Abstract
Efficient maintenance of chromatin structure during passage of RNA polymerase II (Pol II) is critical for cell survival and functioning. Moderate-level transcription of eukaryotic genes by Pol II is accompanied by nucleosome survival, extensive exchange of histones H2A/H2B and minimal exchange of histones H3/H4. Complementary in vitro studies have shown that transcription through chromatin by single Pol II complexes is uniquely coupled with nucleosome survival via formation of a small intranucleosomal DNA loop (Ø-loop) containing the transcribing enzyme. In contrast, transient displacement and exchange of all core histones are observed during intense transcription. Indeed, multiple transcribing Pol II complexes can efficiently overcome the high nucleosomal barrier and displace the entire histone octamer in vitro. Thus, various Pol II complexes can remodel chromatin to different extents. The mechanisms of nucleosome survival and displacement during transcription and the role of DNA-histone interactions and various factors during this process are discussed.
Keywords: chromatin, nucleosome, histones, exchange, RNA polymerase II, transcript elongation
1. Introduction
Gene regulation [1], cell survival [2] and aging [3] in eukaryotes critically depend on efficient maintenance of chromatin structure during passage of RNA polymerase II (Pol II). On many eukaryotic genes, Pol II pauses after transcribing the initial 50–100 bp of the template [4,5,6]. This pause frequently occurs after encountering one of the nucleosomes positioned within the early transcribed region ([7], but also see [8]). Once pol II overcomes the +1 nucleosomal barrier, it can continue transcript elongation over hundreds of kb of predominantly nucleosomal template at a high rate (3–4 kb/min [9]), similar with the rate observed on histone-free DNA in vitro [10,11]. Nucleosomes are not lost from thousands of moderately active genes, although they become more delocalized and less dense at higher transcription rates [6,12,13]. On moderately active genes, histones H2A/H2B are displaced/exchanged at a much higher rate than H3/H4 histones [14,15,16,17,18,19], suggesting that only H2A/H2B are displaced during every round of transcription [20]. At the same time, during intense transcription all core histones are displaced/exchanged at the transcribed regions [14,15,16,17,18,19,21,22]. Thus transient transcription-dependent disruption and recovery of DNA-histone interactions during moderate-level and active transcription occurs through two different mechanisms.
In addition to disruption of DNA-histone interactions, Pol II transcription induces transient perturbation of histone-histone interactions within the nucleosomes in vivo. Non-transcribed, intact nucleosomes do not have reactive SH groups. In contrast, histone H3 SH groups on transcribed genes are accessible to external probes [23,24,25]; the accessibility is higher in nucleosomes containing acetylated histones [26] and closely correlates with ongoing transcription [23,24]. The reactive nucleosomes are predominantly U-shaped [27], suggesting that nucleosomal DNA is unfolded with the associated histones. In agreement with these observations, transcription by Pol II induces appearance of “half-nucleosomal” cleavage periodicity by DNase I in vivo, consistent with formation of unfolded nucleosomes [28].
2. Changes in nucleosome structure relevant for transcription through chromatin
During transcription through chromatin, Pol II forms a tight, highly asymmetric “clump” around DNA [29] that rotates around the helix every 10 bp, making 15 rotations as the enzyme progresses through a nucleosome. Inevitably, all intranucleosomal DNA-histone interactions must be at least transiently disrupted during transcription. In fact, some DNA-histone interactions most likely are disrupted and re-formed several times during transcription through a nucleosome [30,31]. Mapping of intranucleosomal DNA-histone interactions by mechanically “unzipping” DNA from the histone octamer in single nucleosomes [32] identified three regions of strong interactions (I, I′ and II, Fig. 1) that significantly affect the rate and efficiency of Pol II progression through a nucleosome ([33] and see below). An additional DNA region that determines overall affinity of DNA-histone interactions in a sequence-specific way and dictates overall height of the nucleosomal barrier to transcription was identified in transcriptional experiments (region III or polar barrier sequence, PBS; Fig. 1 [34,35]).
Fig. 1.
Positions of nucleosome-specific pauses and strong intranucleosomal DNA-histone interactions within the nucleosome structure. A. The structure of the nucleosome core [104]. The backbone of nucleosomal DNA is shown in white and gray, the H3/H4 tetramer in purple, and the H2A/H2B dimers in green (promoter-distal D-dimer) and blue. B. The path of nucleosomal DNA. The locations of ten base pair intervals (numbered from the lower end of the nucleosome, in direction of Pol II progression (dotted arrow) along nucleosomal DNA) are indicated. Positions of strong, H2A/H2B- and H3/H4-specific DNA-histone interactions I, I′, II [32] and III (polar barrier sequence, PBS [34,35]) are indicated by green/blue and purple lanes, respectively. The regions of related strong nucleosome-specific Pol II pausing [33] are shown by blue and purple arrows.
It has been suggested that Pol II functions as a ratchet. In this model, the rate and efficiency of Pol II movement along nucleosomal DNA depends in part on fluctuations in nucleosome structure [36], most likely uncoiling/recoiling of DNA from/on the histone octamer, respectively (Fig. 2, 1 -> 2′ transition). The rate of transient DNA unwrapping from the octamer is high: “pulses” of unwrapping for 10–50 ms occur with average intervals of 250 ms [37]. Since the time required for bond formation by Pol II in vitro in the absence of additional factors is comparable with dwelling time of the open “window of opportunity” [11], various elongation factors (which increase the rate of bond formation [38]) and factors stabilizing the uncoiled DNA state could greatly affect the rate of transcription through chromatin (see [39] for review). On the other hand, some experiments suggest that the force required to uncoil nucleosomal DNA (10–20 pN [40,41]) is comparable with or lower than the force generated by E. coli RNA polymerase (>25 pN [42]) during transcript elongation. Thus Pol II may have sufficient power to peel DNA away from the octamer surface regardless of DNA “breathing” on the surface of the octamer.
