Abstract
The hemagglutinin (HA) of influenza A(H3N2) virus responsible for the 1968 influenza pandemic derived from an avian virus. On introduction into humans, its receptor binding properties had changed from a preference for avian receptors (α2,3-linked sialic acid) to a preference for human receptors (α2,6-linked sialic acid). By 2001, the avidity of human H3 viruses for avian receptors had declined, and since then the affinity for human receptors has also decreased significantly. These changes in receptor binding, which correlate with increased difficulties in virus propagation in vitro and in antigenic analysis, have been assessed by virus hemagglutination of erythrocytes from different species and quantified by measuring virus binding to receptor analogs using surface biolayer interferometry. Crystal structures of HA–receptor analog complexes formed with HAs from viruses isolated in 2004 and 2005 reveal significant differences in the conformation of the 220-loop of HA1, relative to the 1968 structure, resulting in altered interactions between the HA and the receptor analog that explain the changes in receptor affinity. Site-specific mutagenesis shows the HA1 Asp-225→Asn substitution to be the key determinant of the decreased receptor binding in viruses circulating since 2005. Our results indicate that the evolution of human influenza A(H3N2) viruses since 1968 has produced a virus with a low propensity to bind human receptor analogs, and this loss of avidity correlates with the marked reduction in A(H3N2) virus disease impact in the last 10 y.
Surveillance of influenza viruses is essential for updating vaccines, for tracking the emergence of drug resistant viruses, and for monitoring zoonotic infections. It also gives important insights into the mechanisms of virus evolution. This is particularly the case for interpreting the correlation between antigenic differences and changes in the sialic acid receptor binding properties of the HA glycoprotein. The correlation in these two properties arises because of the close proximity on HA of binding sites for antibodies that neutralize virus infectivity and the sialic acid receptor binding pocket (1), and accounts for the observations that mutations that prevent antibody binding can also result in changes in receptor binding (2–7). Reduction in affinity of human H3N2 viruses for avian receptors since the beginning of the pandemic in 1968 has meant that by the 1990s viruses with reduced ability to agglutinate chicken erythrocytes had emerged (8, 9). Moreover, viruses isolated after 1999 were shown to have reduced affinity for both human and avian receptors, a feature that correlated with their poor growth properties in eggs and different cells in culture (9–14). The evolution of the HA has resulted in at least three key changes that influence receptor binding. Two sequential substitutions occurred at residue 225: in 2001–2002, a substitution Gly-225→Asp was accompanied by a Trp-222→Arg substitution, and in 2004–2005, an Asp-225→Asn substitution was accompanied by the substitution Ser-193→Phe (while maintaining arginine at position 222). Residue 226, a key amino acid in determining receptor specificity (15), also changed twice: before 2001, Leu-226→Val, and in 2004, Val-226→Ile (Fig. S1).
To correlate these amino acid substitutions with the biological properties of the viruses, we have analyzed the receptor binding characteristics of H3N2 viruses isolated between 2001 and 2010, examined changes in their ability to infect cells in culture, and determined the structures of two HAs of virus isolates from 2004 and 2005 in the absence of receptor and complexed with a human receptor analog. The data show that the progressive decrease in binding of these viruses to human receptors from 2000 onward correlates with changes in the efficiencies of infection of cultured cells. Comparison of structural data for HAs of viruses from 1968, 2004, and 2005 explain how particular mutations that affect the conformation of the HA1 220-loop component of the receptor binding site define the receptor binding phenotype of recent H3N2 human influenza viruses.
Results and Discussion
Virus Receptor Binding.
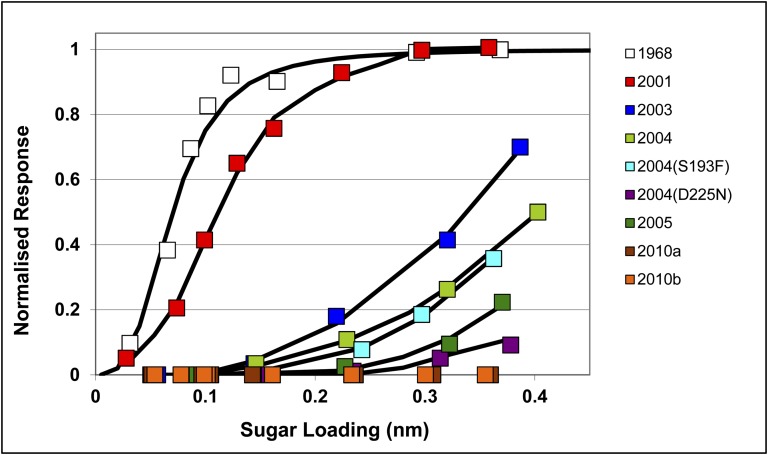
We used surface biolayer interferometry to measure virus binding to human and avian receptor analogs. The results indicate that the avidity of H3N2 viruses for the human receptor analog, α2,6-sialyl lactosamine, decreased over time with a ∼4-fold reduction between 1968 and 2001 and then a further estimated 200-fold reduction in binding over the period of 2001–2004 (Fig. 1). By 2010, viruses failed to bind to human receptor under standard assay conditions, but some very limited binding could be detected at increased virus concentrations (Fig. S2A). In the case of the avian receptor analog, α2,3-sialyl lactosamine, only the 1968 HA showed significant binding under standard conditions; even when a 10-fold increase in virus concentration was used only the 2001 virus showed evidence of binding (Fig. S2B).
