Abstract
Migrating cells possess intracellular gradients of active Rho GTPases, which serve as central hubs in transducing signals from extracellular receptors to cytoskeletal and adhesive machinery. However, it is unknown whether shallow exogenously induced intracellular gradients of Rho GTPases are sufficient to drive cell polarity and motility. Here, we use microfluidic control to generate gradients of a small molecule and thereby directly induce linear gradients of active, endogenous Rac without activation of chemotactic receptors. Gradients as low as 15% were sufficient not only to trigger cell migration up the chemical gradient but to induce both cell polarization and repolarization. Cellular response times were inversely proportional to the steepness of Rac inducer gradient in agreement with a mathematical model, suggesting a function for chemoattractant gradient amplification upstream of Rac. Increases in activated Rac levels beyond a well-defined threshold augmented polarization and decreased sensitivity to the imposed gradient. The threshold was governed by initial cell polarity and PI3K activity, supporting a role for both in defining responsiveness to Rac activation. Our results reveal that Rac can serve as a starting point in defining cell polarity. Furthermore, our methodology may serve as a template to investigate processes regulated by intracellular signaling gradients.
Keywords: cell signaling, synthetic biology, chemotaxis, signal transduction
Directional motility is an intrinsic ability of many eukaryotic cells to migrate to predetermined locations in an efficient manner. Migration bias is tuned by the detection of various guidance cues that often form spatial gradients. The extracellular gradients of diffusing or surface-bound ligands can lead to spatially graded occupancy of extracellular receptor (1). The spatial asymmetry in receptor occupancy is subsequently translated into an intracellular gradient of polarity effectors that can modify cytoskeleton and lead to the development of an asymmetrical cell morphology with functionally distinct front and rear “compartments.” Remarkably, many cell types can accurately detect less than a 5% difference between ligand concentration at the cell front and back (1–3). This exquisite sensitivity suggests that intracellular amplification of extracellular cues may be necessary. Indeed, various groups have demonstrated the existence of local positive feedback loops (4–7) and mutual inhibition between different regulators (8, 9) as likely candidates for response amplification. However, recent studies have also shown that directional motility can still be achieved, albeit less efficiently, when molecules, once thought to be indispensable, involved in putative amplification mechanisms are removed (10, 11). Thus, the various functions of signaling components associated with directed migration still need to be resolved.
An attractive method for resolving the roles of signaling network components in both spatial cue sensing and directed cell motility is direct activation of these components in a spatially constrained and rapid manner, independent of initiation of upstream, receptor-level signaling. Using this principle, a variety of studies have used optical activation to identify the small Rho GTPase Rac (9, 12–16), cofilin (17), thymosin Β4 (18), and calcium (19) as key components that are sufficient to direct cellular motility. However, an important caveat to these studies has been the reliance on highly localized activation that can create artificial regional amplification of target protein activity. In contrast, in more physiological settings, a cell processes a shallow gradient of an external cue into a graded intracellular response, as reflected in polarized effectors (20), including those of the small Rho GTPase family (21–23). Thus, it remains unclear if an induced shallow gradient of an active motility signaling component is capable of reconstituting cell polarization and motility. In particular, it is unknown if such perturbations are sufficient to override or enhance endogenous intracellular signaling of the same component. Finally, localized activation of signaling processes presents considerable challenges to quantitative analysis and coupling to detailed computational models developed to describe more natural, spatially distributed signaling events.
To address these questions, we created microfluidic devices permitting generation of precise gradients of extracellular cues (24) and interfaced them with a rapamycin-induced dimerization system (25). In this system, the addition of rapamycin leads to dimerization of two intracellularly transduced molecular components, FK506 binding protein (FKBP) and the rapamycin binding domain of FKBP-rapamycin binding protein (FRB) (26). Localization and signaling motifs can be linked to either domain, allowing spatial-temporal control of protein function. We used our combined system to study the effects of a rapidly induced intracellular gradient of activated Rac, which is an important regulator of cell polarity (27) and has been shown previously to induce migration when locally activated (9, 12–16).
Results
System Design.
To activate endogenous Rac directly, we introduced two constructs into HeLa cells: a cytoplasm-localized effector unit consisting of YFP-tagged TIAM1, a Rac guanine nucleotide exchange factor (GEF) conjugated to FKBP (YF-TIAM1) and an anchor unit at the cell membrane, Lyn11-FRB (LDR). The introduction of rapamycin dimerizes these modified molecular components, thereby bringing TIAM1 in close proximity to the cell membrane, where it activates endogenous Rac (25) (Fig. 1A). Due to the chemical nature of the activation, this system is amenable to generation of a gradient of Rac activity through microfluidic production of rapamycin gradients. Microfluidic tools have recently been used for the control of complex gradients of extracellular cues (24, 28, 29), cellular localizations (30, 31), and shaping cell morphologies (32, 33).
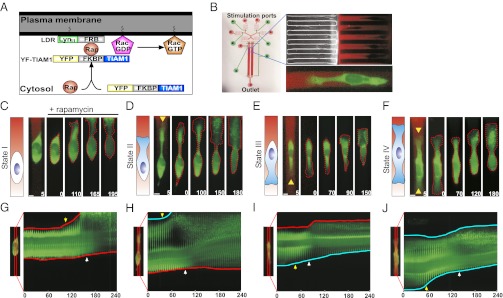
Fig. 1.
Graded activation of Rac directs cellular polarity. (A) Schematic of the mechanism of Rac activation by rapamycin-induced heterodimerization. (B) Microfluidic device used to generate linear gradients of rapamycin with a sample image of the microchannels seeded with individual HeLa cells and the corresponding gradient visualized with Alexa 594 dye. Ports are labeled according to function. The red layer of the device is the fluid flow layer, and the green layer is the control valve layer. Alexa 594 dye is used to visualize the gradient (red) in all subsequent images. A sample image of a cell transfected with the Rac activator, YF-TIAM1, experiencing a gradient of rapamycin is shown below. (C–F) Four polarity states observed after the attachment period with respect to the direction of the imposed rapamycin gradient and associated polarization responses to the gradient of rapamycin. Images are rotated by 90° to aid in visualization. Cartoons illustrate the polarity of the associated state and the direction of the gradient. The green color indicates expression of YF-TIAM1. Yellow arrowheads denote the initial direction of polarity. The rapamycin gradient is shown in the 5-min image and removed in subsequent images for clarity. Red dotted lines highlight evolving changes in cell morphology. Times are given in minutes. (Scale bars, 10 μm.) (G–J) Kymographs taken across cell centers (specified in accompanying image) illustrate the morphological changes of the corresponding cells in C–F over the experimental period. Blue lines trace initially polarized faces, and red lines trace initially unpolarized faces. Yellow arrowheads denote the initial response time, and their location indicates which cell face was the first to change in the gradient. White arrowheads indicate the late polarization time. Times are given in minutes.