Fig. 2.
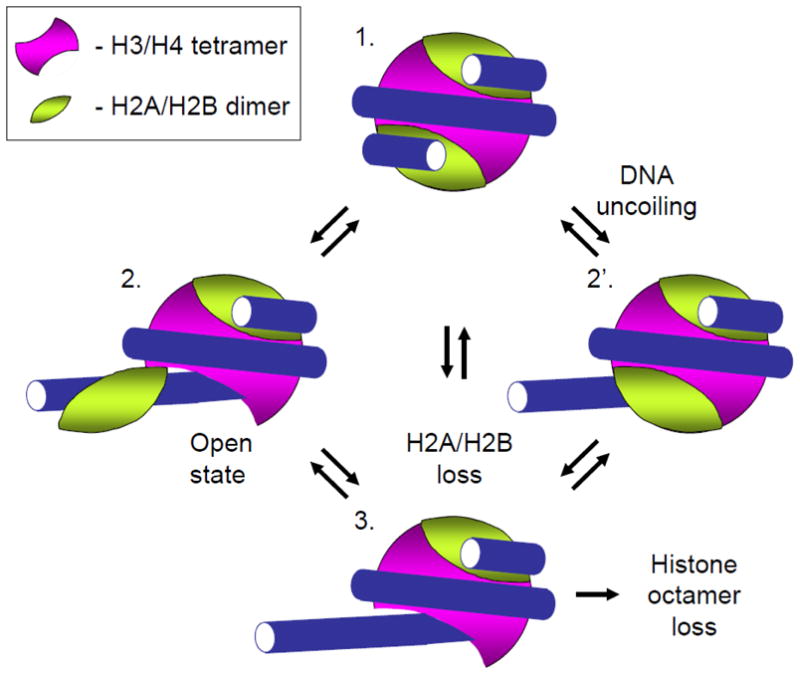
Nucleosome structural changes relevant for transcription through chromatin. The nucleosome structure (1) can be perturbed due to spontaneous DNA uncoiling from the intact octamer (complex 2′ [37]), that increases the accessibility of DNA to protein binding, or by uncoiling of DNA together with the associated H2A/H2B dimer (complex 2 [43]). Formation of complexes 2 and 2′ facilitates dissociation of the H2A/H2B dimer from the nucleosome (complex 3) and dissociation of the entire octamer from DNA that occur during transcription through a nucleosome by Pol II [50].
Another transition in nucleosome structure that likely affects the outcome of transcription through chromatin is spontaneous uncoiling of the ends of nucleosomal DNA with the associated H2A/H2B dimer (Fig. 2, 1 -> 2 transition) that occurs on 0.2–3% of templates [43]. A more dramatic unfolding of the entire octamer together with the associated DNA and its refolding in a nucleosome-like structure that contains right-handed supercoiled DNA (“reversome”) is induced by positive DNA supercoiling [44]. The ability of the histone octamer to unfold and refold together with the associated DNA suggests that nucleosome structure could be adapted to allow traversal during transcription without complete loss of histones from the DNA.
3. Mechanism of single-round transcription through a nucleosome by Pol II
In the majority of recent experiments in vitro dedicated to analysis of the mechanism of transcription through chromatin, mononucleosomal DNA templates and authentic elongation complexes (either promoter-initiated or assembled from synthetic oligonucleotides and purified RNAPses [45]) were employed. The assembled and promoter-initiated elongation complexes (ECs) transcribe chromatin using very similar mechanisms [33,46]. Importantly, these experimental systems recapitulate many important properties of chromatin transcribed by Pol II in vivo (see below). Alternatively, chromatin can be transcribed in vitro using end-initiated elongation complexes [47]. However, it should be noted that after end-initiation the elongation complexes transcribe through chromatin using a different mechanism [48]. Therefore, the data on transcription through chromatin obtained using end-initiated elongation complexes should be reproduced using properly assembled ECs.
The Pol II-specific mechanism of transcription through chromatin in vitro is characterized by a high nucleosomal barrier to transcription [33,49,50], and by displacement of a single H2A/H2B dimer [50,51] that matches the apparent effect of Pol II passage in vivo [17,18,52]. After a single-round of transcription through a nucleosome, the subnucleosome (DNA-bound histone hexamer formed upon release of H2A/H2B dimer from the octamer) survives Pol II passage and remains at the original position on DNA [50]. The Pol II-type mechanism is conserved from yeast to human [33]. A considerably different, Pol III-type mechanism, involves nucleosome translocation from in front of the transcribing enzyme to behind it [35,53,54].
The histone hexamer that survives Pol II transcription never leaves DNA during progression of the enzyme [55]. How can transcription through a nucleosome occur without relocation of the hexamer [50] or dissociation of the histones? The mechanism of this remarkable process has been solved recently (Fig. 3). As Pol II approaches and enters the nucleosome (complex 1), it partially uncoils nucleosomal DNA, primarily behind the enzyme (complex 2). As the enzyme proceeds through the +45 region of nucleosomal DNA (localized ~45 bp from the promoter-proximal boundary of the nucleosome), Pol II encounters areas of strong DNA-histone interactions (regions II and III, Fig. 1). Apparently, uncoiling of the regions II and III of nucleosomal DNA is impossible without formation of a small intranucleosomal DNA loop at the +49 position (the Ø-loop, EC+49, complex 3). Formation of the Ø-loop occurs slowly, causing nucleosome-specific pausing of Pol II at the +45 region (Fig. 1). As the DNA behind Pol II is recoiled on the surface of the octamer, the DNA in front of the complex becomes sterically displaced by Pol II and is partially uncoiled from the octamer (complex 4), allowing further transcription by Pol II (through selective disruption of the downstream DNA-histone interactions) and nucleosome recovery (through sequential re-formation of the original DNA-histone interactions upstream of Pol II) upon further movement of the enzyme (complex 5). During progression of Pol II from the EC+49 to the end of the template an H2A/H2B dimer is displaced from the octamer, thereby forming a hexasome [35,50]. On a fraction of the templates (5–50%) the entire octamer is displaced by Pol II in vitro [36,38,50].