Fig. 1.
Biolayer-interferometry binding curves of influenza viruses (100 pM) to the human receptor analog α2,6-sialyl lactosamine bound to the sensor chip show a large decline in avidity of the virus for receptor over the period of 2001–2010. Viruses used are indicated as follows: 1968, X31 (a high growth reassortant virus carrying the HA and NA genes of A/Aichi/2/68); 2001 A/Toulouse/878/2001; 2003, A/Trieste/2/2003; 2004, A/Finland/486/2004; 2004 (S193F) reverse genetic virus with the HA and NA genes of A/Finland/486/2004 carrying the substitution Ser-193→Phe in HA1; 2004 (D225N), reverse genetic virus with the HA and NA genes of A/Finland/486/2004 with the substitution Asp-225→Asn in HA1; 2005, A/Hong Kong/4443/2005; 2010a, A/Hong Kong/3615/2010; 2010b, A/Esfahan/6117/2010.
Amino acid substitutions in the receptor binding site of the selected viruses used in the binding studies are shown in Table 1. The decrease in receptor binding between 2001 and 2003 and between 2004 and 2005 (Fig. 1) was accompanied by the substitutions Gly-225→Asp and Trp-222→Arg, and then Asp-225→Asn and Ser-193→Phe, respectively. The particular significance of the substitutions observed in the 2005 HA, at residues 193 and 225, was assessed by introducing them independently into the 2004 HA by virus reverse genetics. As shown in Fig. 1, receptor binding by the Asp-225→Asn mutant of the 2004 virus was decreased by ∼10-fold compared with the wild-type 2004 virus and was similar to the level seen with the 2005 virus. In contrast, the impact of the Ser-193→Phe substitution on receptor binding was negligible.
Table 1.
Influenza A(H3N2) properties
| Amino acid position |
Hemagglutination titer* |
Virus plaquing efficiency |
||||||||||
| Virus | 190 | 193 | 222 | 225 | 226 | 227 | Chicken | Turkey | Guinea pig | SIAT | MDCK | Ratio |
| X31 (A/Aichi/2/68) | E | S | W | G | L | S | 2,048 | 4,096 | 1,024 | 2 × 107 | 1 × 107 | 2 |
| A/Toulouse/878/2001 | D | S | W | G | V | S | 4 | 512 | 256 | 3 × 106 | 3 × 106 | 1 |
| A/Trieste/2/2003 | D | S | R | D | V | S | < | 16 | 128 | 1 × 104 | 1 × 103 | 10 |
| A/Finland/486/2004 | D | S | R | D | I | P | < | 512 | 8,192 | 4 × 103 | 2 × 102 | 20 |
| RG/Finland/486/2004 S193F | D | F | R | D | I | P | < | 128 | 1,024 | 8 × 106 | 4 × 105 | 20 |
| RG/Finland/486/2004 D225N | D | S | R | N | I | P | < | < | 512 | 3 × 104 | 4 × 102 | 75 |
| A/Hong Kong/4443/2005 | D | F | R | N | I | P | < | < | 32 | 4 × 104 | 1 × 103 | 40 |
| A/Esfahan/6117/2010 | D | F | R | N | I | P | < | < | 64 | 2 × 104 | 5 × 101 | 400 |
| A/Hong Kong/3615/2010 | D | F | R | N | I | P | < | < | 64 | 2 × 104 | 1 × 101 | 2,000 |
The table shows sequence variation in the vicinity of the receptor binding site of the HA, HA titer with RBCs of different species, and the relative plaque formation on MDCK cells and MDCK-SIAT1 cells.
*The symbol “<” indicates no agglutination of RBCs was observed.
