Majewski J, Nickerson DA, Shendure J, Bamshad MJ, Parboosingh JS. Haploinsufficiency of SF3B4, a Component of the Pre-mRNA Spliceosomal Complex, Causes Nager Syndrome. Am J Hum Genet. 2012;90:925–933. doi: 10.1016/j.ajhg.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauwerse JG, Dixon J, Seland S, Ruivenkamp CA, van Haeringen A, Hoefsloot LH, Peters DJ, Boers AC, Daumer-Haas C, Maiwald R, Zweier C, Kerr B, Cobo AM, Toral JF, Hoogeboom AJ, Lohmann DR, Hehr U, Dixon MJ, Breuning MH, Wieczorek D. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet. 2011;43:20–22. doi: 10.1038/ng.724. [DOI] [PubMed] [Google Scholar]
- Guion-Almeida ML, Zechi-Ceide RM, Vendramini S, Tabith Junior A. A new syndrome with growth and mental retardation, mandibulofacial dysostosis, microcephaly, and cleft palate. Clin Dysmorphol. 2006;15:171–174. doi: 10.1097/01.mcd.0000220603.09661.7e. [DOI] [PubMed] [Google Scholar]
- Lines MA, Huang L, Schwartzentruber J, Douglas SL, Lynch DC, Beaulieu C, Guion-Almeida ML, Zechi-Ceide RM, Gener B, Gillessen-Kaesbach G, Nava C, Baujat G, Horn D, Kini U, Caliebe A, Alanay Y, Utine GE, Lev D, Kohlhase J, Grix AW, Lohmann DR, Hehr U, Bohm D, Majewski J, Bulman DE, Wieczorek D, Boycott KM. Haploinsufficiency of a spliceosomal GTPase encoded by EFTUD2 causes mandibulofacial dysostosis with microcephaly. Am J Hum Genet. 2012;90:369–377. doi: 10.1016/j.ajhg.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Shashi V, Hitomi Y, Schoch K, Shianna KV, McDonald MT, Meisler MH, Goldstein DB. Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet. 2012;49:353–361. doi: 10.1136/jmedgenet-2012-100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Woolford JL., Jr Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr Opin Cell Biol. 2009;21:109–118. doi: 10.1016/j.ceb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Treacher Collins Syndrome Collaborative Group. Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. Nat Genet. 1996;12:130–136. doi: 10.1038/ng0296-130. [DOI] [PubMed] [Google Scholar]
- Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, Hamel BC, Schoenmakers EF, Brunner HG, Veltman JA, van Kessel AG. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- Wieczorek D, Gener B, Gonzalez MJ, Seland S, Fischer S, Hehr U, Kuechler A, Hoefsloot LH, de Leeuw N, Gillessen-Kaesbach G, Lohmann DR. Microcephaly, microtia, preauricular tags, choanal atresia and developmental delay in three unrelated patients: a mandibulofacial dysostosis distinct from Treacher Collins syndrome. Am J Med Genet A. 2009;149A:837–843. doi: 10.1002/ajmg.a.32747. [DOI] [PubMed] [Google Scholar]
- Zentner GE, Hurd EA, Schnetz MP, Handoko L, Wang C, Wang Z, Wei C, Tesar PJ, Hatzoglou M, Martin DM, Scacheri PC. CHD7 functions in the nucleolus as a positive regulator of ribosomal RNA biogenesis. Hum Mol Genet. 2010;19:3491–3501. doi: 10.1093/hmg/ddq265. [DOI] [PMC free article] [PubMed] [Google Scholar]