Fig. 3.

Mechanism of single-round transcription through a nucleosome by Pol II in vitro (modified from [35]). Primary pathway (likely operates in vivo): as Pol II approaches a nucleosome (complex 1), (smaller arrows indicate direction of transcription), upstream nucleosomal DNA is partially uncoiled from the octamer (2). As Poll II proceeds, it encounters strong DNA-histone interactions and pauses immediately upstream of the position +49. As DNA re-coils on the octamer behind the enzyme, a transient Ø-loop is formed at position +49 (3), and Pol II displaces the promoter-distal end of nucleosomal DNA from the octamer surface (4, see Fig. 4), allowing its own further transcription. Strong sequence-specific interactions of H3/H4 histones with the PBS (polar barrier sequence) DNA region (complex 3) can prevent uncoiling of the downstream DNA and cause Pol II arrest. Otherwise Pol II continues transcription and the DNA-histone contacts upstream of the enzyme serve as an anchor to recover the nucleosome behind Pol II (5). An H2A/H2B dimer is lost during transcription (6); alternatively, 5–50% templates lose the entire octamer (complex 6′).
During transcription through a nucleosome by Pol II small intranucleosomal DNA loops containing transcribing enzyme are formed at least twice, at the positions +39 and +49 [35,56]. Formation of the EC+49 complex (the Ø-loop) is critical both for transcription through a nucleosome and hexasome survival [35]. Atomic-resolution modeling of the EC+49 structure suggested that formation of such structures can occur only in one rotational orientation of Pol II, when the bulk of the Pol II molecule faces into solution and there are no steric clashes with core histones [35]. Formation of the complex is facilitated by the 90° DNA bend present in the EC and facing the octamer surface. DNA-histone contacts with ~20-bp DNA region behind the EC are disrupted and replaced by electrostatic Pol II-histone interactions stabilizing the complex. Importantly, formation of EC+49 is incompatible with the presence of the two superhelical coils of nucleosomal DNA; therefore after formation of the EC+49 Pol II likely displaces one DNA coil (shown schematically in Fig. 4). Finally, formation of small intranucleosomal, topologically closed DNA loops constrains rotation of Pol II around DNA and therefore could result in accumulation of unconstrained intranucleosomal DNA supercoiling (Fig. 4). This, in turn, could facilitate unfolding and survival of the histone octamer during transcription.
Fig. 4.

Intranucleosomal DNA Ø-loop containing transcribing Pol II. Small intranucleosomal DNA loops containing transcribing enzyme likely form several times during transcription through a nucleosome [31,35]. Formation of the such loops results in disruption of some DNA-histone interactions, formation of new histone-polymerase interactions, steric interference with the next superhelical coil of nucleosomal DNA (as in Ø-loop [35]) and displacement of ~50-bp DNA region (shown by dashed lines). Formation of small intranucleosomal, topologically closed DNA loops constrains rotation of Pol II around DNA and therefore could result in accumulation of unconstrained DNA supercoiling (positive in front and negative behind the enzyme, respectively [105]). Positive unconstrained DNA supercoiling, in turn, could induce reversible unfolding of nucleosomal DNA that remains associated with core histones [44]. Note: Pol II is not drawn to scale.
4. Overcoming the nucleosomal barrier to transcription
The regions of strong nucleosome-specific pausing (the +15 and +45 regions [33]) are localized 10–20 bp upstream of strong DNA-histone interactions at the regions I and II, respectively (Fig. 1B [32,57]), suggesting that DNA uncoiling from the octamer occurs less frequently within these regions. The shorter dwelling time of the open “window of opportunity”, in turn, causes longer Pol II pausing. Indeed, several single Sin mutations in H3/H4 histones that destabilize DNA-histone interactions in region II [58] selectively affect +45 pausing [56]. Pol II pausing at the +15 and +45 regions is largely sequence-independent as it was detected in nucleosomes formed on various DNA sequences [33,34,59], consistent with the presence of strong, sequence-independent DNA-histone interactions at the regions I and II [32]. Remarkably, the region I′ of strong DNA-histone interactions localized within the promoter-distal half of nucleosomal DNA (Fig. 1B) does not cause any significant Pol II pausing, most likely because during transcription through this region DNA is already uncoiled from the histone octamer in front of Pol II (Fig. 3, complex 5).
More surprisingly, recent functional in vitro studies have identified a novel region of nucleosomal DNA (region III or PBS sequence, Fig. 1B) that affects nucleosome positioning, overall DNA-histone affinity and the +45 pausing in a sequence-specific way [35]. Since the PBS region is positioned far (>50 bp) downstream of the Pol II paused at the +45 region (Fig. 1) it likely affects uncoiling of the promoter-distal end of nucleosomal DNA (Fig. 3 [35]). In agrement with this proposal, mutations in the PBS sequence that decrease DNA-histone affinity allow more efficient transcription through the +45 region by Pol II and dictate the overall height of the nucleosomal barrier to transcription [35]. In several analyzed nucleosomes the PBS-like sequences are localized asymmetrically relative to the nucleosomal dyad [35], dictating formation of a highly polar nucleosomal barrier [33]. The identification of an intranucleosomal PBS region suggests the possibility of sequence-specific regulation of the nucleosomal barrier to transcription. This could be particularly important in the case of nucleosomes encountered by Pol II during the earliest stages of transcript elongation [7].
In summary, the nucleosomal barrier to Pol II is largely dictated by the strength of critical DNA-histone interactions at the intranucleosomal DNA regions I, II and III (PBS) that modulate DNA uncoiling from the octamer and DNA accessibility to transcribing Pol II. Therefore, the intricate sequence of partial DNA uncoiling/recoiling on the surface of the octamer (Fig. 3) largely dictates the position and the strength of the nucleosomal pausing. Once Pol II pauses, the duration of pausing can be further modulated by the probability and extent of Pol II “backtracking” [59].