We applied a previously developed strategy for imposing diffusion-based linear gradients onto cells housed within narrow channels (24, 33). Specifically, the devices contained a series of 6-μm tall microchannels for cell experimentation, flanked by 130-μm tall main flow-through channels (device operation is described in SI Appendix, Fig. S1A and Experimental Setup). Actuation of flow led to the development of a linear gradient across the shallow channels due to uneven stimulus concentration in the flow-through channels (Fig. 1B). The microchannels were designed to be on the order of a cell diameter to relegate cells to a uniaxial phenotype (Fig. 1B). HeLa cells introduced into the microchannels settled into random locations, and after a 3- to 4-h attachment period, they were categorized into different polarity states according to the existence of a lamellipodium (state I, no lamellipodium) and its direction (state II, lamellopodium toward the high side of the gradient; state III, lamellipodium toward the low side of the gradient; and state IV, lamellopodia toward both sides of the gradient) (Fig. 1 C–F). The overall distribution of phenotypes skewed toward cells with a single leading edge lamellipodium, indicating an intrinsic preference for directed motility (SI Appendix, Fig. S1B).
Direct Generation of Active Rac Gradients.
To evaluate HeLa cell responses to synthetic generation of an intracellular gradient of active Rac, we exposed cells to a linear gradient of rapamycin (0–2 nM across the channel or 0.01 nM/μm, yielding front/back concentration differences ranging from ∼15–92% across varied cell lengths) visualized by Alexa 594, a fluorescent dye with a similar molecular weight (Fig. 1B and SI Appendix, Fig. S1C). We chose this concentration range to avoid saturating the FKBP-FRB system with rapamycin [KD = 12 nM (34)]. One consequence of using these concentration values was a slow-down of the response times compared with previously reported values for saturating uniform rapamycin inputs (25). However, the slower response times allowed a better resolution of the effects of gradually accumulating Rac activity and were far below those reported to affect the function of the mammalian target of rapamycin (35). As a control, we compared basal cell motility with and without rapamycin and found that the addition of rapamycin had negligible effects (SI Appendix, Fig. S1 D and E). We tracked both the initial state and subsequent cell responses by imaging over a 4-h time period.
Strikingly, we found that the shallow linear gradient of rapamycin could direct motility of cells in all initial polarity states in the direction up the gradient (Fig. 1 C–F and Movie S1). Unpolarized cells (state I) and bipolar cells (state IV) demonstrated symmetry, breaking with either the establishment of a leading edge or the enhancement of one lamellipodium and retraction of the other, respectively (Fig. 1 C and F and Movie S1). Cells already polarized in the direction of the gradient (state II) exhibited widening and extension of the leading lamellipodium and movement up the gradient (Fig. 1D and Movie S1). Interestingly, cells initially oriented in the direction opposite of the gradient (state III) repolarized (Fig. 1E and Movie S1). In all states, the initiation of migration was followed by a pronounced enhancement of cell polarity (Fig. 1 G–J). The directed cell polarization and migration responses were not observed in untransfected cells (SI Appendix, Fig. S1F) or if the rapamycin gradient was not imposed (SI Appendix, Fig. S1G). For validation of our system, we applied linear gradients of rapamycin to similarly transfected MTLn3 cells, a rat mammary adenocarcinoma line used to assay chemotaxis to EGF in vitro (36) and in vivo (37) (SI Appendix, Fig. S2). MTLn3 cells exhibited initial polarity states similar to those seen in HeLa cells and polarized toward gradients of rapamycin (SI Appendix, Fig. S2). Our data suggest that our system is applicable across multiple cell types and can be used to study signaling pathways regulating chemotaxis.
To verify that the externally imposed rapamycin gradient was translating into a graded change in active Rac across cells, we carried out two control experiments. First, we sought to confirm that the Rac activator, YF-TIAM1, translocated to the membrane in a graded fashion over time. Our results above indicated that translocation of YF-TIAM1 caused ruffling and substantial changes in cell morphology that obscure translocation; therefore, we used a YFP-tagged FKBP (YF) without TIAM1 to assess translocation. YF was previously shown to translocate with similar kinetics to YF-TIAM1 (25), and therefore could serve as a suitable proxy. We tracked the membrane-to-cytoplasm ratio of fluorescence intensities across the length of cells in a gradient of rapamycin over time and found that membrane translocation of YF was increasing significantly in a graded manner, with higher translocation toward the high side of the gradient (SI Appendix, Fig. S3 A–F and Movie S2). The membrane-to-cytoplasm ratio curves increased over time for all states but retained similar slopes (SI Appendix, Fig. S3 D–F). We did not observe a similar response when DMSO was substituted for rapamycin (SI Appendix, Fig. S3 G and H). In both conditions, we did not observe any significant morphological changes, indicating that translocation of FKBP constructs without effectors does not perturb morphology. Having verified that the rapamycin gradient induced graded membrane translocation of FKBP constructs, we used a Raichu Rac FRET sensor (38) to monitor the resulting changes in Rac activity. In these experiments, we used a mCherry-tagged FKBP-TIAM1 (MCHF-TIAM1) to activate Rac. At a basal level before stimulation, state I cells did not show polarized Rac activity (SI Appendix, Fig. S4A), whereas state II and state III cells exhibited higher levels of active Rac at lamellipodia (SI Appendix, Fig. S4 B and C). On introduction of the rapamycin gradient, we observed increases in Rac activity in the direction of the gradient in all cells, whereas state III cells exhibited an additional decrease in Rac activity in the original lamellipodium (SI Appendix, Fig. S4 A–C and Movie S3). We quantified the average Rac activity across cells over time and found that there was a gradual and spatially graded increase in Rac activity in all cell states (SI Appendix, Fig. S4 D–F). In particular, state III cells exhibited a sharp increase in Rac activity in the newly formed protrusion and a decrease in the initial opposite-facing protrusion (SI Appendix, Fig. S4F). As a control, we quantified Rac activity across cells that did not express the Rac activator, MCHF-TIAM1, but had expression of the Rac FRET sensor. Under these conditions, we did not observe any significant changes in Rac activity over time in gradients of rapamycin (SI Appendix, Fig. S4 G and H). Overall, our results suggested that an exogenously applied linear gradient of rapamycin could result in a graded increase in Rac activity, which was sufficient to direct motility and polarization of cells from a variety of preexisting polarity phenotypes.
For further validation of our system, we compared cellular responses to graded rapamycin with responses to uniform rapamycin stimulation. Cells given a uniform stimulus for the entire experimental period displayed extensive uniform flattening with little net motility (SI Appendix, Fig. S5A). This result was in agreement with previous experiments showing that differentiated HL-60 cells exposed to spatially uniform Rac stimulation displayed membrane ruffling around the entire cell periphery (6). For a more detailed comparison of the effects of graded and uniform Rac activation, we exposed state II cells to a rapamycin gradient (0.01 nM/μm) for 2 h, subsequently followed by uniform stimulation (2 nM rapamycin) for 3 h thereafter. As expected, cells polarized and moved in a biased fashion during gradient stimulation; however, on the switch to uniform stimulation, cells started forming protrusions at the rear and showed a decrease in the length of the front (SI Appendix, Fig. S5 B–D and Movie S4). This effect indicated the importance of persistent gradient input but might have also reflected a gradual saturation of rapamycin binding sites after prolonged treatment. We explored the latter possibility by running the converse experiment, exposing cells to spatially uniform rapamycin (2 nM) for 2 h followed by a rapamycin gradient (0.01 nM/μm) for 3 h (SI Appendix, Fig. S5E). Protrusions developed on both sides of cells during uniform stimulation, and subsequently could not be biased by the gradient (SI Appendix, Fig. S5E). However, cells exposed to a lower uniform stimulation (1 nM) were able to polarize toward the ensuing rapamycin gradient (0.01 nM/μm) (SI Appendix, Fig. S5F). In combination, these results indicate that cell polarity can be acutely sensitive to changes in rapamycin gradients as long as the rapamycin exposure does not exceed a threshold level. Below, we explore the effects of the rapamycin dose and exposure time on cell responses in detail. These results suggest that the spatial restriction of Rac activity within a cell is important for maintaining polarization.