None of the viruses isolated in cell culture since 2001 was able to agglutinate chicken erythrocytes through the HA and those isolated from 2005 were unable to bind to turkey erythrocytes (Table 1). Turkey erythrocytes, because of their comparatively greater human receptor content (Table S1), were agglutinated by viruses isolated up to 2004 when the Asp-225→Asn amino acid substitution occurred. The results with the site-specific mutant viruses Asp-225→Asn and Ser-193→Phe, therefore, support the correlation between reduced avidity of virus for human receptor and changes detected in hemagglutination tests. Both assays substantiate the importance of the Asp-225→Asn substitution.
The interferometry data on the receptor binding properties of different H3N2 viruses also correlate with their efficiency of infection of cell lines that differ in receptor expression. Unlike the 1968 and 2001 viruses, those viruses isolated after 2001 had decreased ratios of infection of parental Madin Darby canine kidney (MDCK) cells in comparison with MDCK-SIAT1 (SIAT) cells, which have been modified to express a higher density of α2,6 human receptors. Strikingly, those viruses with the lowest avidity for the human receptor showed the greatest reductions in their ability to infect MDCK versus SIAT cells (Table 1).
Overall Structure.
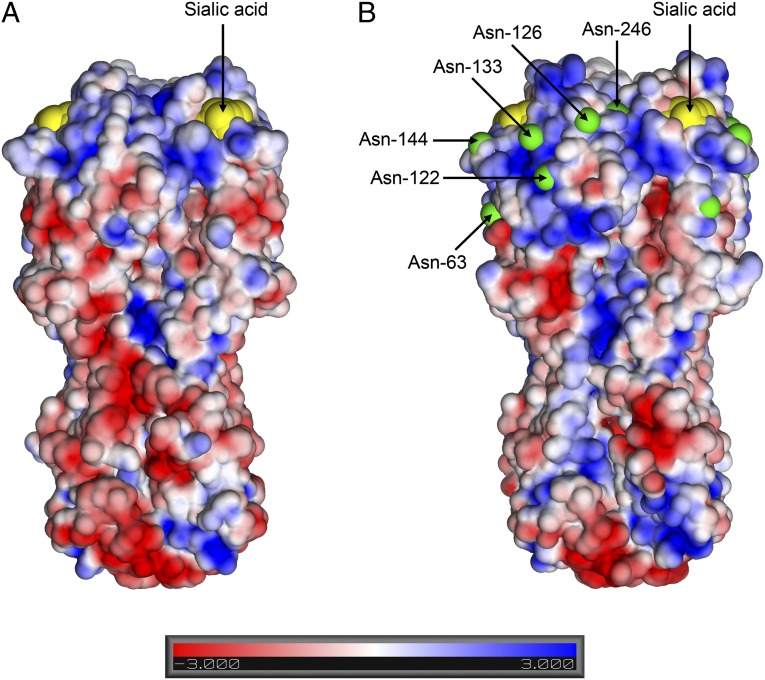
To understand the structural basis of the changes in receptor recognition, the HAs of the 2004 and 2005 viruses were expressed in insect cells and their structures were determined by X-ray crystallography. Diffraction data were obtained for both the HAs alone (apo) and the HA–human receptor analog complexes, and relevant crystallographic statistics are presented in Table S2. The amino acid sequences of the 2004 and 2005 HAs differ by only three amino acids (with 87 and 86% identity, respectively, to the HA of the 1968 virus). The sequence changes since 1968 were primarily on the surface of the membrane-distal domain of HA; consequently, the overall backbone structures are remarkably similar (Fig. 2 and Fig. S3) and validate the frequent use of the 1968 HA structure to locate and assess the antigenic significance of sequence differences in the HAs of viruses isolated since 1968. The sequence changes have resulted in two outstanding consequences: changes in surface charge [the pI has changed from 5.6 (1968) to 8.6 (2004)] and the introduction of six new potential sites for glycosylation (Fig. 2). Electron density for at least part of the carbohydrate moiety is seen at positions 63, 126, 133, and 246. There was the loss of one potential glycosylation site at residue 81. Carbohydrate side chains on HA surfaces have previously been observed to influence the antigenicity of HAs (16, 17), but the additional sites had accumulated during the evolution of HA before the major decline in the virus avidity for the human receptor observed since 2001 (Fig. 1). The significance of the accumulation of surface basic residues, as noted before, is unknown (18); it is, however, striking that HAs from avian viruses of all subtypes have pI values between 5.5 and 6.5, like the 1968 HA, which itself had entered humans only recently from an avian virus source.
Fig. 2.
Crystal structures of H3 HA trimers from 1968 (A) and 2004 (B) viruses shown in a surface electrostatics representation. Negative potential is colored red and positive in blue, and the bound sialic acid moieties from receptor complexes are colored in yellow. Additional potential glycosylation sites present in the 2004 HA, but not 1968 HA, are marked in green and labeled on the structure of the 2004 HA.
