Abstract
Rapid, quantitative Western blotting is a long-sought bioanalytical goal in the life sciences. To this end, we describe a Western blotting assay conducted in a single glass microchannel under purely electronic control. The μWestern blot is comprised of multiple steps: sample enrichment, protein sizing, protein immobilization (blotting), and in situ antibody probing. To validate the microfluidic assay, we apply the μWestern blot to analyses of human sera (HIV immunoreactivity) and cell lysate (NFκB). Analytical performance advances are achieved, including: short durations of 10–60 min, multiplexed analyte detection, mass sensitivity at the femtogram level, high-sensitivity 50-pM detection limits, and quantitation capability over a 3.6-log dynamic range. Performance gains are attributed to favorable transport and reaction conditions on the microscale. The multistep assay design relies on a photopatternable (blue light) and photoreactive (UV light) polyacrylamide gel. This hydrophilic polymer constitutes both a separation matrix for protein sizing and, after brief UV exposure, a protein immobilization scaffold for subsequent antibody probing of immobilized protein bands. We observe protein capture efficiencies exceeding 75% under sizing conditions. This compact microfluidic design supports demonstration of a 48-plex μWestern blot in a standard microscope slide form factor. Taken together, the μWestern blot establishes a foundation for rapid, targeted proteomics by merging exceptional specificity with the throughput advantages of multiplexing, as is relevant to a broad range of biological inquiry.
Keywords: immunoblotting, medical diagnostics, protein microarrays, systems biology, electrophoresis
Western blotting is an indispensable analytical tool, benefiting applications from clinical diagnostics to fundamental questions in the life sciences (1–3). The broad relevance of blotting stems from its highly specific results. Unlike separations or immunological probing alone, blotting reports two physicochemical characteristics: molecular mass and immunoaffinity. Because of this specificity, numerous immunoblotting variants have emerged for measurements of proteins to RNA to biomolecular interactions (2, 4–6). Nevertheless, despite being an information-rich and widely used assay, the Western blot has crucial inadequacies. In particular, limitations in data density and throughput hinder progress for emerging pursuits, including systems approaches to biology. A major shortcoming of Western blotting is the resource-intensive nature of the assay, being comprised of several steps requiring disparate pieces of equipment (Fig. 1A).
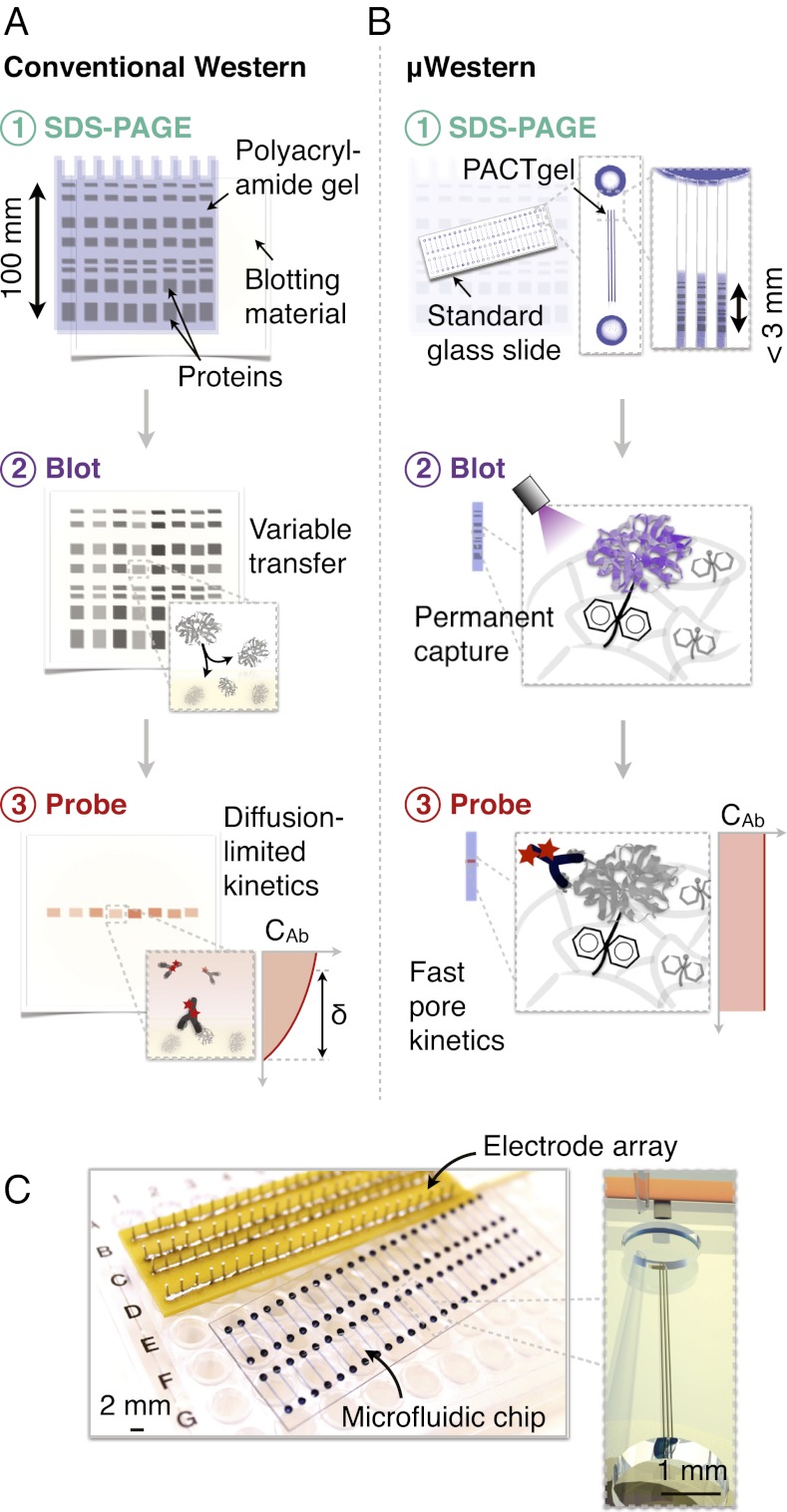
Fig. 1.