Mathematical Modeling of Graded Rac Inputs.
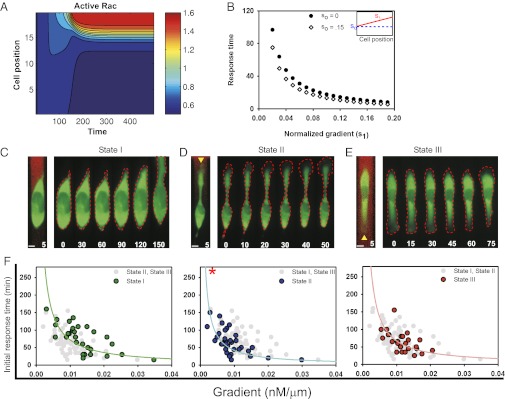
To better understand cell responses to the exogenously imposed graded Rac activation, we developed a mathematical model of cell polarity. The model is based on a simple scheme of Rac-RhoA-Cdc42 small GTPase and phosphoinositide interactions, expanding on earlier modeling studies (39, 40) (modeling details are provided in SI Appendix, Model Supplement). The model made three important qualitative predictions: A spatially graded activation input can trigger initial polarization of Rac activity (Fig. 2 A and B); the timing of that initial Rac polarization is strongly dependent on the input gradient and weakly dependent on the average input (Fig. 2 B and C); and antagonism between the activities of Rac and Rho small GTPases can trigger a phase transition-like change to a substantially more asymmetrical polarization, which can be stably maintained as long as the activity of Rac remains high enough (Fig. 2A). This transition is addressed in more detail below. Model details are provided in SI Appendix, Model Supplement. Our experimental setup allowed direct examination of these predictions.
Fig. 2.
Analysis of the initial response time. (A) Sample kymograph of the mathematical model simulation under graded Rac stimulation. (B) Model response time vs. normalized gradient (s1) for two different values of so (input). (C–E) Kymographs chronicle typical changes in cell morphology seen during early time periods. The first image before each kymograph depicts the gradient (red) that the cell is experiencing as visualized with Alexa 594 dye. The gradient is not shown in resulting images to promote clarity of morphological changes. The green color indicates expression of YF-TIAM1. Red dotted lines highlight evolving cell boundaries. Yellow arrowheads indicate initial polarities. Times are given in minutes. (Scale bars, 10 μm.) (F) Dependence of initial response times on gradient values. States are color-coded: state I, green (n = 29); state II, blue (n = 37); and state III, red (n = 27). In each plot, the colored dots highlight the dependence of that particular state and the gray dots illustrate where the response times of the other states fall. Data are fitted based on simulation results. Spearman correlation coefficient: state I = −0.679, state II = −0.655, and state III = −0.583. Τhe asterisk indicates a statistically significant difference (state II vs. state I, P < 1e-4; state II vs. state III, P < 1e-3) between the state II curve vs. the other states. There is no statistical difference between state I and state III (P = 0.54). Both tests were carried out using an F test.
The Effect of Rac Gradient Steepness on Timing of Cellular Responses.
To validate the model predictions, we next examined the correlates of the initiation of directed cell migration and their dependencies on the local gradient and average value of rapamycin input. HeLa cell responses were characterized by either the retraction of a leading lamellipodium in cells polarized in the direction opposite that of the gradient (state III) or the extension of a preexisting (state II) or new (state I) lamellipodium in the direction of the gradient (Fig. 2 C–E). To evaluate these effects quantitatively, we examined the width and length of the front and rear sides of cells in all states through the entire stimulation period, along with the respective concentrations experienced at each side (SI Appendix, Fig. S6). Cells that exhibited bipolar phenotypes (state IV) were relatively rare (Fig. 1F and SI Appendix, Fig. S1B), and therefore were excluded from subsequent analysis. Our analysis revealed that these initial directed migration response times, in agreement with the model predictions, were indeed inversely dependent on the steepness of the rapamycin gradient across each cell (Fig. 2F). This trend was seen across all polarity states (Spearman correlation coefficients: −0.679, −0.655, and −0.583 for states I, II, and III, respectively). Similar dependencies were also observed in MTLn3 cells (SI Appendix, Fig. S7 A–C). The initial response times of state I cells also showed a discernible but much weaker dependency on the average rapamycin concentration, consistent with the model predictions, whereas cells in states II and III had relatively weaker dependencies (SI Appendix, Fig. S8 A–C) (Spearman correlation coefficients: −0.582, −0.478, and −0.377 for states I, II, and III, respectively). State II cells had consistently shorter initial response times for any given gradient steepness compared with those in states I and III (Fig. 2F and SI Appendix, Fig. S9) (F test: state II vs. state I, P < 0.0001; state II vs. state III, P < 0.001), whereas state I and III cells behaved similarly (state I vs. state III, P = 0.54). For example, when comparing the average initial response times at a gradient of 0.01 nM/μm ± 0.001, state II cells had an average initial response time of 51 min, whereas state I cells had an average initial response time of 98 min and state III cells had an average initial response time of 71 min. These results suggest that cells are able to make migration decisions rapidly in the presence of Rac activity gradients overlapping with their initial polarization state. Together, our data demonstrate that the magnitude of active Rac gradients can influence the timing of the onset of directed cell motility.
In addition to the initiation of biased cell migration, spatially graded Rac activation eventually triggered a striking enhancement in cell polarization, with a substantial enlargement of the directed leading lamellipodium (Fig. 3A). This unexpected morphological change was consistent with the model prediction that crossing a threshold of Rac activity triggers a rapid, strong, and stable polarization, akin to a phase transition. According to the model, a gradual variation of Rac GEF levels causes an abrupt transition from a moderately polarized to highly polarized state, expressed mathematically as a bifurcation in the model response (Fig. 3B). We hypothesized that the use of low rapamycin concentrations allowed us to observe this effect by gradually titrating intracellular Rac GEF and Rac activity levels, ultimately leading to this strong polarization. The gradual stimulus-induced Rac GEF build-up was expected to follow a simple mathematical representation of accumulation of Rac GEF concentration over the duration, t, of exposure to rapamycin:
where k is the constant defining tripartite rapamycin–FKBP–FRB complex formation. This expression allowed us to test the hypothesized existence of a Rac activity threshold mediating enhanced polarization.