Receptor Binding Site.
Given that the 1968 HA has significantly higher affinity for human receptor than 2004 and 2005 HAs (Fig. 1), we compared the structures of the human receptor complexes formed by all three HAs.
In the 2004 HA complex, the receptor analog is well defined in electron density maps, and adopts a folded-back conformation, with an α2,6-cis linkage between sialic acid and the adjoining galactose-2 (Fig. 3A and Fig. S4). The conformation of the human receptor, and the interactions it forms with the HA, are generally similar to those seen in its complex with the 1968 HA (19). However, unlike the 1968 HA, changes in the structure of the 220-loop of the 2004 HA occur on receptor binding that appear to facilitate the interaction (Fig. 3A).
Fig. 3.
The structure of the receptor binding site of H3 HAs in complex with human receptor analogs. (A) Overlap of apo (gray) and human receptor complex of 2004 HA (protein in green; receptor in yellow) showing the hydrogen bond interactions between Asp-225 and the 3-hydroxyl of galactose-2 made possible by the altered conformation of the 220-loop in the receptor complex. (B) Similar overlap to A of the apo (gray) and human receptor complex of 2005 HA (protein in blue; receptor in yellow), illustrating their similarity. In the 2005 complex, only the sialic acid moiety of the receptor can be built. (C) Overlap of the human receptor complexes of the 1968 HA (colored magenta) and 2004 HA (colored green). Key differences between the two, including the greater distance between the hydrophobic Ile-226 and the bridging carbon of the receptor in the 2004 complex (4.5 versus 3.8 Å) and the presence of Glu-190 in the 1968 structure versus Asp-190 in the 2004 structure, are indicated. Selected interactions are shown that are additional to the highly conserved hydrogen bonds formed between the sialic acid carboxylate and the OH of Ser-136 and main-chain amide 137, and between main-chain carbonyl 135 and the N of the acetamido substituent. The selected interactions shown are as follows: (A) Ser-137 OH indirect through H2O to sialic acid carboxylate, Ser-137 OH direct to sialic acid carboxylate, and Thr-135 OH indirect through H2O to sialic acid 4-OH. (B) Ser-136 OH indirect through H2O to sialic acid carboxylate. Thr-135 OH indirect through H2O to sialic acid 4-OH.
In contrast, the electron density for the receptor in the 2005 HA complex is both weaker and less well defined, consistent with poorer binding (Fig. 1). Only the sialic acid of the sialyllactosamine can be modeled, with direct hydrogen bonds, as in the 2004 HA, between its carboxylate and Ser-136 and Ser-137 (Fig. 3B). All other interactions with the sialic acid appear to be made via water molecules.
Comparison of the 1968 and 2004 HA complexes with the human receptor analog reveals a number of features that probably account for the marked reduction in affinity for receptor over time. In particular, whereas there are no significant conformational differences between the apo and receptor-bound forms of the 1968 HA, in the 2004 HA–human receptor analog complex, Asp-225 forms a salt bridge with Arg-222 and is shifted 1.5 Å closer to the bound receptor where it is able to form a hydrogen bond with the 3-hydroxyl of galactose-2 (Fig. 3A). Furthermore, the peptide carbonyl of Asp-225 in the complex is located about 1.3 Å closer to the receptor, forming a hydrogen bond with the 4-hydroxyl of galactose-2. The energetic cost of these rearrangements of the 220-loop, to better suit binding to receptor, is expected to reduce the affinity of the interaction. In the 1968 HA the side chain of Leu-226 is closer to C-6 of galactose-2 (3.8 Å) than Ile-226 (4.5 Å) in the corresponding 2004 HA–receptor complex (Fig. 3C). In addition, Glu-190 in the 1968 HA protrudes into the receptor binding pocket toward the glycerol substituent of sialic acid, such that the 9-hydroxyl of the glycerol substituent makes two additional hydrogen bonds with His-183 and Glu-190. In contrast, the shorter side chain of Asp-190 in the 2004 HA does not reach into the pocket sufficiently to interact with the glycerol substituent of sialic acid.
Thus, the observations that the 220-loop of the 1968 HA is already positioned for receptor binding, that there is a closer hydrophobic contact between the C-6 of galactose-2 and Leu-226 of 1968 HA, and that two additional hydrogen bonds are formed by 1968 HA with the glycerol substituent of sialic acid, are consistent with the higher affinity of the 1968 HA for the human receptor.