Single-microchannel μWestern assay design enables high device density formats. Aspects of scale, reagent use, blotting efficiency, and probe-binding kinetics are illustrated by comparative schematics for the conventional (A) and μWestern (B) assays (δ indicates a diffusion boundary layer thickness). The microfluidic workflow is comprised of: (i) analyte stacking and SDS-PAGE within the PACTgel matrix; (ii) band capture (“blotting”) onto the benzophenone-decorated PACTgel in response to UV light (as opposed to transfer to a separate sheet of hydrophobic material in conventional Western blotting); (iii) removal of SDS by brief electrophoretic washing and electrophoretic introduction of fluorescently labeled primary and (optionally) secondary detection antibodies specific to the target. Finally, excess probe is electrophoretically driven out of each device and peak intensities determined by fluorescence micrograph analysis. (C) Modular interfacing of standard microscope slide-sized chips with a scalable electrode array accommodating 48 blots per chip in triplicate (144 microchannels).
In the first stage of Western blotting, protein sizing, samples are analyzed by denaturing PAGE. During PAGE, proteins electromigrate through a polyacrylamide sieving gel, allowing determination of molecular mass. Once protein size is determined, the protein separation is incubated with antibodies (probing), thus allowing detection of interactions. Target identity is then established by linking immunoaffinity information to molecular mass. Although conceptually straightforward, substantial preparation and manual intervention are required. To prepare for probing, the sized proteins are transferred from the sieving gel to a blotting membrane. The blotting membrane immobilizes the protein separation and is then subjected to a series of handling steps, including blocking of nonspecific membrane interactions through coating with a dummy protein (e.g., BSA), incubation with antibodies to accomplish the probing step, and washing before obtaining assay readout. Several fundamental limitations impede performance, including slow mass transport. In fact, antibody probing often requires an overnight incubation period to compensate for diffusional limitations on antibody equilibration with antigen captured on the blotting material. Moreover, material and reagent consumption is extensive. A single 12-lane Western blot requires ∼300 mL of buffer and, most importantly, 1 μg of each detection antibody per analyte of interest. Because of the power of the assay and in light of these deficiencies, Western blotting requires continued innovation to improve throughput, minimize resource use, and advance analytical sensitivity and dynamic range (7).
Alternative single-step assays have emerged to overcome conventional Western blotting drawbacks, yet none afford the specificity of the two-pronged blotting assay (8–13). Primarily relying on spatially encoded antibody-probing, single-readout assays have been developed for ultrahigh sensitivity yet remain inherently vulnerable to nonspecific bias. Cross-reactivity is especially challenging for biological matrices and availability of specific antibodies can be limiting (10, 14, 15). The inability of single-step assays to guarantee specificity in complex biological samples underpins the need for targeted immunoblotting methods coupled to protein separation processes (16).
Recent innovation in protein analysis tools recognizes the crucial protein separation stage and seeks to retain separation-based information. In an approach using slab gels, Ciaccio et al. achieved remarkable miniaturization and scale-up of the Western blot workflow through a novel combination of highly multiplexed fluid handling and a large format slab gel (14). However, the approach conceded a loss in PAGE separation resolution because of elimination of sample stacking by isotachophoresis (ITP), a sample preconcentration step standard in conventional slab-gels that requires pore size and buffer chemistry discontinuities in the polyacrylamide sieving matrix. Furthermore, the workflow retained the conventional membrane transfer and antibody-probing paradigm. In a commercial approach using capillary electrophoresis, downstream membrane electrotransfer was replaced with photoactivated capture of proteins onto the inner wall of the capillary (17, 18). The capillary platform streamlined and automated Western blotting, but suffered from low protein-capture efficiencies of ∼0.01% (104-fold lower than membrane electrotransfer) and 3- to 5-h run times. In our own approaches using microfluidics, integration overcame some of the macroscale shortcomings, but we accepted either substantial complexity in interfacing and device architecture or did not implement the most widely used separation approach—protein sizing—as the first assay stage (7, 19).
In this study, we introduce scalable, automated μWesterns uniting protein sizing and antibody probing in a single microfluidic platform. The precision and control offered by microfluidic integration and photoresponsive materials achieves advances not realized previously, including development of rapid 48 concurrent μWesterns on a standard microscope slide footprint, multiplexed analysis of three protein targets per blot, and quantitation over a linear dynamic range of 3.6 logs with 50-pM lower limits of detection. We apply the μWestern to multiplexed protein analyses of complex proteinaceous samples, including crude cell lysate and crude human sera. Results suggest that purely microfluidic technologies are a viable means to imbue core analytical tools with automation, quantitative capability, and speed; thus paralleling advances that have positioned protein microarrays for high-throughput proteomics duty.
Results and Discussion
μWestern Device Design and Assay Operation.
Performance advances in slab gel-based Western blotting are limited by fundamental processes, including high transport timescales, variable blotting transfer, and diffusion-limited kinetics (Fig. 1A). To overcome these limits on macroscale performance, we explored a microscale Western blot (Fig. 1B, μWestern blot). We hypothesized that miniaturization applied in conjunction with photopatternable, light-responsive polymers would enable high analytical performance. We used this combined approach to integrate the distinct stages of the canonical Western blot into a simple, passively actuated microchannel. Specifically, the μWestern blot is comprised of isotachophoretic sample stacking during sample injection, weight-based separation of denatured protein analytes through the widely used SDS-PAGE, and in situ immunoblotting with fluorescently labeled primary and secondary antibodies (Fig. 1B). We first describe the microfluidic design strategy followed by details of the μWestern assay operation, experimental observations, and resulting performance.