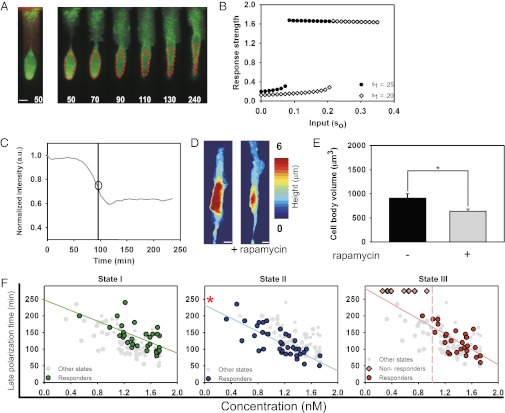
Fig. 3.
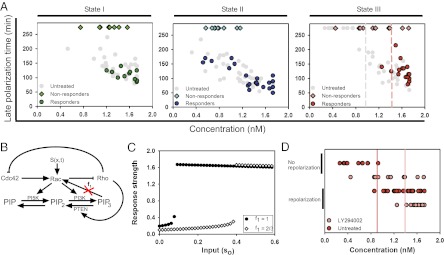
Analysis of the late response time. (A) Kymograph depicts the change in the lamellipodium directed by the gradient and dimming seen in the cell body during the late polarization time. The image preceding the kymograph illustrates the gradient (red), visualized with Alexa 594, received by the cell. The green color visualizes expression of YF-TIAM1. Red dotted lines indicate where the fluorescent values in C are taken from. Times are given in minutes. (Scale bar, 10 μm.) (B) Simulations show response strength vs. input (so) for two gradient levels (s1); note bifurcations at distinct so values. Response strength is defined as the ratio of Rac activity at the front vs. the back in the model cell. (C) Intensity of the cell body normalized to the initial time point. Intensity values are taken as the mean of the fluorescence intensity of the area enclosed by the red trace in A. The drop line indicates the late polarization time, with a circle highlighting the inversion of response used to define this time. (D) Three-dimensional reconstruction of confocal slices of the same cell taken before and after rapamycin addition. The “post” cell image was taken 240 min after treatment. (Scale bars, 10 μm.) (E) Cell body volume before and after rapamycin addition. The data show the mean of n = 9 cells, and error bars show the SEM. The asterisk denotes a statistically significant difference (P = 0.019), using a two-sided Student’s t test. (F) Late response time as a function of mean concentration: state I, green (n = 29); state II, blue (n = 37); and state III, red (n = 27). The response times of other states are superimposed on each plot in gray. The pink drop line in the state III plot demarcates the separation point between unresponsive cells and responsive cells. Pearson correlation coefficient of linear regressions: state I = −0.608, state II = −0.783, and state III = −0.698. The red asterisk denotes a statistically significant difference (state II vs. state I, P < 1e-4; state II vs. state III, P < 1e-4) between the y intercept of the linear regression for state II vs. the y intercepts of the regression data from other states. There is no significant difference between the y intercept of state I vs. state III (P = 0.13). Both statistical analyses were carried out using an ANCOVA test.
One of the immediate consequences of the expansion of the lamellipodium in the direction of the rapamycin gradient was an apparent dimming of the YFP fluorescence signal from the cell body when observed with a wide-field epifluorescence microscope (Fig. 3 A and C). The dimming of the fluorescence intensity was likely due to a redistribution of cytoplasmic volume from the cell body to the expanding lamellipodium and an increase in the translocation of YF–TIAM1 complexes to the membrane (Fig. 3 D and E). We thus used the fluorescence intensity of the cell body as a metric for the timing of late polarization. Our analysis indicated that the late polarization time exhibited a linear dependency on the average local rapamycin concentration (Fig. 3F) (Pearson correlation coefficients: −0.608, −0.783, and −0.698 for states I, II, and III, respectively), in agreement with model predictions. MTLn3 cells also showed similar dependencies on mean rapamycin concentrations (SI Appendix, Fig. S7 D–F). A weaker dependence on the sharpness of the rapamycin gradient, consistent with the model, was also detected in these experiments (SI Appendix, Fig. S8 D–F) (Pearson correlation coefficients: 0.103, −0.511, and −0.228 for states I, II, and III, respectively). As with observations of early cell responses, state II cells reached late polarization significantly faster than cells in other states for a given rapamycin concentration (Fig. 3F and SI Appendix, Fig. S10) [analysis of covariance (ANCOVA) test of y intercept: state II vs. state I, P < 0.0001; state II vs. state III, P < 0.0001], whereas the difference between the other two states was negligible (state I vs. state III, P = 0.13). We also found that a subpopulation of state III cells exposed to lower concentrations of rapamycin completely failed to reach late polarization (Fig. 3F). In combination, these results suggested the existence of a threshold for late polarization, which is variable across individual cells and dependent on the initial polarity state.
Transient Graded Rac Activation.
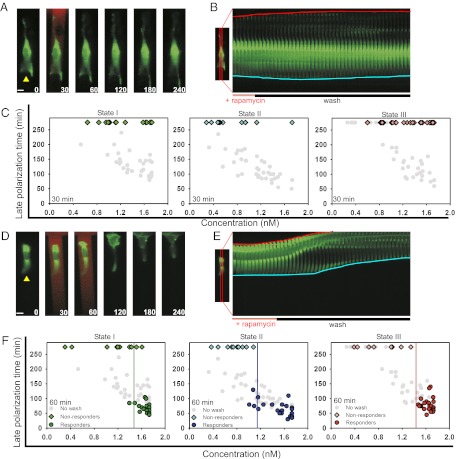
Eq. 1 suggests that the duration of rapamycin exposure is as critical as the local rapamycin concentration in exceeding the polarization threshold. Based on in vitro estimates of rapamycin–FKBP–FRB complex formation, the characteristic binding time estimated for 1 nM rapamycin is in the range of tens of minutes (34). To test this prediction, we varied the time interval of rapamycin stimulation, taking into account the earliest polarization time observed for the rapamycin concentrations tested (i.e., 30 min) and the estimated equilibration time above. We exposed cells to transient rapamycin gradients of 0.01 nM/μm for 30 min or 1 h, followed by perfusion of the devices with rapamycin-free media for the rest of the experiment. We found no cells undergoing late polarization after a 30 min rapamycin gradient exposure (Fig. 4 A–C and Movie S5); however, after a 1-h stimulation, we found subsets of cells in all polarity states able to undergo late polarization provided that they were exposed to sufficiently high concentrations of rapamycin (Fig. 4 D–F and Movie S6). The polarization responses were morphologically similar to those seen earlier during continuous stimulation (Fig. 4 D and E and Movie S6), with the timing to late polarization indistinguishable from that observed for continuous stimulation (Fig. 4F). Responding cells continued to polarize even after the stimulus was withdrawn, suggesting fixation of the induced polarity and migration states (Fig. 4 D–E and Movie S6) in contrast to their loss during the transition to spatially homogeneous Rac activation (SI Appendix, Fig. S5A). This behavior was in agreement with model predictions of a stable polarization beyond a Rac activity threshold, which enables cells to maintain a strongly polarized state whose direction is based on but not continuously informed by the input gradient. Using a classification algorithm to separate responding and nonresponding cells, we found that the minimum concentrations needed to elicit a response varied across the polarity states (Fig. 4F). In agreement with the earlier observation of distinct responses in state II cells, cells in this state possessed the lowest response threshold (Fig. 4F) (state I = 1.5 nM, state II = 1.1 nM, and state III = 1.4 nM). These data further support the existence of a well-defined initial polarity-dependent Rac activation threshold essential for the rapid and profound induced changes in polarized cell morphology.