Site-directed mutagenesis experiments indicate the importance of the Asp-225→Asn substitution for the decreased affinity for human receptor of the 2005, compared with the 2004 virus (Fig. 1). In the 2005 HA structure, the side-chain carbonyl of Asn-225 forms a hydrogen bond with Arg-222 but the side-chain amino group cannot form the hydrogen bond with the hydroxyl of galactose-2 observed in the 2004 complex. Moreover, the backbone conformation of the 220-loop in both the apo and receptor complexed forms of the 2005 HA are the same and similar to the apo form of the 2004 HA. It appears likely, therefore, that the loss of the hydrogen bond interaction between residue 225 and galactose-2 of the receptor means that, in the 2005 HA, there is insufficient binding energy to facilitate the conformational change in the 220-loop. A further manifestation of the weaker binding of receptor by the 2005 HA is that the sialic acid sits less deeply in its binding pocket (Fig. 3B and Fig. S4). Consequently, receptor interactions only involve sialic acid in the 2005 structure and are likely to be weaker than those involved in complexes formed by earlier H3 HAs.
Biological Consequences of Receptor Binding Variation.
There are potentially important consequences of changes in receptor binding properties for virus isolation, for antigenic characterization, and for vaccine production. Viruses with decreased affinity for cellular receptors are under increased selection pressure when propagated in tissue culture cells and in hens' eggs for vaccine production (7). Indeed, the reduced avidity of the HA for the receptor is likely to be a key factor that promotes the observed ability of the virus neuraminidase to substitute for the HA in receptor binding by viruses propagated in certain cell lines (20). Difficulties with antigenic characterization derive largely from differences in the behavior of viruses in the hemagglutination assays that are central to antigenic analyses, leading to the need to use guinea pig erythrocytes in standard hemagglutination and hemagglutination inhibition assays for recent H3N2 viruses (21).
Biologically, the most important consequence of amino acid substitutions that result in decreased affinity for receptors is their effect on the efficiency of infection and transmission. Despite the changes in receptor binding that have been observed, and the widespread immunity to H3N2 viruses generally, these viruses continue to circulate and have predominated in North America in the 2010/2011 influenza season and in the 2011/2012 season in Europe (22, 23). Nevertheless, it is striking that the severity of infections has declined since the major epidemics of 1968–1970, 1975–1976, 1989–1990, 1994–1995, and 1999–2000 (24), and this may be linked to the marked reduction in receptor binding affinity in the recent years of this 44-y-long pandemic.
Materials and Methods
Cells.
SIAT cells, kindly provided by M. Matrosovich (Philipps Universität, Marburg, Germany), stably expressing human CMP-N-acetylneuraminate:β-galactoside α-2,6-sialyltransferase for enhanced expression of sialic acid α2–6Gal-terminated oligosaccharides (25) and parental MDCK cells were propagated at 37 °C with 5% (vol/vol) CO2 in DMEM (Sigma; D6429), supplemented with 10% (vol/vol) heat-inactivated FCS, antibiotics [penicillin (100 U⋅mL−1), streptomycin (100 μg⋅mL−1)], and, for the SIAT cells, 1 mg⋅mL−1 G418 sulfate (Geneticin; Invitrogen).
Viruses.
Wild-type influenza viruses used in this study were originally isolated from clinical samples by World Health Organization (WHO) National Influenza Centres and retrieved from stocks held in the WHO Collaborating Centre for Reference and Research on Influenza, National Institute for Medical Research (London, United Kingdom). Viruses were propagated in either MDCK or SIAT cells, and viruses were used following a final passage through SIAT cells.
Recombinant Viruses.
Recombinant viruses were generated by reverse genetics, as described by Hoffmann et al. (26). Both HA and NA genes were amplified with primers (sequences are available on request) and cloned into the vector pHW2000. Site-specific mutagenesis of cDNA clones was performed using a QuikChange Site-Directed Mutagenesis kit (Stratagene). Plasmids were transfected into cocultured 293T and SIAT cells and recovered virus was propagated in SIAT cells.
Hemagglutination Assays.
Hemagglutination assays were performed according to standard methods using suspensions of guinea pig [1.0% (vol/vol)], chicken (0.5%), or turkey (0.5%) red blood cells (RBCs) from Matrix, TCS, and the Health Protection Agency Centre for Infection, respectively, under procedures regulated by the Animals (Scientific Procedures) Act 1986. Hemagglutination titers were determined in the presence 20 nM oseltamivir carboxylate to circumvent possible agglutination of RBCs mediated by the virus neuraminidase (20). For lectin hemagglutinin assays, the concentrations of the various bloods were standardized at 1% (vol/vol) to enable direct comparison between species. Maackia amurensis II and Sambucus nigra lectins were from Vector Laboratories.