We used single glass microchannels to facilitate high-density integration of μWestern blots within the footprint of a standard microscope slide (Fig. 1C). As such, each μWestern is comprised of a pair of access wells linked by three parallel microchannels (technical triplicates, each 70-μm wide × 10-μm deep) with electrical connectivity provided by an electrode array. To facilitate integration of the μWestern stages in the single microchannel geometry, we designed a unique photoactive gel with tunable porosity (PACTgel) that is both photopatternable and light-responsive. Photopatterning of the sieving gel is needed for reproducible control of the gel interface position along the channel axis and, hence, assay repeatability (Fig. 1B). A light-responsive material functionality was sought to allow the resulting photopatterned gel to switch from a molecular sieving gel during sizing to a blotting polymer for subsequent antibody probing. To introduce these new functionalities, we used two spectrally distinct chemical mechanisms. First, a riboflavin-driven photopolymerization strategy was used to photopattern the material (using 470-nm light, see Materials and Methods). Second, a spectrally distinct benzophenone-driven photo-immobilization strategy allows the sieving matrix to form covalent bonds with proteins in the gel (via UV excitation) (Fig. 1B and Materials and Methods). Benzophenone is incorporated in the polyacrylamide gel via a methacrylamide comonomer (BPMAC; N-[3-[(4-benzoylphenyl)formamido]propyl] methacrylamide). As a corollary outcome, the light-activated mechanism of the benzophenone-functionalized, hydrophilic gel means that no separate blocking steps are needed after protein immobilization and before antibody probing. Owing to the PACTgel functionality, the μWestern design is compatible with 48-sample throughput within a standard 1-inch by 3-inch microscope slide footprint (Fig. 1C).
Stage 1: Single Microchannel Protein Sizing.
In seeking a broadly relevant protein separation assay, we adapted the widely used Tris•glycine SDS-PAGE system of Laemmli (1, 20). Importantly, the assay requires a transient-ITP buffer arrangement and a large-to-small pore-size discontinuity a short distance along the microchannel axis to yield high-resolution protein sizing (Fig. 2A). For the gel discontinuity, we used an open channel-to-7.5%T sieving PACTgel interface at ∼400 μm into the microchannel (SI Materials and Methods). Transparency mask lithography of the photopatternable PACTgel was used to define the gel interface. Photopatterning yields fine control of the interface position (coefficient of variation, CV, of 3.5%, n = 60). During the ITP stacking phase, a diffuse plug of protein injected at the microchannel entrance is electrophoretically compacted into a ∼200-μm zone before electromigration across the sharp sieving gel interface. As is also shown in Fig. 2A, protein electromigration through the gel interface transitions ITP to SDS-PAGE, as the protein stack slows down substantially and the trailing glycine electrolyte overspeeds the stack (1, 20). In characterizing ITP, we observe reduced injection dispersion and >twofold sample stacking factors. The total mass of injected sample scales linearly with the length of the sample loading region. Thus, initial sample distributions spanning hundreds of micrometers to several millimeters can be stacked into final zones of equivalent width in the completed separations, achieving stacking factors over a broad range that can be tuned simply by manipulating the length of the open channel region upstream of the separation gel. In seeking to understand the sample enrichment limits in this system, the length of the sample loading region was extended to 4 mm and the sample loading times were increased to 30 s, yielding as high as ∼100-fold stacking factors (Fig. S1). These ITP-enabled enrichment and stacking qualities are important to obtaining high-separation resolution in subsequent sizing, as well as competitive limits of antigen detection. Interestingly, we observe that band ordering in the ITP stack is not necessarily governed by molecular mass, thus dynamic band reordering is often detected during the brief transition from ITP to PAGE (see band “x” in Fig. 2A).
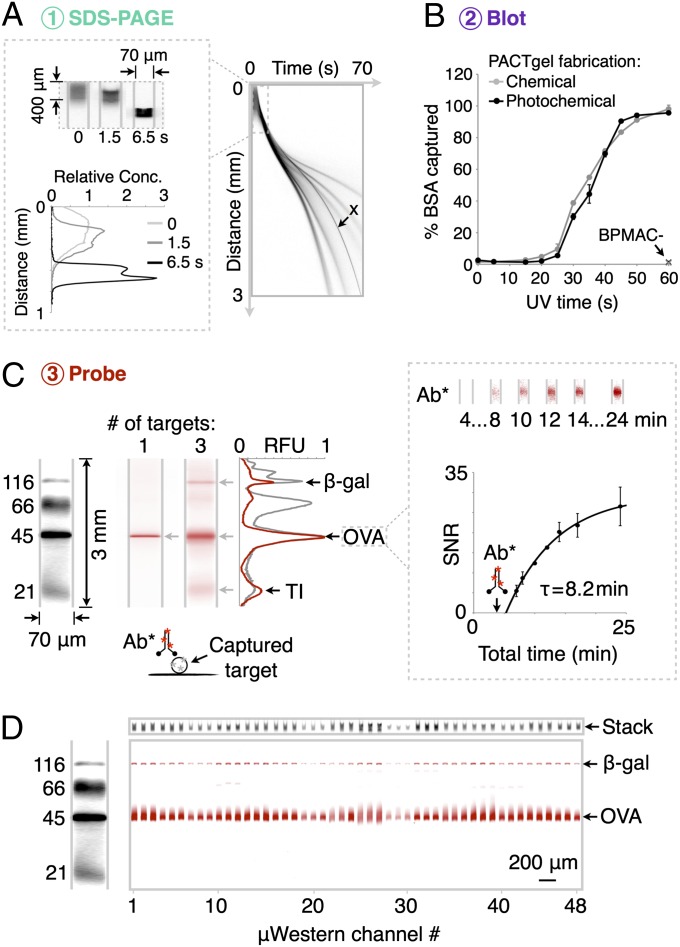
Fig. 2.