Fig. 4.
Overcoming a Rac activity threshold determines the late polarization time. (A and D) Time series of representative cells stimulated with a gradient of rapamycin for 30 min and 60 min, respectively. Yellow arrowheads indicate the initial direction of cell polarity. Times are given in minutes. (Scale bars, 10 μm.) (B and E) Kymographs taken from the center of cells (specified in accompanying image) illustrate morphological changes seen in A and D, respectively. Blue lines track the initially polarized cell face, and red lines track the opposite face. Below the kymograph is the experimental scheme used to stimulate the cell. (C and F) Late polarization time for 30-min and 60-min stimulation periods for all three states, respectively. For 30 min: state I, green (n = 17); state II, blue (n = 13); and state III, red (n = 31). For 60 min: state I, green (n = 32); state II, blue (n = 37); and state III, red (n = 32). Diamonds indicate cells that did not respond within the experimental time frame (240 min), and circles indicate responsive cells. Gray dots in each plot represent the late polarization times for cells in each state from Fig. 3F. The drop lines demarcate a threshold between responding and nonresponding cells.
Inhibition of Upstream Activators.
Direct activation of Rac allows bypassing of many signaling species commonly thought to be either upstream of Rac or involved in a regulatory feedback with this molecule. A well-studied example of such a molecule is phosphatidylinositol 3,4,5-triphosphate (PIP3) (6, 41, 42). In chemoattractant gradients, PI3K is recruited to the plasma membrane and phosphorylates the abundant phosphatidylinositol 4,5-bisphosphate to yield PIP3 (2, 43). Due to spatial regulation of PI3K recruitment, PIP3 is often enriched at the front areas of migrating cells (43), displaying an intracellular gradient that is sharper than the gradient of the extracellular chemoattractant (2, 44). It is thought that PI3K can influence cell guidance through its interaction with small GTPases and actin, but the mechanism of these interactions and the resultant role of PI3K in regulating chemotaxis are still under investigation (10, 11). If, as sometimes assumed, PI3K is upstream of Rac activation in chemotactic signaling systems, its perturbations are not expected to lead to alteration of cell responses to rapamycin-based Rac GEF stimulation. If, on the other hand, PI3K forms a feedback loop with Rac (4, 6, 13) or otherwise enables Rac-mediated outputs, cell responses might be affected by its inhibition, with the change in cell behavior potentially suggesting the mechanism of PI3K regulatory involvement. We used a pharmacological inhibitor of PI3K, LY294002, to inhibit PI3K activity during stimulation with a gradient of rapamycin. As a control, we first observed the effects of LY294002 with and without rapamycin on basal cell motility and found no significant effect (SI Appendix, Fig. S1 D and E). After induction of the graded stimulus, in contrast to the responses of cells in which PI3K was not perturbed, we observed large subsets of cells exhibiting no response, both in terms of the initial and late polarization, for all three initial polarity states across various gradients and concentrations (Fig. 5A, SI Appendix, Fig. S11, and Movie S7). Interestingly, the cells that did undergo the initial migration and late polarization responses did so with the same kinetics as observed in the absence of PI3K perturbation (Fig. 5A). This was consistent with the model prediction that a decrease in simulated strength of the PI3K-mediated feedback to Rac could lead to an increased threshold for cell responsiveness, requiring a sharper effective internal Rac activity gradient for cells to respond (Fig. 5 B and C). As a consequence, the stochastic differences in internal states of the cells, defining cell sensitivity to the graded signaling input, can lead to a greater degree of cell population separation into responding and nonresponding cells, without affecting the timing of responses in responding cells.
Fig. 5.
PI3K modulates Rac-mediated polarization. (A) Late polarization time dependence against mean concentration with LY294002 treatment. State I, green (n = 21); state II, blue (n = 31); and state III, red (n = 27). Diamonds represent nonresponder cells, and circles represent responding cells. Gray dots on each plot illustrate the late polarization times seen in Fig. 3F. The gray drop line in the state III cell response plot represents the previous response threshold between nonresponding and responding untreated cells, and the colored line represents the shifted threshold following LY294002 treatment. (B) Model schematic with the red “X” indicating that feedback from PIP3 (f1) is decreased during the subsequent simulations. (C) Simulations of response strength vs. signal strength for different feedback levels (in all previous simulations, f1 = 1). (D) Response threshold for state III cells with and without LY294002 treatment, respectively. The drop lines indicate the response threshold.
The results in Fig. 3F suggested the existence of a relatively high (re)polarization threshold for state III cells. Thus, we explored whether there would be synergy between this threshold and the increase in polarization threshold caused by inhibition of PI3K. We found that this threshold was indeed shifted in the presence of PI3K inhibition to a higher rapamycin concentration level (Fig. 5D) (1.0 nM for untreated state III vs. 1.4 nM for LY294002-treated state III). These results thus support the notion that PI3K can serve to sensitize cells to spatially graded Rac activation, allowing them to exceed polarization and repolarization thresholds more readily.
Discussion
The results presented in this report argue that directly induced, spatially graded membrane translocation of a Rac activator, TIAM1, can trigger unambiguous polarization and directed movement of cells aligned with the direction of the stimulation gradient. TIAM1 is a specific Rac GEF (45), and another key regulator of polarity, CDC42, is considered upstream of Rac (46); therefore, we attribute our observed phenotypes to be originating from direct Rac activation. The gradients of the inducer of Rac activation, the exogenously added rapamycin, can be effective with values as low as 15% across the cell length, with the rates of cellular responses to the stimulation being defined by the gradient steepness. The results suggest that even mild initial Rac GEF gradients can trigger strongly polarized cell responses, potentially providing insights into the levels at which graded inputs can be amplified in the signaling network. However, the response kinetics can be enhanced with sharper input gradients (Fig. 2F) or if these gradients are amplified upstream of Rac activation.
We propose that rapamycin-induced graded Rac activation can induce qualitatively similar polarization responses to those seen in chemoattractant gradients. For example, state I cells exhibit initially low, mostly homogeneous Rac activity, followed by the induction of high Rac activity at a newly formed front when a gradient of rapamycin is applied. This pattern of Rac activity is similar to that seen in neutrophils polarizing to a gradient of f-Met-Leu-Phe (47). Additionally, state III cells repolarize when given sufficiently high gradients of rapamycin by forming a new front at the rear and retracting the previous front. This behavior is seen when chemoattractant gradients are presented at the rear of polarized cells through microfluidics or a micropipette. Chemoattractant-induced repolarization can be seen in neutrophils (48), social amoebae (49), and breast cancer cells (36). Given that rapamycin-induced graded Rac activation can mimic polarization behaviors seen with chemoattractant gradients, we believe that polarization can be defined at the level of Rac, or at least starting from the level of Rac.
