Abstract
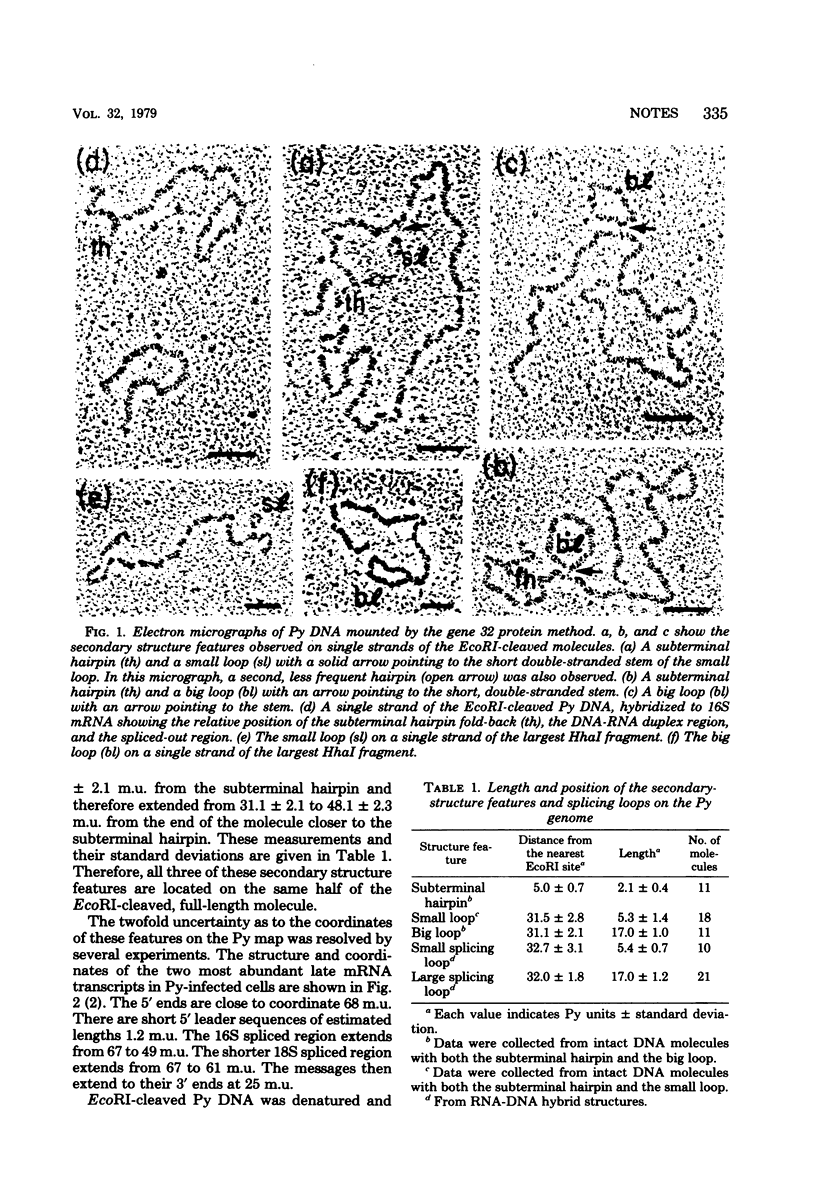
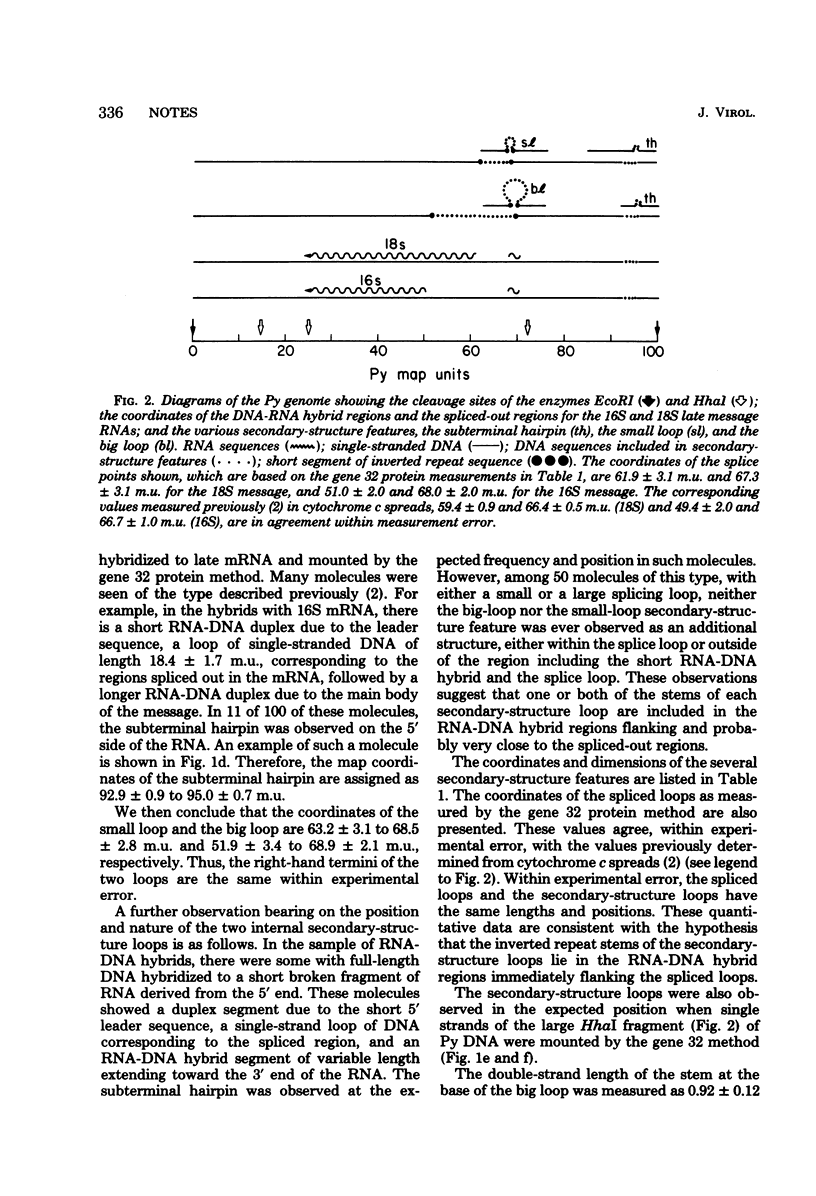
Three reproducible secondary-structure features were observed on single strands of polyoma virus DNA mounted for electron microscopy by the T4 gene 32 protein technique: (i) a hairpin fold-back extending from 92.9 +/- 0.8 to 95.0 +/- 0.7 map units; (ii) a small loop extending from 63.2 +/- 3.1 to 68.5 +/- 2.8 map units; and (iii) a big loop extending from 51.9 +/- 2.3 to 68.9 +/- 2.1 map units. Both loops are bounded by inverted repeat stems of length 40 +/- 20 base pairs. The stem sequences around 68.5 and 68.9 of the large and small loops overlap, either partially or completely. Several lines of evidence indicate that the inverted repeat stems of the two secondary-structure loops lie in the regions of polyoma virus DNA flanking and probably very close to the sequences that are spliced out in the formation of the late 16S and 18S messages, whereas the hairpin fold-back appears to map at a splicing point of an early message. These structures may therefore be important for the processing of the primary transcripts to form the early and late messages.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Hsu M. T., Jelinek W. R. Mapping of inverted repeated DNA sequences within the genome of simian virus 40. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1631–1634. doi: 10.1073/pnas.74.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor H., Wu M., Baran N., Davidson N. Electron microscopic mapping of RNA transcribed from the late region of polyoma virus DNA. J Virol. 1979 Oct;32(1):293–303. doi: 10.1128/jvi.32.1.293-303.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C. K., Hearst J. E. Mapping of sequences with 2-fold symmetry on the simian virus 40 genome: a photochemical crosslinking approach. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1363–1367. doi: 10.1073/pnas.74.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Davidson N. Use of gene 32 protein staining of single-strand polynucleotides for gene mapping by electron microscopy: application to the phi80d3ilvsu+7 system. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4506–4510. doi: 10.1073/pnas.72.11.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Roberts R. J., Davidson N. Structure of the inverted terminal repetition of adenovirus type 2 DNA. J Virol. 1977 Feb;21(2):766–777. doi: 10.1128/jvi.21.2.766-777.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Steitz J. A. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3593–3597. doi: 10.1073/pnas.75.8.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]