Compact μWestern with integrated high-resolution SDS-PAGE, blot, and detection. (A) SDS-PAGE of fluorescently labeled six protein ladder (black), complete in 60 s (4× magnification; band weights are 155, 98, 63, 40, 32, and 21 kDa). Channel aspect ratios are adjusted to produce gel-like images (see dimensions). (B) Capture efficiency of BSA (± SD, n = 3) for PACTgels fabricated chemically or photochemically. (C, Left) Multiplexed μWestern readout (red) in 40-min total assay times using primary antibodies for (i) OVA, and (ii) β-gal, OVA, and TI; all at 1 μM. (Right) Fluorescence micrographs and plot of SNR (± SD, n = 3) for electrophoretic introduction of red fluorescent primary antibody (Ab*) to OVA band at 4 min total assay time (arrow). (D) Forty-eight concurrent μWesterns of the four-protein fluorescent ladder probed for OVA and β-gal targets (1 μM each) with unlabeled primary and red fluorescent secondary antibodies in 60-min total assay time. At top, total injected (“stack”) fluorescence on weight marker spectral channel at the end of the ITP phase of SDS-PAGE acts as loading control.
Given this observation, we next sought to confirm that the separation mechanism in the PACTgel is indeed governed by protein molecular mass differences. Analysis of protein migration in the PACTgel yields a log-linear molecular mass versus migration distance relationship over the 20- to 150-kDa analyte range (R2 > 0.98 for ladder proteins) (Fig. S2), thus confirming this characteristic of SDS-PAGE (21). Protein peaks with molecular mass differences of >19% were resolvable (separation resolution Rs ≥ 1) (Fig. S3), with resolution similar to conventional slab-gel Western blotting. Although only uniform pore-size gels are studied here, the PACTgel pore size distributions are tunable, thus allowing enhanced resolution over specific weight ranges of interest (22–24). Importantly, the first sizing stage is observed to complete in compact 3-mm separation distances in separation times of ∼60 s. Consequently, the duration of the sizing step in μWestern blotting compares favorably to the 40–90 min required for macroscale protein sizing. The high-performance of protein-sizing benefits from the favorable scaling of electrophoretic transport with miniaturization.
Stage 2: In-Chip Protein Blotting by Photocapture.
Next, to permit antibody-based probing of the sized protein bands, we immobilized each species on the PACTgel polymer using UV irradiation of the entire separation channel (Fig. 2B and Fig. S4). UV exposure activates benzophenone groups to undergo hydrogen abstraction and covalent coupling to nearby biomolecules by a free radical mechanism (19, 25). First, we compared protein capture efficiencies for both chemically and photochemically fabricated PACTgels (i.e., gels without and with riboflavin) to determine the impact of the riboflavin-driven polymerization mechanism on the UV-initiated protein capture. In both cases, characterization of fluorescence retained on the PACTgel after photocapture and electrophoretic washout reveals a sigmoidal dependence of fluorescently labeled BSA capture efficiency on UV exposure time (Fig. 2B). The capture time courses for chemically and photochemically initiated PACTgel formulations show ∼100% BSA capture when UV exposure times are > 45 s. As negative controls, PACTgel formulations lacking benzophenone were studied and exhibited negligible protein blotting (Fig. 2B).
Interestingly, PACTgel protein capture efficiencies are significantly higher than those previously reported by our group for gels operating under nondenaturing isoelectric focusing conditions (1.3–13%) (19). We hypothesized that the capture efficiency improvement for SDS-PAGE stems from the denatured state of the target proteins. Denaturation likely exposes buried protein residues to the sieving matrix, thus promoting hydrophobic interactions between the unfolded analytes and the PACTgel benzophenone groups (19). Furthermore, the reduced requirement for protein solubilizing agents (especially detergents) in SDS-PAGE compared with isoelectric focusing likely reduces steric (among other) barriers to efficient coupling between proteins and the PACTgel. Second, we observed low capture efficiencies for exposure times of <20 s. We hypothesized that this is an initial inhibitory phase, perhaps caused by scavenging of reactive benzophenone sites by dissolved oxygen before a productive phase of analyte capture onto the PACTgel (26). In sum, we conclude that photopatterning using riboflavin and blue light does not compromise subsequent UV-mediated protein photocapture by the benzophenone-decorated polyacrylamide gels.
Next, we examined capture efficiencies for a wider range of proteins under the 45-s UV exposure conditions that lead to highly productive capture. We now considered only the photochemically fabricated gels of interest. We observed appreciable immobilization of a set of test proteins on the PACTgels: 97.5 ± 0.7%, 93.1 ± 3.4%, and 75.2 ± 0.8% for β-galactosidase (β-gal, 116 kDa), ovalbumin (OVA, 45 kDa), and trypsin inhibitor (TI, 21 kDa), respectively (all ± SD, n = 3). These capture efficiencies rival conventional electrotransfer blotting efficiencies on polymer membranes (27). Furthermore, the nearly complete protein capture in the photoactive bulk polyacrylamide gels is an orders-of-magnitude improvement over reported capture efficiencies for photoactive inner capillary surfaces [0.01% for GFP (17)]. We observed similar capture efficiencies for protein concentrations up to 100 pg⋅nL−1 (∼109 proteins nL−1 or 0.1 mg⋅mL−1), a capacity attributed to an estimated benzophenone site density of ∼1012 nL−1 of the gel structure. Thus, the 3D reactive gel offers a high volumetric density of binding sites. This large number of binding sites distributed throughout the channel volume enables efficient photocapture in < 60 s. In contrast, membrane electrotransfer in conventional bench top Western blotting requires 90 min to complete. This rapid capture kinetic of the μWestern is critical, first to yield low overall assay durations. Second, the rapid kinetic is essential to maintain performance; this is because diffusional band broadening erodes both SDS-PAGE separation resolution and analytical sensitivity of subsequent probing given the small interpeak displacement distances and peak widths in the microfluidic format (19, 28).