The research platform described here enabled the screening of the effects of a slow variation in the total cellular Rac activity. Both a simple model describing a feedback-based interplay between small GTPases in a cell and the corresponding experimental observations support the unique finding that a rapid and pronounced transition to a much stronger degree of polarization can occur if Rac activity exceeds a threshold level. This threshold was found to be strongly affected by the initial polarization status of the responding cell. Cells initially polarized in the direction of the applied gradient have lower response thresholds, on average, than cells that are unpolarized or polarized in the opposite direction. Moreover, only a fraction of cells initially polarized away from the gradient responded to the gradient of Rac activator. These results are consistent with the following view supported by the model: An existing endogenous gradient of Rac activity in state II cells would lead to a smaller difference between the maximum local initial Rac activity within a cell and the polarization threshold value.
Our results further suggest that signal processing upstream of Rac activation in the context of chemoattractant stimulation may limit the degree of total Rac activation, and thus the ability of the cell to reach the threshold controlling the transition into the strongly polarized state. Thus, a single ligand may not induce such a transition. However, the threshold might potentially be reached and exceeded, given multiple inputs converging on Rac activation, which may be affected by cell type-specific peculiarities of the signaling apparatus, such as basal levels of Rac activation and the expression of the signaling proteins.
The rapamycin stimulation system described here also allows a more detailed study of the interplay between Rac activation and activity of other signaling species, including those that might be involved in various feedback interactions. This analysis is akin to the more common epistasis assays, but with subtler phenotypes related more closely to gradient sensing responses. In particular, our analysis suggested that PI3K interplay with Rac activation, although consistent with the recently proposed formation of an AND gate in terms of the response (6), in which both inputs are necessary to induce directed migration, acts more specifically by controlling the threshold of cell responsiveness to Rac activity gradients. Whereas PI3K inhibition does not prevent the ability of the cells to undergo directed cell polarization or migration responses, it can strongly reduce the fraction of cells capable of these responses within the same set of experimental conditions.
The analysis here represents a more general framework extensible to other rapamycin-activatable signaling molecules (25, 50, 51), as well as to other cell types and multicellular systems. Furthermore, the effects of gradients of other proteins engineered to be sensitive to small membrane-permeable molecules, such as ATP analogs (52) and imidazole (53), could also be analyzed to refine our understanding of the mechanisms of cell responses to graded intracellular signaling activity. As also demonstrated in this report, such efforts could help develop qualitatively and quantitatively improved mathematical and computational models of gradient sensing and chemotaxis phenomena, extending common approaches to these processes. We suggest that as the repertoire of methods for direct control of cellular events increases, microfluidics-based tools will play an important role in exploitation of these methods in cell navigation research.
Materials and Methods
Modeling.
As described in SI Appendix, Model Supplement, we have developed a simple model based primarily on the antagonistic relationship between the two small Rho family GTPases involved in polarized cell migration, Rac and Rho, modulated by Cdc42, based on sequential model selection. The model was implemented as a system of partial differential equations for Rho GTPases in one space dimension and was explored numerically using MATLAB (MathWorks). Details are provided in SI Appendix, Model Supplement.
Device Fabrication.
Microfluidic chips were created using a two-layer soft lithography process (54). Polydimethylsiloxane (RTV615; Momentive) was used to create molds for the devices as described previously (55). To increase the height of the flow-through channels, the membrane between the control layer and flow layer was removed. Before an experiment, each device was treated with HCl, cleaned with 70% (vol/vol) ethanol, and allowed to bond to a clean 22-mm × 40-mm coverslip (Fisher).
Cell Culture and Transfection.
HeLa cells were maintained in DMEM with 10% (vol/vol) FBS and 1% penicillin streptomycin (Gibco). MTLn3 cells were cultured in α-minimum essential medium supplemented with 5% (vol/vol) FBS and 1% penicillin streptomycin (Gibco). Both cell lines were kept in a 37 °C and 5% CO2 environment during culture and in experiments. MTLn3 cells were kindly provided by the laboratory of Jeffrey Segall (New York City, NY). The constructs, YF-TIAM1, YF, and LDR, were transfected into cells using Fugene HD (Roche) per the manufacturer’s recommendations. The Raichu Rac FRET probe was kindly provided by the laboratory of Michiyuki Matsuda (Kyoto, Japan) and was transfected in a similar manner. During FRET experiments, MF-TIAM1 was used in place of YF-TIAM1. MF-Tiam1 has a mCherry fluorophore in place of YFP and was used to avoid spectral overlap with the FRET probe.
Imaging.
Microfluidic experimental imaging was performed using an inverted Zeiss Axiovert 200M epifluorescence microscope at 37 °C and 5% CO2, coupled to a Cascade II:1024 EMCCD camera (Photometrics) using a 40×, 1.3-N.A. oil immersion objective (Zeiss). The microscope was driven by Slidebook software (Intelligent Imaging Innovations). Images were taken every 5 min in the YFP channel using a 494-nm excitation filter and 530-emission filter (Semrock), and Alexa 594 dye was imaged using a 572-nm excitation filter and 628-emission filter (Semrock) over a 4-h period. A spectral 2D template autofocus algorithm was used between images to account for any focus fluctuations. To correct for uneven illumination, all images were normalized with the following correction C = (I − D/F − D) * M, where C is the corrected image, I is the initial image, D is the darkfield image, F is the flatfield image, and M is the mean of difference between flatfield and darkfield images. The flatfield and darkfield images were taken as averages of multiple images. FRET images were taken using a cyan fluorescent protein (CFP) excitation filter (Semrock), an appropriate dichroic (Semrock), and YFP/CFP emission filters (Semrock).Volume analysis was performed on an inverted Zeiss Axiovert 200 spinning disk confocal microscope, coupled to a CCD camera (Hamamatsu) using a 40× objective (Zeiss). The microscope was driven by Metamorph 7.5 imaging software (Molecular Devices). YFP excitation was trigged with an argon laser (CVI Melles Griot) which was fiber-coupled (OZ Optics) to the spinning disk confocal unit (CSU10; Yokogawa) mounted with a YFP dichroic mirror (Semrock) and an appropriate YFP filter (Chroma Technology).
Analysis of Gradient and Mean Values of Rapamycin.
All analyses were performed using custom-written codes in MATLAB 2007b (MathWorks). Cell-based data (e.g., cell length, centroid) were obtained from the YFP images, whereas gradient-based data were obtained from the Alexa 594 dye images. Cells were segmented from the YFP channel based on intensity. The signal-to-noise ratio was sufficiently high to preclude the use of more sophisticated segmentation techniques. Once cell boundaries were determined from the segmentation, concentration lines spanning the width of the channel were generated from the front and back of the cell to obtain local concentration data. The concentration lines were restricted to the width and length of an individual channel by manually selecting the boundaries of the channel from the Alexa 594 dye image. For any cell that extended out of the channel, the concentration lines were restricted to the respective ends of the channel to avoid spurious measurements associated with the height difference between the arms of the flow channels and the cross-channels. Dye intensities were extrapolated to the intensities at the respective ends of the channel to determine concentration.