5. Histone survival and displacement during transcription
Since even transient complete histone displacement from DNA leads to fast and irreversible nucleosome disruption in the absence of histone chaperones [60], the efficiency of nucleosome survival during transcription in vitro is largely determined by the ability of core histones to remain associated with DNA. Remarkably, ~50–95% of nucleosomes survive single-round transcription in vitro by purified Pol II [36,38,50]; nucleosomes remaining on the template lose only one H2A/H2B dimer and are converted into a histone hexamer bound at the original position on DNA [50]. More efficient transcription and recovery of the hexamer in vitro occur in the presence of ATP-dependent chromatin remodeler RSC and the histone chaperone NAP1 [61].
Most likely, loss/survival of histones occurs during the critical transition after Pol II pausing at the +45 region [35]. This transition involves coordinated re-coiling of upstream DNA (partially displaced by Pol II) on the octamer to form the Ø-loop (allowing nucleosome survival), and uncoiling of downstream DNA (allowing further transcription, Fig. 3). Therefore it has been proposed that transcription past the +45 pausing region is coupled with nucleosome survival [35]. Indeed, the Sin mutations that destabilize DNA-histone interactions at the region II [58] and decrease the efficiency of +45 pausing also promoted more efficient displacement of all histones [56]. Furthermore, the absence of the promoter-distal D-dimer in the nucleosome results in coordinated relief of the +45 pausing [35] and the loss of histones [62]. The derivative concept suggests that inefficient formation or resolution of the Ø-loop can cause histone loss or strong nucleosome-specific pausing, respectively.
In particular, the proximal H2A/H2B P-dimer is likely essential for both nucleosome survival and efficient transcription because removal of the P-dimer eliminates a DNA-binding site upstream of the elongation complex paused at the +45 region (Fig. 3) and therefore is expected to considerably destabilize the Ø-loop. Indeed, displacement of the P-dimer leads to arrest of Pol II within the nucleosome [35]. In contrast, removal of the distal D-dimer results in release of the promoter-distal end of nucleosomal DNA into solution, facilitates formation of the Ø-loop and transcription through the nucleosome [35]. Thus two nearly identical H2A/H2B dimers play different roles during transcription through a nucleosome and their loss is expected to have different consequences for transcription in vivo (see below).
The effect of the absence of the promoter-proximal H2A/H2B P-dimer is particularly striking. One might imagine intuitively that removing histones from the nucleosome would necessarily facilitate traversal by pol II, but in fact the opposite result is observed: in hexasomes lacking the P-dimer, pol II arrest increases [35]. The central point is that without the ability to form the critical zero-loop intermediate, the critical steric clash of Pol II with downstream histone-DNA contacts does not occur, downstream DNA is not driven away from the octamer surface and continued traversal by Pol II is blocked. This result is in strong contrast to just-discussed effect of removing the D-dimer on transcription. Thus two nearly identical H2A/H2B dimers play different roles during transcription through a nucleosome and their loss is expected to have different consequences for transcription in vivo [35]. Since both H2AH2B dimers are exchanged during Pol II transcription in vivo [52], the P-dimer must be immediately replaced to avoid arrest of transcribing Pol II.
Transcription is accompanied by formation of “open” intermediates, with large regions of nucleosomal DNA displaced by Pol II and extensive solvent-accessible surface of the histone hexamer (Fig. 3). The open octamer surface may bind to any DNA available nearby, leading to nucleosome translocation in cis (to the same DNA molecule) or in trans. Indeed, nucleosome translocation was observed during transcription by Pol II [36,55,63], provided that an extensive histone-free DNA region (≫150 bp) is available upstream of transcribed nucleosome [55]. However nucleosomes formed on shorter (250–300 bp) templates remain at the original position on the DNA during Pol II transcription [50,55]. In contrast, constitutive nucleosome translocation was observed during transcription of even very short (160–200 bp) templates by bacteriophage SP6 RNA polymerase (RNAP) that uses the Pol III-type mechanism of transcription through chromatin [64]. It has been proposed that the different nucleosome fates during transcription by the Pol II- and Pol III-type mechanisms (nucleosome survival vs. translocation, respectively) could be explained by the lower efficiency of formation of the “survival” intermediate (the Ø-loop) in the latter case [31,35]. Thus the fate of nucleosomes on transcription depends on the nature of transcribing enzyme and the length of the templates.
The fate of nucleosomes on transcription in vitro also depends on the rate of transcription through chromatin and the number of transcribing Pol II. An increase in transcription rate from 0.5 to 1 bp s−1 favors displacement of the octamer from DNA [63], presumably due to more efficient uncoiling of nucleosomal DNA from the octamer by the faster enzyme.
The effect of multiple transcribing Pol II complexes on the fate of nucleosomes on transcription depends on the spacing between the enzymes. Transcription of nucleosomes by closely spaced, tandem Pol II complexes in vitro occurs more efficiently than by single complexes, likely due to less efficient backtracking of the leading complex [62,65]. Transcription by tandem complexes and by single complexes in vitro produce similar outcomes (hexasome survival with ~50% efficiency) [50,62]. Since tandem polymerases efficiently overcome the nucleosome barrier and maintain histone association with DNA, a pair of Pol II complexes transcribing in tandem could serve as a “pioneering” complex, performing initial remodeling of gene body during the first round of transcription on an activated gene (see below). Alternatively, single transcribing complexes could be paused at the early transcribed region of a transcribed gene, waiting for a second Pol II complex to arrive and allow further transcription [62].