Stage 3: Probing.
The final assay stage is in situ antibody probing of the immobilized, sized proteins. In the μWestern, probes are electrophoresed through and along the length of the microchannel by an applied electric field. The approach ensures that probes sample positions along the entire length of the protein-decorated PACTgel. Results for probing of OVA with a red fluorescently labeled antibody are shown in Fig. 2C. Antibody was electrophoretically introduced 4 min after the start of the assay and required ∼1 min to migrate through the gel pores to the immobilized OVA band. We observed negligible red signal away from the OVA peak, suggesting that probe retention arises from specific interactions with immobilized protein targets. Building on the single protein probing result, Fig. 2C reports simultaneous probing of three analytes in a single microchannel using a three-antibody mixture applied in one electrophoretic step. Again, negligible off-target signal was detected. As the number of antibodies present in the probing mixture increases (see five-probe mixture in Fig. S5), multiplexing on one spectral channel becomes limited by increasing background arising from overlap of minor components in each target protein. The use of spectrally distinct dyes is supported by the current platform, pushing the conceptual multiplexing limit to ∼5n analytes per blot, where n is the number of dyes that can be imaged without significant spectral bleed-through. Between-device peak area CVs for identical samples probed simultaneously for OVA and β-gal were 25% each, with the ratio of peak areas varying with a CV of 14.7%. Use of internal migration controls allows data comparison across channels and chips.
Next, we sought to develop an ultrarapid μWestern blot. In designing this assay, we used dynamic imaging of fluorescent antibody probe accumulation at the site of captured analytes (Fig. 2C). This dynamic imaging mode yields a primary antibody probing time constant of 8.2 min for a 1-μM OVA band captured on the PACTgel after SDS-PAGE. A probe band signal-to-noise ratio (SNR) of >10 was recorded for a 10-min total assay time. We ascribed the rapid probing kinetics to the electrokinetic through-pore probe delivery strategy that leverages favorable overall reaction kinetics that are minimally impeded by surface boundary layer diffusion resistances (19). Dynamic monitoring of target peak SNR enables the assay to trade-off between readout signal and assay time. Finally, we sought to introduce a broadly relevant μWestern using unlabeled primary antibodies and fluorescently labeled secondary antibodies, as is common in conventional slab-gel Western blotting (Fig. 2D). In the 48-channel μWestern device, we probed both OVA and β-gal with unlabeled primary antibodies specific to the target. We also probed with secondary fluorescently labeled antibody probes and obtained the endpoint readout after electrophoretic washout of excess probes. This dual antibody approach gave higher SNRs over the full separation range (19) but still maintained relatively rapid assay times of less than 60 min.
High-Sensitivity and Quantitative Measurements for Proteinaceous Biospecimens.
Next, we sought to ascertain the robustness of the μWestern to analysis of biological samples. In the first study, we applied the μWestern to measurement of the transcription factor NFκB (p105, p50) in lysate from an NFκB-transfected 293T cell line (Fig. 3A). Western blotting is commonly applied to protein measurement in lysate (e.g., protein signaling studies). An unlabeled primary and a fluorescently labeled secondary antibody were used for immunoprobing. Assays of NFκB transfected lysate and untransfected negative controls yielded similar probing patterns in on-chip and conventional formats (Fig. 3A). We included conventional GAPDH probing as μWestern loading and biological controls; the total injected zone fluorescence of the ladder proteins serves as a convenient alternative loading control. With important implications to antibody screening frameworks and systems biology, we observed total assay operation to consume <1 ng of each antibody, in contrast with ∼1 μg consumed in conventional Western blotting.
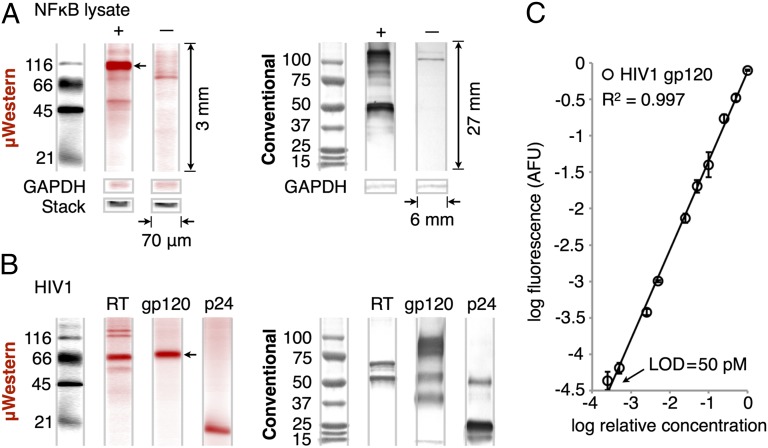
Fig. 3.