Cell Tracking.
Cell velocity was tracked by obtaining the coordinates of cell nuclei using a semiautomated script written in MATLAB.
Quantification of the Membrane Distribution of YFP-FKBP.
The translocation of YFP-FKBP to the membrane was quantified by taking the ratio of intensities at the cell periphery compared with the intensity in the cytoplasm. First, the cytoplasm and membrane were segmented by intensity thresholding from cell images, and a subsequent morphological erosion was used to obtain each component separately. To obtain the membrane intensity across the cell length, the maximum value of each row of pixels in the membrane segmentation was taken. The cytoplasm intensity was obtained by taking the mean value of each row of pixels from the cytoplasm segmentation. Ratio values across the length of the cell were normalized between 0 and 1 and smoothed with a 10-point moving average. To aggregate data from multiple cells, time 0 was chosen as 30 min before visible translocation and subsequent profiles were normalized to values at time 0. For control cells, time 0 was chosen at times comparable to cells positioned in similar locations during rapamycin experiments.
FRET Analysis.
FRET analysis was done according to a previously described protocol (56). Briefly, FRET images were analyzed by first subtracting background from each individual CFP and YFP FRET image (CFP excitation, YFP emission). Images were thresholded and subsequently aligned with a discrete Fourier transform (DFT) registration algorithm (57). The FRET ratio was calculated by dividing the YFP FRET image by the registered CFP image pixel by pixel. Final images were Gaussian-filtered to remove noise. To obtain the distribution of FRET activity across the cell length, the mean value of each row of pixels composing the ratio image was taken and final values were normalized between 0 and 1 for the cell length and smoothed with a 10-point moving average. To aggregate data from multiple cells, time 0 was chosen as 30 min before the appearance of morphological changes in accompanying MCHF-TIAM1 images. FRET profile data at later time points were normalized to time 0. For control cells, time 0 was chosen based on MCHF-TIAM1 images from comparably located stimulated cells.
Volume Analysis.
Cell heights were determined by first taking confocal Z slices of cells before and after rapamycin treatment. Afterward, the cell body was segmented and superimposed to obtain the final image. The cell volume was determined by taking a region of interest (ROI) in the cell body from 3D reconstructions carried out using MATLAB. The number of pixels in the ROI was then converted to micrometers based on precalibration of the slice height.
Measurement of Initial Response Time and Late Polarization Time.
All measurements were performed using MATLAB. To determine the length of a cell’s front and back, the cell nucleus was tracked by manually fitting an ellipse to the nucleus image and taking the centroid of the fitted ellipse as the position of the nucleus. Nuclei were clearly distinguishable in all YFP images analyzed due to exclusion of the construct. Cell front and back lengths were calculated by taking the coordinates of the front and rear of cells from segmented images and calculating the distance to the nucleus position (SI Appendix, Fig. S6). To assay the width of the front and rear of each cell, the distance between the rightmost and leftmost coordinates of the cell front (top 10% of pixels) and rear (bottom 10% of pixels) was calculated (SI Appendix, Fig. S6). The initial response time was taken as the time to reach 20% of the total magnitude of the first morphological change toward the gradient (SI Appendix, Fig. S9). The initial response time was taken at the 50% level for MTLn3 cells due to the faster kinetics associated with their responses. To find the late polarization time, the decrease in fluorescence of the cell body was measured as a function of time. The fluorescence intensity was determined in ROIs, chosen automatically based on the end coordinates of the cell (front and back) (SI Appendix, Fig. S10). The ROIs were then further eroded by several pixels to avoid any effects from the cell membrane. The late polarization time was taken as the time to reach 50% of the full fluorescence intensity drop in the ROI from the peak intensity value.
Population Separation.
Thresholds between nonresponding cells and responding cells were determined using quadratic discriminant analysis (“classify” function) in MATLAB. The two populations, along with their corresponding concentrations, were input into the function as training data, and a separation point was generated from a given vector of concentrations.
Statistical Analysis.
Statistical analysis was carried out with Sigmaplot (Systat) and Prism (GraphPad) software. Experimental results were expressed as means with error bars equal to SEM. Comparisons between two groups were carried out with a two-sided Student t test when assumptions of normality were fulfilled. For comparisons that did not satisfy the assumption of normality, a Mann–Whitney rank sum test was used. Comparisons were deemed to be significant if P values were <0.05. The comparison between the y-axis intercepts of the linear regressions of the relevant data points was carried out using an ANCOVA test. First, the difference between slopes was compared; if the difference was insignificant, a comparison between the y-axis intercepts was performed. To compare fitted curves, an F test was conducted using the standard procedure.
Supplementary Material
Acknowledgments
L.E.-K. and W.R.H. were supported by Natural Sciences and Engineering Research Council discovery and accelerator supplement grants. This work was supported by National Institutes of Health (NIH) Grants GM092930 (to T.I. and B.L.) and GM072024, GM084332, and CA15578 (to A.L. and B.L.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Author Summary on page 21194 (volume 109, number 52).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210295109/-/DCSupplemental.