In contrast, encounter of a previously transcribed nucleosome (converted to hexasome) by the trailing Pol II complex in vitro results in displacement of all histones, most likely due to formation of less stable Ø-loop [62]. The data argue that histone displacement/exchange observed in vivo during intense transcription [14,15,16,17,18,19,21,22] occurs in part due to close spacing between the transcribing complexes.
In summary, numerous studies show that the “minimal” experimental systems used in transcriptional experiments described above and typically containing mononucleosomes formed on DNA fragments recapitulate many important properties of Pol II-transcribed chromatin. In particular, survival of histones H3/H4 and displacement of H2A/H2B observed on moderately transcribed genes were detected during single-round transcription in vitro [50,62]. Even fine effects of single Sin mutations in histones H3/H4 on nucleosome survival during transcription were recapitulated [56], suggesting that this experimental system can be used for further dissection of the underlying mechanism. Finally, the displacement of all histones observed during intense transcription in vivo occurs in the “minimal” system during transcription by multiple Pol II complexes [62].
6. Transcription through chromatin in vivo
The model proposed for transcription through chromatin (Fig. 3) explains numerous properties of chromatin transcribed in vivo (Fig. 5) and makes several important predictions. Thus on moderately Pol II-transcribed genes exchange of H3/H4 histones occurs at least 20-fold slower than that of H2A/H2B (see Introduction). The observed hexasome survival at the original position during transcription through a nucleosome in vitro [50,55] and structures of the intermediates formed during this process [35] suggest that H3/H4 are not exchanged in vivo because they are never fully displaced from the DNA during Ø-loop-mediated nucleosome survival. Since the majority of eukaryotic genes are transcribed at moderate levels, transcription-dependent exchange of bulk H3/H4 histones is considerably slower than exchange of H2A/H2B [66].
Fig. 5.
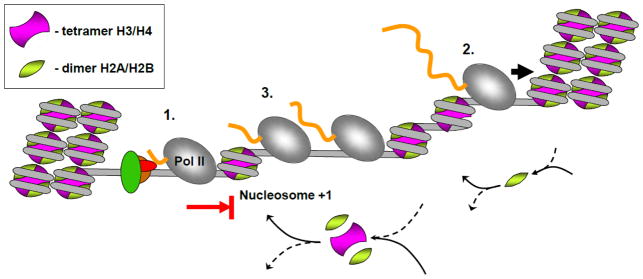
Mechanism of transcription through chromatin by Pol II. After initiation of transcription by Pol II, the enzyme can be paused within the first nucleosome positioned on the gene (1). After overcoming the initial nucleosomal barrier, at a low Pol II density transcription is accompanied by transient displacement/exchange of H2A/H2B dimer(s); the nucleosome structure is recovered before arrival of the next Pol II complex (2). This Pol II-specific mechanism allows survival of H3/H4 histones and their modifications on DNA during transcription. At a higher density Pol II complexes encounter hexasomes that are missing H2A/H2B dimer(s) (3). In this case an unstable intermediate with a smaller number of DNA-histone contacts is formed, resulting in eviction of the histone hexamer from DNA [62]; therefore all core histones are evicted and exchanged (3). Thus proximity of Pol II complexes to each other dictates the fate of nucleosomes on transcription.
Nucleosome survival at the original position during Pol II transcription is likely to be essential property of the mechanism. During transcription of a eukaryotic gene using the alternative, Pol III-type mechanism, extensive displacement/exchange of all core histones would occur because after several rounds of transcription, the histone octamer would be transferred upstream and ultimately occlude the previously nucleosome-free promoter (or will be removed by remodelers) [35,53]. Since the H3/H4 histones contain the majority of the sites for post-translational modifications [67], the Pol II-type mechanism could be specifically adopted to allow maintenance of epigenetic marks during transcription of the entire eukaryotic genome that occurs at least once during the cell cycle [68,69]. The importance of nucleosome survival at the original positions during Pol II transcription suggests that the changes of nucleosome positioning observed during moderate-level transcription are not induced by transcription per se, but rather by ATP-dependent remodelers associated with the transcribed genes [8,70].
The high rate of Pol II progression through chromatin in vivo [9] suggests that there are mechanisms facilitating transcription through the high nucleosomal barrier observed in vitro. Indeed, transcribed genes are associated with numerous factors including ATP-dependent chromatin remodelers, histone modifications and histone chaperones (reviewed in [71]). The associated histone modifications include acetylation, methylation, and ubiquitylation; the enzymes introducing these modifications are often associated with elongating Pol II [72,73,74,75,76]. Most likely, histone acetylation facilitates transcription through chromatin and transcription-dependent histone exchange, while methylation results in recruitment of histone deacetylases that reestablish the inactive chromatin state after transcription [71,77,78]. In particular, histone tail-modifying acetyltransferases (HATs) PCAF and Elp3 specifically interact with the elongating, phosphorylated form of Pol II [72,79], and NuA3 interacts with elongation factor FACT both in vivo and in vitro [80]. In addition to modifications of histone tails, acetylation of H3K56 (within the globular domain) is associated with transcribing Pol II [81]. Histone chaperones FACT and Asf1 are associated with transcribed genes and facilitate nucleosome disassembly and reassembly during transcript elongation [82,83]. FACT has higher affinity to H2A/H2B dimers than to H3/H4 tetramers [51,84,85] and facilitates transcription through chromatin in vitro [33,51,86,87]. H2B ubiquitylation and the presence of chromatin remodeler Chd1 are required for Spt16 recruitment to the transcribed genes, efficient Pol II transcription and reassembly of nucleosomes [88,89,90].
As the rate of transcription is increased, the distance between transcribing Pol II complexes becomes shorter and displacement/exchange of all histones are observed (see Introduction). The results of the in vitro experiments with multiple Pol II complexes transcribing nucleosomal templates suggest that trailing Pol II complexes may encounter the hexasome formed after previous round of transcription, before the H2A/H2B dimer re-binds to the hexasome (Fig. 5). In this case all core histones are displaced from DNA in vitro [62]. This model explains both the lack of exchange of H3/H4 histones at moderate transcription and their exchange on actively transcribed genes. Furthermore, the model provides an explanation for the gradual dependence of the rates of histone displacement/exchange on the efficiency of transcription [17,91,92,93] because it suggests that the extent of histone displacement would be inversely proportional to average distance between transcribing Pol II complexes. Note that on some highly inducible genes partial transcription-independent loss of all core histones has been observed [94,95].