Validation of μWestern for cell lysate and purified proteins. (A) Sixty-minute μWestern of 0.5 mg/mL transfected 293T lysate probed for NFκB with unlabeled primary and fluorescently labeled secondary antibodies (red). Untransfected negative control lysate and loading controls (GAPDH and total injected fluorescence, “stack”) are included. (Right) The corresponding conventional 6- to 8-h Western blot readouts for visual comparison. Note relative dimensions of the conventional blot. (B) Forty-minute μWestern of purified HIV proteins (reverse-transcriptase, 200 nM; gp120, 200 nM; p24, 1 μM) after probing targets with fluorescently labeled primary antibodies (red). (C) Standard curve for gp120 over the 50 pM to 200 nM range (± SD, n = 3) constructed from peak areas of the band indicated by an arrow in B. See Fig. S6 for standard curve of the NFκB p105 peak indicated by an arrow in A.
In a second study to assess assay relevance, we applied our μWestern assay to several purified HIV proteins (Fig. 3B). HIV confirmatory diagnosis—presently a process that requires a central laboratory and hours to complete—relies on Western blotting. The μWestern and slab gel Western blot results agree to within 12% for the mass of the major bands of viral reverse transcriptase and the envelope glycoprotein gp120 (Table S1). For the smallest protein, capsid protein p24, a 25% error in the measured weights is attributed to the reduced performance of SDS-PAGE at the lower mass end of the sizing range, which is incurred regardless of format. Nevertheless, precision in molecular mass prediction on-chip gives within-device CVs of <2% (n = 3 for each) across the entire mass range. Minor bands for both gp120 (56 kDa; 40 kDa) and p24 (49 kDa) observed only on conventional Western blots are attributed to differences between the macro- and microscale workflows, including differences in blotting efficiency, in the SDS-PAGE and probing buffer systems, and in the degree of analyte renaturation before immunoprobing. Potentially relevant to point-of-care μWestern applications, the total wash and transfer buffer requirement is ∼300 μL, compared with 300 mL consumed in the conventional assay.
In both of the above studies we sought to realize high-sensitivity measurement while enabling a capacity for quantitation in μWestern blotting. For both NFκB and gp120 we observe quantitative antibody signal readout over a linear dynamic range of up to 3.6 logs, on par with expected performance for macroscale counterparts (Fig. 3C and Fig. S6). Using ITP to obtain a high stacking-factor enrichment, we measured a lower limit of detection of 50 pM for the latter analyte. This lower detection limit is comparable to enzyme-amplified chemiluminescent readouts in conventional blots. Considered another way, the detection capacity translates into measurement of 12 pg per 2-μL sample or a total mass of 17 fg of gp120 per sample injection volume. These results compare favorably to recent low-mass sensitivity (340 fg) slab gel Western blots (14). The mass limits also suggest that we can detect the equivalent of ∼4,000 virus particles on a gp120 basis (4–35 copies per virion) or as few as ∼20 particles for p24 (5,000 copies per virion) (29). As relevant to single-cell proteomics, the mass detection limit of ∼80,000 molecules we demonstrate is within the 104–106 molecule range expected for signaling proteins in single mammalian cells (30).
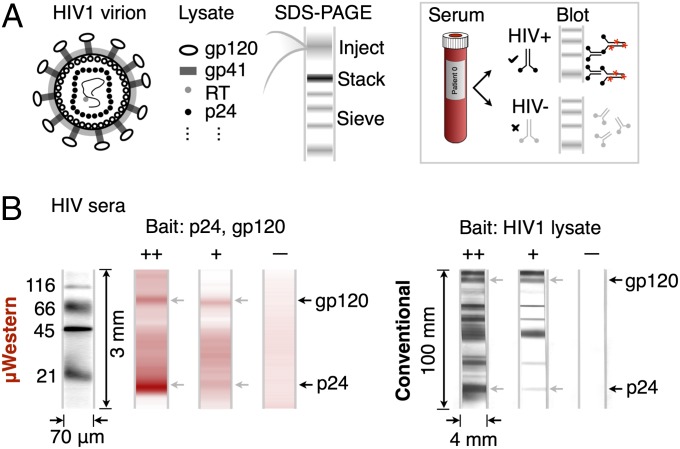
Building on the above HIV antigen study, we conducted a third study in which we sought to measure HIV antibodies directly in human sera. Currently, HIV diagnosis employs a conventional Western blot as the final (confirmatory) assay, following a positive ELISA-based screening result (15). In a 6- to 18-h workflow, an HIV viral lysate is subjected to SDS-PAGE and immunoblotting (Fig. 4A). Diluted patient serum is incubated with a nitrocellulose strip carrying the HIV protein bands. Any HIV-reactive antibodies in the serum bind to specific HIV proteins on the strip. A positive result is indicated if two or more of the p24, gp41 and gp120/160 bands exhibit reactivity at least as intense as that of the p24 band on a blotting strip subjected to a weakly reactive control serum (15). We translated the confirmatory HIV diagnostic assay to the μWestern by assaying human sera against purified gp120 and p24 HIV proteins (Fig. 4B). A mixture of these antigens was subjected to the μWestern assay, and the first probing step performed with 1:100 diluted human serum. Specific serum reactivity to each “bait” protein was determined using a fluorescently labeled secondary antibody directed to human IgG on the PACTgel. The resulting dose–response was consistent with the expected antibody titer in each of three sera (strongly reactive, weakly reactive, nonreactive), in accordance with guidelines for determining HIV infection in humans (15).
Fig. 4.
Sixty-minute μWestern for HIV antibody detection in human sera. (A) Conventional confirmatory HIV diagnostic assay schematic. (B) Reactivity of 1:100-diluted strongly reactive (++), weakly reactive (+), and nonreactive control (–) human sera to gp120 (200 nM) and p24 (1 μM) “bait” proteins revealed by fluorescently labeled secondary antibody to human IgG (red). At right, the conventional 6- to 18-h HIV Western blot, with gp120- and p24-reactive bands indicated by arrows. The conventional blot employs whole HIV lysate, whereas the μWestern uses specific HIV antigens, accounting for the additional reactive bands visible in the conventional blot.