References
- 1.Parent CA, Devreotes PN. A cell’s sense of direction. Science. 1999;284(5415):765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 2.Servant G, et al. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. 2000;287(5455):1037–1040. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zigmond SH. Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors. J Cell Biol. 1977;75(2 Pt 1):606–616. doi: 10.1083/jcb.75.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiner OD, et al. A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat Cell Biol. 2002;4(7):509–513. doi: 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sasaki AT, et al. G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J Cell Biol. 2007;178(2):185–191. doi: 10.1083/jcb.200611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inoue T, Meyer T. Synthetic activation of endogenous PI3K and Rac identifies an AND-gate switch for cell polarization and migration. PLoS ONE. 2008;3(8):e3068. doi: 10.1371/journal.pone.0003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niggli V. A membrane-permeant ester of phosphatidylinositol 3,4, 5-trisphosphate (PIP(3)) is an activator of human neutrophil migration. FEBS Lett. 2000;473(2):217–221. doi: 10.1016/s0014-5793(00)01534-9. [DOI] [PubMed] [Google Scholar]
- 8.Iijima M, Devreotes PN. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109(5):599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 9.Wu YI, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461(7260):104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoeller O, Kay RR. Chemotaxis in the absence of PIP3 gradients. Curr Biol. 2007;17(9):813–817. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson GJ, et al. PI(3)Kgamma has an important context-dependent role in neutrophil chemokinesis. Nat Cell Biol. 2007;9(1):86–91. doi: 10.1038/ncb1517. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, He L, Wu YI, Hahn KM, Montell DJ. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol. 2010;12(6):591–597. doi: 10.1038/ncb2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoo SK, et al. Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev Cell. 2010;18(2):226–236. doi: 10.1016/j.devcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461(7266):997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Umeda N, Ueno T, Pohlmeyer C, Nagano T, Inoue T. A photocleavable rapamycin conjugate for spatiotemporal control of small GTPase activity. J Am Chem Soc. 2011;133(1):12–14. doi: 10.1021/ja108258d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein-protein interactions in live cells using light. Nat Biotechnol. 2009;27(10):941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh M, et al. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304(5671):743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- 18.Roy P, et al. Local photorelease of caged thymosin beta4 in locomoting keratocytes causes cell turning. J Cell Biol. 2001;153(5):1035–1048. doi: 10.1083/jcb.153.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng JQ. Turning of nerve growth cones induced by localized increases in intracellular calcium ions. Nature. 2000;403(6765):89–93. doi: 10.1038/47501. [DOI] [PubMed] [Google Scholar]
- 20.Ridley AJ, et al. Cell migration: Integrating signals from front to back. Science. 2003;302(5651):1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 21.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305(5690):1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 22.Kraynov VS, et al. Localized Rac activation dynamics visualized in living cells. Science. 2000;290(5490):333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 23.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440(7087):1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 24.Paliwal S, et al. MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast. Nature. 2007;446(7131):46–51. doi: 10.1038/nature05561. [DOI] [PubMed] [Google Scholar]
- 25.Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2(6):415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivera VM, et al. A humanized system for pharmacologic control of gene expression. Nat Med. 1996;2(9):1028–1032. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- 27.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279(5350):509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 28.Li Jeon N, et al. Neutrophil chemotaxis in linear and complex gradients of interleukin-8 formed in a microfabricated device. Nat Biotechnol. 2002;20(8):826–830. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- 29.Herzmark P, et al. Bound attractant at the leading vs. the trailing edge determines chemotactic prowess. Proc Natl Acad Sci USA. 2007;104(33):13349–13354. doi: 10.1073/pnas.0705889104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Carlo D, Wu LY, Lee LP. Dynamic single cell culture array. Lab Chip. 2006;6(11):1445–1449. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 31.Yin Z, Noren D, Wang CJ, Hang R, Levchenko A. Analysis of pairwise cell interactions using an integrated dielectrophoretic-microfluidic system. Mol Syst Biol. 2008;4:232. doi: 10.1038/msb.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faure-André G, et al. Regulation of dendritic cell migration by CD74, the MHC class II-associated invariant chain. Science. 2008;322(5908):1705–1710. doi: 10.1126/science.1159894. [DOI] [PubMed] [Google Scholar]
- 33.Irimia D, Charras G, Agrawal N, Mitchison T, Toner M. Polar stimulation and constrained cell migration in microfluidic channels. Lab Chip. 2007;7(12):1783–1790. doi: 10.1039/b710524j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banaszynski LA, Liu CW, Wandless TJ. Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc. 2005;127(13):4715–4721. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- 35.McMahon LP, Choi KM, Lin TA, Abraham RT, Lawrence JC., Jr The rapamycin-binding domain governs substrate selectivity by the mammalian target of rapamycin. Mol Cell Biol. 2002;22(21):7428–7438. doi: 10.1128/MCB.22.21.7428-7438.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailly M, Yan L, Whitesides GM, Condeelis JS, Segall JE. Regulation of protrusion shape and adhesion to the substratum during chemotactic responses of mammalian carcinoma cells. Exp Cell Res. 1998;241(2):285–299. doi: 10.1006/excr.1998.4031. [DOI] [PubMed] [Google Scholar]
- 37.Wyckoff JB, Segall JE, Condeelis JS. The collection of the motile population of cells from a living tumor. Cancer Res. 2000;60(19):5401–5404. [PubMed] [Google Scholar]
- 38.Komatsu N, et al. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell. 2011;22(23):4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dawes AT, Edelstein-Keshet L. Phosphoinositides and Rho proteins spatially regulate actin polymerization to initiate and maintain directed movement in a one-dimensional model of a motile cell. Biophys J. 2007;92(3):744–768. doi: 10.1529/biophysj.106.090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marée AFM, Grieneisen VA, Edelstein-Keshet L. How cells integrate complex stimuli: The effect of feedback from phosphoinositides and cell shape on cell polarization and motility. PLOS Comput Biol. 2012;8(3):e1002402. doi: 10.1371/journal.pcbi.1002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aoki K, Nakamura T, Inoue T, Meyer T, Matsuda M. An essential role for the SHIP2-dependent negative feedback loop in neuritogenesis of nerve growth factor-stimulated PC12 cells. J Cell Biol. 2007;177(5):817–827. doi: 10.1083/jcb.200609017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stephens L, Milne L, Hawkins P. Moving towards a better understanding of chemotaxis. Curr Biol. 2008;18(11):R485–R494. doi: 10.1016/j.cub.2008.04.048. [DOI] [PubMed] [Google Scholar]
- 43.Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95(1):81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- 44.Janetopoulos C, Ma L, Devreotes PN, Iglesias PA. Chemoattractant-induced phosphatidylinositol 3,4,5-trisphosphate accumulation is spatially amplified and adapts, independent of the actin cytoskeleton. Proc Natl Acad Sci USA. 2004;101(24):8951–8956. doi: 10.1073/pnas.0402152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Worthylake DK, Rossman KL, Sondek J. Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature. 2000;408(6813):682–688. doi: 10.1038/35047014. [DOI] [PubMed] [Google Scholar]
- 46.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81(1):53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 47.Gardiner EM, et al. Spatial and temporal analysis of Rac activation during live neutrophil chemotaxis. Curr Biol. 2002;12(23):2029–2034. doi: 10.1016/s0960-9822(02)01334-9. [DOI] [PubMed] [Google Scholar]
- 48.Irimia D, et al. Microfluidic system for measuring neutrophil migratory responses to fast switches of chemical gradients. Lab Chip. 2006;6(2):191–198. doi: 10.1039/b511877h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meier B, et al. Chemotactic cell trapping in controlled alternating gradient fields. Proc Natl Acad Sci USA. 2011;108(28):11417–11422. doi: 10.1073/pnas.1014853108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suh B-C, Inoue T, Meyer T, Hille B. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 2006;314(5804):1454–1457. doi: 10.1126/science.1131163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fegan A, White B, Carlson JCT, Wagner CR. Chemically controlled protein assembly: Techniques and applications. Chem Rev. 2010;110(6):3315–3336. doi: 10.1021/cr8002888. [DOI] [PubMed] [Google Scholar]
- 52.Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407(6802):395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 53.Qiao Y, Molina H, Pandey A, Zhang J, Cole PA. Chemical rescue of a mutant enzyme in living cells. Science. 2006;311(5765):1293–1297. doi: 10.1126/science.1122224. [DOI] [PubMed] [Google Scholar]
- 54.Unger MA, Chou H-P, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288(5463):113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 55.Joanne Wang C, et al. A microfluidics-based turning assay reveals complex growth cone responses to integrated gradients of substrate-bound ECM molecules and diffusible guidance cues. Lab Chip. 2008;8(2):227–237. doi: 10.1039/b713945d. [DOI] [PubMed] [Google Scholar]
- 56.Hodgson L, Shen F, Hahn K. Current Protocols in Cell Biology. New York: Wiley; 2001. Biosensors for characterizing the dynamics of Rho family GTPases in living cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guizar-Sicairos M, Thurman ST, Fienup JR. Efficient subpixel image registration algorithms. Opt Lett. 2008;33(2):156–158. doi: 10.1364/ol.33.000156. [DOI] [PubMed] [Google Scholar]