Paused Pol II complexes are observed immediately downstream of thousands of human, yeast and Drosophila promoters, often close to the +1 nucleosome [4,5,6]. In the case of human genes the large majority of transcripts are shorter than ~100 nt, suggesting that the early stage of transcript elongation constitutes a major rate-limiting step in gene regulation in vivo. Given the typical location for these stalled polymerases (the active center is localized 30–50 bp downstream of the start site [96]), it is reasonable to suggest that induction of transcription on such genes would lead to accumulation of two closely spaced Pol II elongation complexes. Such tandem polymerases (Pol II “dimer”) would overcome the +1 nucleosome or other elongation blocks (e.g. DNA-bound proteins) more efficiently than a single Pol II complex [62,65,97,98] and would displace only H2A/H2B histones from DNA [62]. These results argue that the Pol II “dimer” could serve as a “pioneering” Pol II. Indeed, nucleosomes present a strong barrier to single complexes of transcribing Pol II in vitro [33,49,50] and perhaps on some inactive genes in vivo [6,99]. According to this concept, after gene activation the “pioneering” Pol II dimer would be able to overcome the +1 barrier and efficiently transcribe the entire gene. During the initial round of transcription chromatin structure is perturbed and histones become more accessible to covalent modifications characteristic for transcribed genes [100,101]. Such histone modifications could then serve as a transcriptional “memory” [77,102]. Furthermore, it has been shown that intergenic transcription is required for chromatin remodeling and subsequent transcriptional activation of large domains of chromatin in development [103].
The data described above suggest that Pol II transcription in vivo can lead to several quite different outcomes (Fig. 5). After initiation of transcription, Pol II can be paused within one of nucleosomes positioned on the early transcribed region of a gene (part 1, Fig. 5). The height of the nucleosomal barrier to transcription could be dependent on the underlying DNA sequence [33,35]. After overcoming the initial nucleosomal barrier (perhaps with help of second Pol II complex [62,65], ATP-dependent chromatin remodelers [34] or specific histone chaperones like FACT [51]), Pol II can efficiently proceed through chromatin. At low- to moderate-level transcription by single, widely spaced complexes progression of Pol II is characterized by displacement/exchange of only H2A/H2B dimer(s) [14,15,16,17,18,19] and hexasome survival, likely mediated through formation of small intranucleosomal DNA loops (part 2 of Fig. 5 [35]). During moderate transcription, the dimers re-bind to the hexasomes before the next Pol II complex arrives; therefore each complex encounters complete nucleosomes and there is no functional cooperation between the complexes. This is consistent with the observation that the loss of certain histone chaperones which bind H2A/H2B dimer also leads to loss of nucleosomes within transcription units [1]. This Pol II-specific mechanism allows survival of H3/H4 histones and their modifications on DNA during transcription. As the transcription rate is increased, the distance between transcribing Pol II complexes becomes shorter, and trailing complexes of Pol II may encounter the hexasome formed after previous round of transcription, before the H2A/H2B dimer re-binds to the hexasome (part 3 of Fig. 5). In this case an unstable intermediate with a smaller number of DNA-histone contacts is formed, resulting in eviction of the histone hexamer from DNA in vitro [62]; therefore all core histones are evicted/exchanged in vivo [14,15,16,17,18,19,21,22].
Highlights.
Chromatin recovery during transcription is essential for cell viability
Chromatin recovery occurs differently during moderate and intense transcription
Original histones remain on DNA during moderate-level transcription
Histone survival during moderate transcription involves DNA looping
Multiple transcribing complexes displace histones during intense transcription
Acknowledgments
This work was supported by NIH GM58650 and Government of the Russian Federation (order #220) grants to V.M.S. and NSF 0743298 grant to D.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008;6:e277. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martens JA, Wu PY, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005;19:2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. Elevated histone expression promotes life span extension. Mol Cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, Gilmour DS, Albert I, Pugh BF. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- 11.Izban MG, Luse DS. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- 12.Koerber RT, Rhee HS, Jiang C, Pugh BF. Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol Cell. 2009;35:889–902. doi: 10.1016/j.molcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wirbelauer C, Bell O, Schubeler D. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 2005;19:1761–1766. doi: 10.1101/gad.347705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz BE, Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiriet C, Hayes JJ. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 2005;19:677–682. doi: 10.1101/gad.1265205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 18.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 20.Kulaeva OI, Gaykalova DA, Studitsky VM. Transcription through chromatin by RNA polymerase II: Histone displacement and exchange. Mutat Res. 2007;618:116–129. doi: 10.1016/j.mrfmmm.2006.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katan-Khaykovich Y, Struhl K. Splitting of H3–H4 tetramers at transcriptionally active genes undergoing dynamic histone exchange. Proc Natl Acad Sci USA. 2011;108:1296–1301. doi: 10.1073/pnas.1018308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen TA, Allfrey VG. Rapid and reversible changes in nucleosome structure accompany the activation, repression, and superinduction of murine fibroblast protooncogenes c-fos and c-myc. Proc Natl Acad Sci USA. 1987;84:5252–5256. doi: 10.1073/pnas.84.15.5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen TA, Smith MM, Le SY, Sternglanz R, Allfrey VG. Nucleosome fractionation by mercury affinity chromatography. Contrasting distribution of transcriptionally active DNA sequences and acetylated histones in nucleosome fractions of wild-type yeast cells and cells expressing a histone H3 gene altered to encode a cysteine 110 residue. J Biol Chem. 1991;266:6489–6498. [PubMed] [Google Scholar]