Nucleotide Sequence Analyses.
Nucleotide sequences of HA and NA genes in viruses, cDNA clones, and recombinant viruses were determined using ABI Prism BigDye terminator cycle sequencing kits and an ABI-3730XL DNA analyzer. The sequences of all of the viruses in this study were verified before use and have been deposited in the Global Initiative on Sharing All Influenza Data (GISAID) database (Table S3).
Plaque Assays.
Plaque assays were performed on SIAT or unmodified MDCK cells using 1.2% (wt/vol) Avicel and 2 µg⋅mL−1 trypsin in six-well plates (27).
Quantification of Viruses and Biolayer Interferometry Receptor Binding Assay.
Viruses propagated in SIAT cells in serum-free DMEM containing 2 µg⋅mL−1 TPCK-treated trypsin (Sigma; T1426) were concentrated by centrifugation at 100,000 × g for 1 h, resuspended in PBS, and quantified by solid-phase ELISA using a mouse monoclonal antibody against the nucleoprotein (NP) of influenza A virus (a gift from Centers for Disease Control and Prevention), goat anti-mouse horseradish peroxidase conjugate, and 3,3′,5,5′-tetramethyl-benzidine. The concentrations of the viruses, standardized by comparison with a virus of known concentration, were calculated from the estimated NP contents. The number of NP molecules per virion was calculated based on the influenza virus genome consisting of 13,588 nucleotides and the binding of 23 nucleotides per NP monomer (28). Virus binding to receptor analogs was measured on an Octet RED instrument (ForteBio). Biotinylated α2,3- and α2,6-linked sialyl lactosamine sugars (2,3SLN and 2,6SLN, respectively) linked to a polyacrylamide backbone of 30-kDa polymers containing 20 mol % sugar and 5 mol % biotin were from Lectinity. Sugars were loaded onto streptavidin biosensors (ForteBio) at 0.01–0.5 µg⋅mL−1 for 5 min in 150 mM NaCl, 10 mM Hepes, pH 7.4, 3 mM EDTA, 100 μM oseltamivir carboxylate (Roche), and 0.005% Tween 20, and then virus was added at either 100 pM or 1 nM in the same buffer. Association was measured over 30 min at 25 °C. Equilibrium responses for virus binding were plotted as a function of the amount of sugar immobilized on the biosensor calculated from the response during the sugar loading step.
Expression and Purification of 2004 and 2005 H3-HAs.
Arg-329→Gln substitutions were introduced into the HA genes of A/Finland/486/2004 and A/Hong Kong/4443/2005 that had been cloned into vector pHW2000 by QuikChange PCR mutagenesis (Agilent). The cDNA encoding residues 1–504 (HA0) of the mature Arg-329→Gln A/Finland/486/2004 and A/Hong Kong/4443/2005 HA proteins were subcloned, using In-fusion cloning (Clontech), between the BamHI and NotI sites of a modified pAcGP67A (BD Biosciences) vector (pHAEM). The expressed proteins carried a C-terminal extension: SGRENLYFQGGGGSGYIPEAPRDGQAYVRKDGEWVLLSTFLGHHHHHH, where the italicized sequence is the recognition site for TEV protease cleavage, the underlined sequence is a trimerization foldon (29), and the sequence in bold is a hexa-His tag.
Recombinant baculovirus was generated by cotransfection of BaculoGold Bright Linearized Baculovirus DNA (BD) and pHAEM into Sf9 cells. Following virus amplification, large-scale expression was performed with 2.5-L Sf9 cell cultures. Cells were removed by centrifugation 72 h after infection, and supernatant was concentrated and loaded onto a Talon cobalt column (Clontech). Fractions containing HA were pooled and dialyzed against 50 mM NaCl, 10 mM Tris⋅HCl, pH 8.0, buffer. Dialyzed protein was concentrated and treated with trypsin [10:1 HA/trypsin (wt/wt ratio); 4 °C 16 h] to remove the foldon and His-tag. The reaction was stopped by adding soybean trypsin inhibitor and the HAs were subjected to further purification by gel filtration chromatography using a Superdex-200 16/60 column (GE) in 150 mM NaCl, 10 mM Tris⋅HCl, pH 7.0, buffer. The gel-filtered protein was buffer exchanged into 50 mM NaCl,10 mM Tris⋅HCl, pH 7.0, and concentrated to 15 mg⋅mL−1 for crystallization.
Crystallization of H3-HAs.