Although promising, ongoing development of integrated serum processing (31) and optimization of assay conditions is underway to reduce nonspecific background in the HIV μWestern. Electrophoretic transport of antibody probes through the dense PACTgel matrix produces nonspecific staining in the low-nanomolar probe concentration range. This staining is strongly dependent on PACTgel pore size, and to a lesser extent on the degree of UV exposure during capture of separated antigens (Fig. S7). As the gel pore size decreases, large antibody probes can become irreversibly immobilized. Favorable to assay performance, however, we observed little impact of pore size on blotting background in the 7–8%T range that realizes optimal SDS-PAGE performance. A less dramatic increase in probing background as a function of UV exposure time during antigen capture may be caused by decreased effective pore size because of the formation of accessory gel-gel cross-links, or because of the production of a hydrophobic benzopinacol reaction product (26) that may encourage antibody immobilization within activated PACTgels. In any case, nonspecific probing is only two- to threefold higher in BPMAC+ PACTgels than in BPMAC− control gels of the same nominal porosity following UV exposure sufficient to cause near-quantitative antigen capture.
Specific to the HIV assay, we primarily attribute the background evident in the micrograph data of Fig. 4B to this basal nonspecific probing caused by off-target immobilization of human IgGs present in serum at high concentrations (∼10–15 mg/mL), as well as by immobilization of the secondary antibody probe within the PACTgel matrix. To further enhance the assay performance, ongoing efforts are focused on minimizing nonspecific background through on-chip sample cleanup, optimization of probe antibody concentrations, and alternative approaches for probe introduction and washout. After fully scrutinizing clinical specificity and sensitivity performance, we see potential for a 60-min stand-alone, rapid confirmatory HIV diagnostic for near-patient application.
Conclusions
Protein measurement tools that are high-throughput yet afford high-specificity quantitation hold great promise for advances across a swath of inquiry, from systems biology to clinical medicine. The studies detailed herein introduce a fully microfluidic Western blot that modernizes and automates conventional Western blotting. Because of the precision and control offered by microfluidic integration, we achieve advances in four key aspects of analytical performance: exceptional protein blotting efficiency with near complete analyte capture, accelerated run times as all steps from sample separation to probing are completed in 10–60 min, small device footprint (800-fold smaller device area compared with conventional gel lane), and superb reagent economy with a 103-fold reduction in antibody and buffer requirements over conventional Western blot. In an advance over conventional capability, the μWestern yields quantitative readouts from multiplexed analyte probing in a single sample and from 48-blot microchips. As is important to myriad protein measurements, the μWestern achieves desired limits of detection across numerous specifications, including starting sample concentration (low picomolar), starting sample total mass and volume (picograms per 2 μL of sample) and, finally, total detected mass (tens of femtograms of material per injected volume). To ensure relevance, we validated our μWestern for purified proteins, crude cell lysate, and crude human sera.
Looking forward, success in microfluidic integration of disparate separation, blotting, and immunoprobing stages into a unified workflow presents an exciting opportunity for “quantitative western blot microarrays.” Such approaches may eventually rival the throughput capacity of protein microarrays yet retain a currently missing and crucial separation step. As relevant to personalized medicine, our flexible electrophoretic blotting strategy is likely amenable to diverse probing (e.g., lectin) and gel-staining strategies needed for characterization of protein posttranslational modifications (32), characterization that is difficult with spatially encoded immunoreagents alone. Intriguingly, stable PACTgel analyte capture offers the ability to archive and reanalyze μWestern chips. Taking these data together, we see promise for challenging analytical applications in systems biology, cancer biology, and infectious disease diagnostics.
Materials and Methods
Microfluidic Assay Instrumentation.
Wet etching of microchannels in borosilicate glass, high-voltage control, fluorescence microscopy, and UV exposure system details are in SI Materials and Methods. Microfluidic channels were functionalized with acrylate-terminated self-assembled monolayers and PACTgels fabricated as detailed in SI Materials and Methods.
μWestern Protocol.
Sample preparation, loading, separation, capture, and probing along with specific imaging protocols are detailed in SI Materials and Methods.
Reagents and Samples.
BPMAC monomer was synthesized in-house and verified by 1H NMR and mass spectrometry as previously described (19). Purified proteins, antibodies and fluorescence labeling protocols are described in SI Materials and Methods.
Benchmark Analysis.
Conventional bench top Western blotting was performed as detailed in SI Materials and Methods.
Data Acquisition and Analysis.
Whole-channel imaging at 10× was conducted via stitching of adjacent, overlapping CCD images in ImageJ (National Institutes of Health, Bethesda, MD) to produce full gel-channel images and electropherograms (SI Materials and Methods). Assay limits of detection were defined as the nominal antigen concentrations at which probed peak SNRs were no smaller than 5 under optimal fluorescence imaging conditions.
Supplementary Material
Acknowledgments
A.J.H. is a Department of Defense National Defense Science and Engineering Graduate research fellow and a 2013 Siebel Scholar. A.E.H. is an Alfred P. Sloan research fellow (chemistry). This study was supported in part by National Institutes of Health’s New Innovator Award 1DP2OD007294 (to A.E.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207754110/-/DCSupplemental.