- 25.Prior CP, Cantor CR, Johnson EM, Littau VC, Allfrey VG. Reversible changes in nucleosome structure and histone H3 accessibility in transcriptionally active and inactive states of rDNA chromatin. Cell. 1983;34:1033–1042. doi: 10.1016/0092-8674(83)90561-5. [DOI] [PubMed] [Google Scholar]
- 26.Walia H, Chen HY, Sun JM, Holth LT, Davie JR. Histone acetylation is required to maintain the unfolded nucleosome structure associated with transcribing DNA. J Biol Chem. 1998;273:14516–14522. doi: 10.1074/jbc.273.23.14516. [DOI] [PubMed] [Google Scholar]
- 27.Bazett-Jones DP, Mendez E, Czarnota GJ, Ottensmeyer FP, Allfrey VG. Visualization and analysis of unfolded nucleosomes associated with transcribing chromatin. Nucleic Acids Res. 1996;24:321–329. doi: 10.1093/nar/24.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee MS, Garrard WT. Transcription-induced nucleosome ‘splitting’: an underlying structure for DNase I sensitive chromatin. EMBO J. 1991;10:607–615. doi: 10.1002/j.1460-2075.1991.tb07988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 30.Studitsky VM, Walter W, Kireeva M, Kashlev M, Felsenfeld G. Chromatin remodeling by RNA polymerases. Trends Biochem Sci. 2004;29:127–135. doi: 10.1016/j.tibs.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Bednar J, Studitsky VM, Grigoryev SA, Felsenfeld G, Woodcock CL. The nature of the nucleosomal barrier to transcription: direct observation of paused intermediates by electron cryomicroscopy. Mol Cell. 1999;4:377–386. doi: 10.1016/s1097-2765(00)80339-1. [DOI] [PubMed] [Google Scholar]
- 32.Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat Struct Mol Biol. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 34.Gaykalova DA, Nagarajavel V, Bondarenko VA, Bartholomew B, Clark DJ, Studitsky VM. A polar barrier to transcription can be circumvented by remodeler-induced nucleosome translocation. Nucleic Acids Res. 2011;39:3520–3528. doi: 10.1093/nar/gkq1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, Studitsky VM. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat Struct Mol Biol. 2009;16:1272–1278. doi: 10.1038/nsmb.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325:626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 38.Luse DS, Spangler LC, Ujvari A. Efficient and rapid nucleosome traversal by RNA polymerase II depends on a combination of transcript elongation factors. J Biol Chem. 2011;286:6040–6048. doi: 10.1074/jbc.M110.174722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luse DS, Studitsky VM. The mechanism of nucleosome traversal by RNA polymerase II: roles for template uncoiling and transcript elongation factors. RNA Biol. 2011;8:581–585. doi: 10.4161/rna.8.4.15389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brower-Toland B, Wacker DA, Fulbright RM, Lis JT, Kraus WL, Wang MD. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J Mol Biol. 2005;346:135–146. doi: 10.1016/j.jmb.2004.11.056. [DOI] [PubMed] [Google Scholar]
- 41.Mihardja S, Spakowitz AJ, Zhang Y, Bustamante C. Effect of force on mononucleosomal dynamics. Proc Natl Acad Sci USA. 2006;103:15871–15876. doi: 10.1073/pnas.0607526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 43.Bohm V, Hieb AR, Andrews AJ, Gansen A, Rocker A, Toth K, Luger K, Langowski J. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2011;39:3093–3102. doi: 10.1093/nar/gkq1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bancaud A, Wagner G, Conde ESN, Lavelle C, Wong H, Mozziconacci J, Barbi M, Sivolob A, Le Cam E, Mouawad L, Viovy JL, Victor JM, Prunell A. Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol Cell. 2007;27:135–147. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 45.Kireeva ML, Komissarova N, Waugh DS, Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J Biol Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- 46.Walter W, Kireeva ML, Studitsky VM, Kashlev M. Bacterial polymerase and yeast polymerase II use similar mechanisms for transcription through nucleosomes. J Biol Chem. 2003;278:36148–36156. doi: 10.1074/jbc.M305647200. [DOI] [PubMed] [Google Scholar]
- 47.Dedrick RL, Chamberlin MJ. Studies on transcription of 3′-extended templates by mammalian RNA polymerase II. Parameters that affect the initiation and elongation reactions. Biochemistry. 1985;24:2245–2253. doi: 10.1021/bi00330a019. [DOI] [PubMed] [Google Scholar]
- 48.Liu YV, Clark DJ, Tchernajenko V, Dahmus ME, Studitsky VM. Role of C-terminal domain phosphorylation in RNA polymerase II transcription through the nucleosome. Biopolymers. 2003;68:528–538. doi: 10.1002/bip.10302. [DOI] [PubMed] [Google Scholar]
- 49.Izban MG, Luse DS. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991;5:683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- 50.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 51.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 52.Thiriet C, Hayes JJ. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 2005;19:677–682. doi: 10.1101/gad.1265205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Studitsky VM, Clark DJ, Felsenfeld G. A histone octamer can step around a transcribing polymerase without leaving the template. Cell. 1994;76:371–382. doi: 10.1016/0092-8674(94)90343-3. [DOI] [PubMed] [Google Scholar]
- 54.Studitsky VM, Kassavetis GA, Geiduschek EP, Felsenfeld G. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science. 1997;278:1960–1963. doi: 10.1126/science.278.5345.1960. [DOI] [PubMed] [Google Scholar]
