The recent outbreak of Shiga toxin-producing Escherichia coli in Europe resulted in 3,816 cases in Germany (1), with a smaller outbreak in France resulting in 24 cases (2). Grad et al. identified 19 single nucleotide polymorphisms (SNPs) in seven isolates from the French outbreak but only two SNPs in four isolates from the German outbreak (3). They conclude that the diversity in the small French outbreak is much larger than in the German outbreak. However, we argue that this conclusion is not justified.
First, in their analysis, Grad et al. discarded all SNPs found solely in the reference genome TY2482 (4) as sequencing errors, but our careful reanalysis identifies two genuine SNPs in TY2482. This doubles the number of SNPs in the few German isolates examined. Second, we found 18 SNPs in eight genomes from strains isolated from Swedish patients who had visited Germany during the outbreak (sequenced with Illumina; coverage >250 and all SNPs validated by PCR and Sanger sequencing). Thus, there is a similar genomic diversity in isolates originating from Germany as seen in the French isolates (3). Furthermore, synonymous substitutions accounted for 31.2% of SNPs in coding regions and another 11.1% at intergenic sites in the German outbreak strains, which is very similar to 36% and 14.3%, respectively, for the French strains. We conclude that the average number of SNPs per genome and their substitution patterns are similar in the French and German outbreaks. This argues against the hypothesis that mutation rates vary in the two bacterial populations, as proposed by Grad et al. (3).
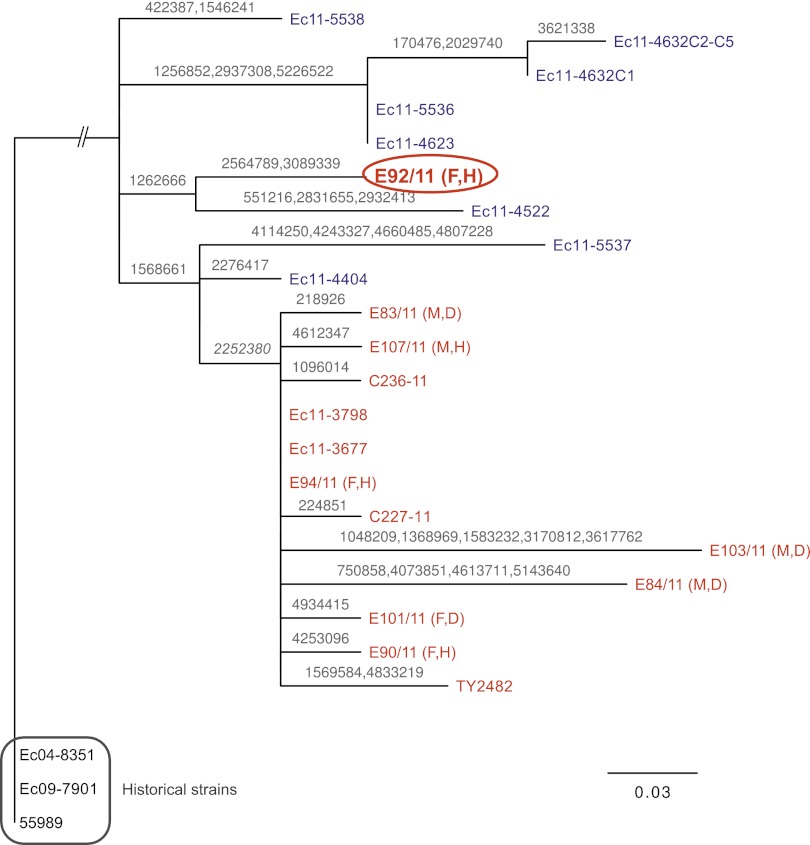
Grad et al. further suggested that all German strains went through a bottleneck and form a clade nested within the diversity of the French strains (3). This hypothesis is based on the presence of a single SNP identified in all German strains and some French strains (3). However, the Swedish isolate E92/11 does not contain this particular SNP, but instead shares an SNP with the French isolate Ec11-4522 (Fig. 1). All other Swedish strains, including E107/11, contain the SNP specific for the German strains. This difference is notable because E92/11 and E107/11 were isolated from patients who had traveled together to Germany. They and several others in the traveling group developed disease symptoms shortly before the peak of the outbreak, suggesting that they all consumed the same infected sprouts.
Fig. 1.
Phylogeny of the French (blue) and German (red) outbreak strains. The tree was inferred by maximum likelihood using an alignment of the polymorphic positions within the outbreak strains. Numbers on branches inside the outbreak clade indicate SNP positions. Letters beside the Swedish–German strains refer to sex and symptoms (H, hemolytic uremic syndrome; D, diarrhea). Strain E92/11 was obtained from the only fatal case reported in Sweden. Strains Ec04-8351, Ec09-7901, and 55989 were isolated independently of the outbreak and used to root the tree. Raw reads for all strains sequenced in this study have been deposited in the Sequence Read Archive at the National Center for Biotechnology Information under study no. SRP008458.
These findings strongly suggest that the French and German outbreaks originated from a common source that contained at least two different genotypes. Thus, the German outbreak sequences are not simply nested in the French ones as proposed by Grad et al. (3). We favor the hypotheses that (i) there was an uneven distribution of genotypes in the infected seeds or (ii) some genotypes underwent clonal expansion after the seeds were distributed (3). However, biased sampling also cannot be ruled out: 29% of the patient population (seven of 24) has been sampled in the French outbreak, compared with only 0.3% (13 of ∼4,000) in the German outbreak. A broader sampling, particularly of German strains, is clearly needed.
Acknowledgments
We thank K. Näslund for her technical support and the Genomic Sequencing Platform at SciLifeLab (Uppsala, Sweden) for Illumina sequencing. This work was supported by Swiss National Science Foundation Grant PA00P3_131491 (to L.G.) and grants from the Swedish Research Council (to L.E. and S.G.E.A) and Science for Life Laboratory (to S.G.E.A.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Frank C, et al. HUS Investigation Team Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med. 2011;365(19):1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 2.King LA, et al. Outbreak of Shiga toxin-producing Escherichia coli O104:H4 associated with organic fenugreek sprouts, France, June 2011. Clin Infect Dis. 2012;54(11):1588–1594. doi: 10.1093/cid/cis255. [DOI] [PubMed] [Google Scholar]
- 3.Grad YH, et al. Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011. Proc Natl Acad Sci USA. 2012;109(8):3065–3070. doi: 10.1073/pnas.1121491109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rohde H, et al. E. coli O104:H4 Genome Analysis Crowd-Sourcing Consortium Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N Engl J Med. 2011;365(8):718–724. doi: 10.1056/NEJMoa1107643. [DOI] [PubMed] [Google Scholar]