Diffraction-quality crystals were generated following the addition of 5,000 U of Endo H (NEB) to 1.5 mg of purified protein and incubation at room temperature for 16 h to remove glycans. After deglycosylation, Finland and Hong Kong H3 HA crystals routinely diffracting to 1.85 Å were obtained from 200 mM KCl, 100 mM Hepes, pH 7.5, 30% (wt/vol) pentaerythritol propoxylate (5/4 PO/OH). To generate HA–receptor complexes, HA crystals were soaked in crystallization solution supplemented with 60 mM receptor analogs 2,3SLN, 2,6SLN, or LS-tetrasaccharide c (LSTc) (Dextra) for 16 h. Crystals of the 1968 X31 H3 hemagglutinin were regenerated following the published protocol (30), and crystals were soaked in crystallization solution supplemented with 12 mM LSTc for 16 h. All crystals were frozen directly from the drop and diffraction data collected using synchrotron beamlines at DIAMOND.
Structure Determination.
Diffraction data were indexed and integrated in iMOSFLM (31); the space group was verified by the CCP4 program Pointless (32) before being scaled by Scala (32). Molecular replacement was performed in Phaser (33) using the H3 X31 (1968) HA structure (30) as the search model, and one HA molecule was located by molecular replacement giving a solvent content of 61%. Structures were subsequently built in Coot (34) and refined in Refmac (35) with TLS parameters generated by the TLS Motion Determination server (http://skuld.bmsc.washington.edu/~tlsmd/) (36, 37) and 5% reflections of the merged dataset were set aside for calculating free R factor (Rfree). Model quality was validated using MolProbity (molprobity.biochem.duke.edu/) (38). Crystallographic statistics are summarized in Table S2.
Supplementary Material
Acknowledgments
We thank members of the National Institute for Medical Research WHO Collaborating Centre for Reference and Research on Influenza team for their assistance during the course of these studies and Lesley Haire for assistance with crystallization. This work was funded by the Medical Research Council through programmes U117585868, U117512723, U117584222, and U117570592.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2YPG, 2YP2–2YP5, 2YP7, 2YP8, and 2YP9); and the gene data reported in this paper have been deposited in the Global Initiative on Sharing All Influenza Data database, http://platform.gisaid.org/epi3/ (accession nos. EPI242151, EPI397685http://gisaid.org/EPI/397686http://gisaid.org/EPI/397687–EPI397688, EPI302190, and EPI287110).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218841110/-/DCSupplemental.
References
- 1.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 2.Underwood PA, Skehel JJ, Wiley DC. Receptor-binding characteristics of monoclonal antibody-selected antigenic variants of influenza virus. J Virol. 1987;61(1):206–208. doi: 10.1128/jvi.61.1.206-208.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniels PS, et al. The receptor-binding and membrane-fusion properties of influenza virus variants selected using anti-haemagglutinin monoclonal antibodies. EMBO J. 1987;6(5):1459–1465. doi: 10.1002/j.1460-2075.1987.tb02387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniels RS, et al. Antigenic analyses of influenza virus haemagglutinins with different receptor-binding specificities. Virology. 1984;138(1):174–177. doi: 10.1016/0042-6822(84)90158-2. [DOI] [PubMed] [Google Scholar]
- 5.Aytay S, Schulze IT. Single amino acid substitutions in the hemagglutinin can alter the host range and receptor binding properties of H1 strains of influenza A virus. J Virol. 1991;65(6):3022–3028. doi: 10.1128/jvi.65.6.3022-3028.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das SR, et al. Fitness costs limit influenza A virus hemagglutinin glycosylation as an immune evasion strategy. Proc Natl Acad Sci USA. 2011;108(51):E1417–E1422. doi: 10.1073/pnas.1108754108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hensley SE, et al. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science. 2009;326(5953):734–736. doi: 10.1126/science.1178258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medeiros R, Escriou N, Naffakh N, Manuguerra JC, van der Werf S. Hemagglutinin residues of recent human A(H3N2) influenza viruses that contribute to the inability to agglutinate chicken erythrocytes. Virology. 2001;289(1):74–85. doi: 10.1006/viro.2001.1121. [DOI] [PubMed] [Google Scholar]
- 9.Nobusawa E, Ishihara H, Morishita T, Sato K, Nakajima K. Change in receptor-binding specificity of recent human influenza A viruses (H3N2): A single amino acid change in hemagglutinin altered its recognition of sialyloligosaccharides. Virology. 2000;278(2):587–596. doi: 10.1006/viro.2000.0679. [DOI] [PubMed] [Google Scholar]
- 10.Stevens J, et al. Receptor specificity of influenza A H3N2 viruses isolated in mammalian cells and embryonated chicken eggs. J Virol. 2010;84(16):8287–8299. doi: 10.1128/JVI.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu B, Zhou H, Chan W, Kemble G, Jin H. Single amino acid substitutions in the hemagglutinin of influenza A/Singapore/21/04 (H3N2) increase virus growth in embryonated chicken eggs. Vaccine. 2006;24(44–46):6691–6693. doi: 10.1016/j.vaccine.2006.05.062. [DOI] [PubMed] [Google Scholar]