- 55.Kulaeva OI, Studitsky VM. Mechanism of histone survival during transcription by RNA polymerase II. Transcr. 2010;1:85–88. doi: 10.4161/trns.1.2.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsieh FK, Fisher M, Ujvari A, Studitsky VM, Luse DS. Histone Sin mutations promote nucleosome traversal and histone displacement by RNA polymerase II. EMBO Rep. 2010;11:705–710. doi: 10.1038/embor.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 58.Muthurajan UM, Bao Y, Forsberg LJ, Edayathumangalam RS, Dyer PN, White CL, Luger K. Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J. 2004;23:260–271. doi: 10.1038/sj.emboj.7600046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 60.Feng HP, Scherl DS, Widom J. Lifetime of the histone octamer studied by continuous-flow quasielastic light scattering: test of a model for nucleosome transcription. Biochemistry. 1993;32:7824–7831. doi: 10.1021/bi00081a030. [DOI] [PubMed] [Google Scholar]
- 61.Kuryan BG, Kim J, Tran NN, Lombardo SR, Venkatesh S, Workman JL, Carey M. Histone density is maintained during transcription mediated by the chromatin remodeler RSC and histone chaperone NAP1 in vitro. Proc Natl Acad Sci U S A. 2012;109:1931–1936. doi: 10.1073/pnas.1109994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kulaeva OI, Hsieh FK, Studitsky VM. RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc Natl Acad Sci USA. 2010;107:11325–11330. doi: 10.1073/pnas.1001148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bintu L, Kopaczynska M, Hodges C, Lubkowska L, Kashlev M, Bustamante C. The elongation rate of RNA polymerase determines the fate of transcribed nucleosomes. Nat Struct Mol Biol. 2011;18:1394–1399. doi: 10.1038/nsmb.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Studitsky VM, Clark DJ, Felsenfeld G. Overcoming a nucleosomal barrier to transcription. Cell. 1995;83:19–27. doi: 10.1016/0092-8674(95)90230-9. [DOI] [PubMed] [Google Scholar]
- 65.Jin J, Bai L, Johnson DS, Fulbright RM, Kireeva ML, Kashlev M, Wang MD. Synergistic action of RNA polymerases in overcoming the nucleosomal barrier. Nat Struct Mol Biol. 2010;17:745–752. doi: 10.1038/nsmb.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Araki R, Fukumura R, Sasaki N, Kasama Y, Suzuki N, Takahashi H, Tabata Y, Saito T, Abe M. More than 40,000 transcripts, including novel and noncoding transcripts, in mouse embryonic stem cells. Stem Cells. 2006;24:2522–2528. doi: 10.1634/stemcells.2006-0005. [DOI] [PubMed] [Google Scholar]
- 70.Gkikopoulos T, Schofield P, Singh V, Pinskaya M, Mellor J, Smolle M, Workman JL, Barton GJ, Owen-Hughes T. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science. 2011;333:1758–1760. doi: 10.1126/science.1206097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Selth LA, Sigurdsson S, Svejstrup JQ. Transcript Elongation by RNA Polymerase II. Annu Rev Biochem. 2010;79:271–293. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- 72.Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P, Svejstrup JQ. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- 73.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 Histone Methyltransferase Functions through the Phosphorylated Carboxyl-terminal Domain of RNA Polymerase II. J Biol Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 75.Schaft D, Roguev A, Kotovic KM, Shevchenko A, Sarov M, Neugebauer KM, Stewart AF. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 2003;31:2475–2482. doi: 10.1093/nar/gkg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 78.Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 79.Cho H, Orphanides G, Sun X, Yang XJ, Ogryzko V, Lees E, Nakatani Y, Reinberg D. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.John S, Howe L, Tafrov ST, Grant PA, Sternglanz R, Workman JL. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 2000;14:1196–1208. [PMC free article] [PubMed] [Google Scholar]
- 81.Schneider J, Bajwa P, Johnson FC, Bhaumik SR, Shilatifard A. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J Biol Chem. 2006;281:37270–37274. doi: 10.1074/jbc.C600265200. [DOI] [PubMed] [Google Scholar]
- 82.Saunders A, Werner J, Andrulis ED, Nakayama T, Hirose S, Reinberg D, Lis JT. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301:1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- 83.Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 84.Stuwe T, Hothorn M, Lejeune E, Rybin V, Bortfeld M, Scheffzek K, Ladurner AG. The FACT Spt16 “peptidase” domain is a histone H3–H4 binding module. Proc Natl Acad Sci USA. 2008;105:8884–8889. doi: 10.1073/pnas.0712293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Winkler DD, Muthurajan UM, Hieb AR, Luger K. The histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J Biol Chem. 2011 doi: 10.1074/jbc.M111.301465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 87.Ujvari A, Hsieh FK, Luse SW, Studitsky VM, Luse DS. Histone N-terminal tails interfere with nucleosome traversal by RNA polymerase II. J Biol Chem. 2008;283:32236–32243. doi: 10.1074/jbc.M806636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 89.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 90.Lee JS, Garrett AS, Yen K, Takahashi YH, Hu D, Jackson J, Seidel C, Pugh BF, Shilatifard A. Codependency of H2B monoubiquitination and nucleosome reassembly on Chd1. Genes Dev. 2012;26:914–919. doi: 10.1101/gad.186841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kristjuhan A, Svejstrup JQ. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 93.Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao J, Herrera-Diaz J, Gross DS. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol Cell Biol. 2005;25:8985–8999. doi: 10.1128/MCB.25.20.8985-8999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Epshtein V, Toulme F, Rahmouni AR, Borukhov S, Nudler E. Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J. 2003;22:4719–4727. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Epshtein V, Nudler E. Cooperation between RNA polymerase molecules in transcription elongation. Science. 2003;300:801–805. doi: 10.1126/science.1083219. [DOI] [PubMed] [Google Scholar]
- 99.Sullivan EK, Weirich CS, Guyon JR, Sif S, Kingston RE. Transcriptional activation domains of human heat shock factor 1 recruit human SWI/SNF. Mol Cell Biol. 2001;21:5826–5837. doi: 10.1128/MCB.21.17.5826-5837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 101.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 103.Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol Cell. 2000;5:377–386. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- 104.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 105.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]