References
- 1.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 2.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA. 1979;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurien BT, Scofield RH. Protein Blotting and Detection: Methods and Protocols. New York: Springer; 2009. [Google Scholar]
- 4.Southern E. Southern blotting. Nat Protoc. 2006;1(2):518–525. doi: 10.1038/nprot.2006.73. [DOI] [PubMed] [Google Scholar]
- 5.Alwine JC, Kemp DJ, Stark GR. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci USA. 1977;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Y, Li Q, Chen XZ. Detecting protein-protein interactions by Far western blotting. Nat Protoc. 2007;2(12):3278–3284. doi: 10.1038/nprot.2007.459. [DOI] [PubMed] [Google Scholar]
- 7.He M, Herr AE. Automated microfluidic protein immunoblotting. Nat Protoc. 2010;5(11):1844–1856. doi: 10.1038/nprot.2010.142. [DOI] [PubMed] [Google Scholar]
- 8.Lequin RM. Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA) Clin Chem. 2005;51(12):2415–2418. doi: 10.1373/clinchem.2005.051532. [DOI] [PubMed] [Google Scholar]
- 9.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289(5485):1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 10.Spurrier B, Ramalingam S, Nishizuka S. Reverse-phase protein lysate microarrays for cell signaling analysis. Nat Protoc. 2008;3(11):1796–1808. doi: 10.1038/nprot.2008.179. [DOI] [PubMed] [Google Scholar]
- 11.Gaster RS, et al. Matrix-insensitive protein assays push the limits of biosensors in medicine. Nat Med. 2009;15(11):1327–1332. doi: 10.1038/nm.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rissin DM, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28(6):595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu H, et al. Global analysis of protein activities using proteome chips. Science. 2001;293(5537):2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 14.Ciaccio MF, Wagner JP, Chuu CP, Lauffenburger DA, Jones RB. Systems analysis of EGF receptor signaling dynamics with microwestern arrays. Nat Methods. 2010;7(2):148–155. doi: 10.1038/nmeth.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control (CDC) Interpretation and use of the Western blot assay for serodiagnosis of human immunodeficiency virus type 1 infections. MMWR Morb Mortal Wkly Rep. 1989;38(Suppl 7):1–7. [PubMed] [Google Scholar]
- 16.Ramaswamy A, et al. Application of protein lysate microarrays to molecular marker verification and quantification. Proteome Sci. 2005;3:9. doi: 10.1186/1477-5956-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Neill RA, et al. Isoelectric focusing technology quantifies protein signaling in 25 cells. Proc Natl Acad Sci USA. 2006;103(44):16153–16158. doi: 10.1073/pnas.0607973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan AC, et al. Nanofluidic proteomic assay for serial analysis of oncoprotein activation in clinical specimens. Nat Med. 2009;15(5):566–571. doi: 10.1038/nm.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes AJ, Lin RKC, Peehl DM, Herr AE. Microfluidic integration for automated targeted proteomic assays. Proc Natl Acad Sci USA. 2012;109(16):5972–5977. doi: 10.1073/pnas.1108617109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ornstein L. Disc electrophoresis—I Background and theory. Ann N Y Acad Sci. 1964;121:321–349. doi: 10.1111/j.1749-6632.1964.tb14207.x. [DOI] [PubMed] [Google Scholar]
- 21.Herr AE, Singh AK. Photopolymerized cross-linked polyacrylamide gels for on-chip protein sizing. Anal Chem. 2004;76(16):4727–4733. doi: 10.1021/ac049686u. [DOI] [PubMed] [Google Scholar]
- 22.Hou C, Herr AE. Ultrashort separation length homogeneous electrophoretic immunoassays using on-chip discontinuous polyacrylamide gels. Anal Chem. 2010;82(8):3343–3351. doi: 10.1021/ac100182j. [DOI] [PubMed] [Google Scholar]
- 23.Hughes AJ, Herr AE. Quantitative enzyme activity determination with zeptomole sensitivity by microfluidic gradient-gel zymography. Anal Chem. 2010;82(9):3803–3811. doi: 10.1021/ac100201z. [DOI] [PubMed] [Google Scholar]
- 24.Lo CT, Throckmorton DJ, Singh AK, Herr AE. Photopolymerized diffusion-defined polyacrylamide gradient gels for on-chip protein sizing. Lab Chip. 2008;8(8):1273–1279. doi: 10.1039/b804485f. [DOI] [PubMed] [Google Scholar]
- 25.Dormán G, Prestwich GD. Benzophenone photophores in biochemistry. Biochemistry. 1994;33(19):5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- 26.Schneider MH, Tran Y, Tabeling P. Benzophenone absorption and diffusion in poly(dimethylsiloxane) and its role in graft photo-polymerization for surface modification. Langmuir. 2011;27(3):1232–1240. doi: 10.1021/la103345k. [DOI] [PubMed] [Google Scholar]
- 27.Jungblut P, Eckerskorn C, Lottspeich F, Klose J. Blotting efficiency investigated by using two-dimensional electrophoresis, hydrophobic membranes and proteins from different sources. Electrophoresis. 1990;11(7):581–588. doi: 10.1002/elps.1150110709. [DOI] [PubMed] [Google Scholar]
- 28.Tia SQ, He M, Kim D, Herr AE. Multianalyte on-chip native Western blotting. Anal Chem. 2011;83(9):3581–3588. doi: 10.1021/ac200322z. [DOI] [PubMed] [Google Scholar]
- 29.Swanson CM, Malim MH. SnapShot: HIV-1 proteins. Cell. 2008;133(4):742–, 742.e1. doi: 10.1016/j.cell.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Milo R, Jorgensen P, Moran U, Weber G, Springer M. BioNumbers—The database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010;38(Database issue):D750–D753. doi: 10.1093/nar/gkp889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Apori AA, Herr AE. Homogeneous immunosubtraction integrated with sample preparation enabled by a microfluidic format. Anal Chem. 2011;83(7):2691–2698. doi: 10.1021/ac103219x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He M, Novak J, Julian BA, Herr AE. Membrane-assisted online renaturation for automated microfluidic lectin blotting. J Am Chem Soc. 2011;133(49):19610–19613. doi: 10.1021/ja207963f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.