- 12.Oh DY, Barr IG, Mosse JA, Laurie KL. MDCK-SIAT1 cells show improved isolation rates for recent human influenza viruses compared to conventional MDCK cells. J Clin Microbiol. 2008;46(7):2189–2194. doi: 10.1128/JCM.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asaoka N, et al. Low growth ability of recent influenza clinical isolates in MDCK cells is due to their low receptor binding affinities. Microbes Infect. 2006;8(2):511–519. doi: 10.1016/j.micinf.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Owen RE, et al. Alterations in receptor binding properties of recent human influenza H3N2 viruses are associated with reduced natural killer cell lysis of infected cells. J Virol. 2007;81(20):11170–11178. doi: 10.1128/JVI.01217-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogers GN, et al. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304(5921):76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 16.Skehel JJ, et al. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc Natl Acad Sci USA. 1984;81(6):1779–1783. doi: 10.1073/pnas.81.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abe Y, et al. Effect of the addition of oligosaccharides on the biological activities and antigenicity of influenza A/H3N2 virus hemagglutinin. J Virol. 2004;78(18):9605–9611. doi: 10.1128/JVI.78.18.9605-9611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arinaminpathy N, Grenfell B. Dynamics of glycoprotein charge in the evolutionary history of human influenza. PLoS One. 2010;5(12):e15674. doi: 10.1371/journal.pone.0015674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisen MB, Sabesan S, Skehel JJ, Wiley DC. Binding of the influenza A virus to cell-surface receptors: Structures of five hemagglutinin-sialyloligosaccharide complexes determined by X-ray crystallography. Virology. 1997;232(1):19–31. doi: 10.1006/viro.1997.8526. [DOI] [PubMed] [Google Scholar]
- 20.Lin YP, et al. Neuraminidase receptor binding variants of human influenza A(H3N2) viruses resulting from substitution of aspartic acid 151 in the catalytic site: A role in virus attachment? J Virol. 2010;84(13):6769–6781. doi: 10.1128/JVI.00458-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barr IG, et al. Epidemiological, antigenic and genetic characteristics of seasonal influenza A(H1N1), A(H3N2) and B influenza viruses: Basis for the WHO recommendation on the composition of influenza vaccines for use in the 2009-2010 Northern Hemisphere season. Vaccine. 2010;28(5):1156–1167. doi: 10.1016/j.vaccine.2009.11.043. [DOI] [PubMed] [Google Scholar]
- 22.World Health Organization Recommended composition of influenza vaccines for use in the 2012 southern hemisphere influenza season. Wkly Epidemiol Rec. 2011;86(42):457–468. [PubMed] [Google Scholar]
- 23.World Health Organization Recommended composition of influenza virus vaccines for use in the 2012–2013 northern hemisphere influenza season. Wkly Epidemiol Rec. 2012;87(10):83–95. [PubMed] [Google Scholar]
- 24.Fleming DM, Elliot AJ. Lessons from 40 years’ surveillance of influenza in England and Wales. Epidemiol Infect. 2008;136(7):866–875. doi: 10.1017/S0950268807009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matrosovich M, Matrosovich T, Carr J, Roberts NA, Klenk HD. Overexpression of the alpha-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J Virol. 2003;77(15):8418–8425. doi: 10.1128/JVI.77.15.8418-8425.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci USA. 2000;97(11):6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matrosovich M, Matrosovich T, Garten W, Klenk HD. New low-viscosity overlay medium for viral plaque assays. Virol J. 2006;3:63. doi: 10.1186/1743-422X-3-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruigrok RW. 1998. Structure of influenza A, B, and C viruses. Textbook of Influenza, eds Nicholson KG, Webster RG, Hay AJ (Blackwell Science, Oxford), pp 29–42.
- 29.Stevens J, et al. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303(5665):1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 30.Watowich SJ, Skehel JJ, Wiley DC. Crystal structures of influenza virus hemagglutinin in complex with high-affinity receptor analogs. Structure. 1994;2(8):719–731. doi: 10.1016/s0969-2126(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 31.Leslie AGW, Powell HR. Processing diffraction data with MOSFLM. NATO Sci Ser II Math. 2007;245:41–51. [Google Scholar]
- 32.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 33.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 4):439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 37.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Cryst. 2006;39:109–111. [Google Scholar]
- 38.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.