Abstract
The accuracy of the 3′-end processing by cleavage and polyadenylation is essential for mRNA biogenesis and transcription termination. In yeast, two poorly conserved neighboring elements upstream of cleavage sites are important for accuracy and efficiency of this process. These two RNA sequences are recognized by the RNA binding proteins Hrp1 and Rna15, but efficient processing in vivo requires a bridging protein (Rna14), which forms a stable dimer of hetero-dimers with Rna15 to stabilize the RNA–protein complex. We earlier reported the structure of the ternary complex of Rna15 and Hrp1 bound to the RNA processing element. We now report the use of solution NMR to study the interaction of Hrp1 with the Rna14–Rna15 heterodimer in the presence and absence of 3′-end processing signals. By using methyl selective labeling on Hrp1, in vivo activity and pull-down assays, we were able to study this complex of several hundred kDa, identify the interface within Hrp1 responsible for recruitment of Rna14 and validate the functional significance of this interaction through structure-driven mutational analysis.
Keywords: mRNA processing, mRNA transport, protein-RNA, methyl-TROSY, modeling
Eukaryotic messenger RNA precursors (pre-mRNA) undergo extensive cotranscriptional processing before their transport to the cytoplasm and subsequent translation (1, 2). The processing events include 5′-end capping, splicing to remove intronic sequences and cleavage and polyadenylation at the 3′-end (3, 4). Correct 3′-end processing is necessary for mRNA biogenesis and transcription termination, preserves RNAs from degradation (5), promotes its export to the cytoplasm (6), enhances translation (7) and promotes transcriptional activation (4, 8). Cleavage and polyadenylation can be uncoupled in vitro but in vivo they are connected with each other and with transcription termination. Defects in 3′-end processing have also been associated with several diseases in humans (9), but this process may be even more critical in yeast that has shorter intergenic regions and much rarer alternative splicing (10, 11). In this organism, efficient mRNA transport is mostly dependent on polyadenylation rather than splicing.
In yeast, the 3′-end processing reaction is executed by a complex machine containing more than 20 proteins (4) which share significant similarities with human 3′-end processing proteins. The entire yeast 3′-end processing complex is divided into two subcomplexes, called cleavage factor I (CF I) and cleavage and polyadenylation factor (CPF) (4). CF I binds to the pre-mRNA upstream of the cleavage signal within the pre-mRNA, whereas CPF binds at the cleavage site as well as upstream and downstream of the cleavage site. Several sequences within yeast pre-mRNAs, which are surprisingly poorly conserved, direct the recruitment of cleavage and polyadenylation factors (12, 13). In the 5′–3′ direction, they include AU-rich efficiency element (EE) responsible for polyadenylation efficiency; A-rich positioning element (PE) critical for precise 3′-end processing; the poly(A) cleavage site, and upstream and downstream U-rich enhancer elements (UUE and DUE).
The CF I complex is further divided into two smaller subcomplexes: CF IA and CF IB. CF IA consists of Rna14, Rna15, Clp1, and Pcf11, whereas CF IB includes only the RNA-binding protein Hrp1. The entire complex binds to AU-rich upstream sequences and to the A-rich downstream elements of pre-mRNA transcripts. Hrp1, the only component of CF IB, is a 534-aa-long protein necessary for proper 3′-end cleavage in yeast (14), although no homologs of Hrp1 have been reported in mammals or metazoans. It contains two centrally located RNA binding domains of the RRM type, followed by a C-terminal domain with several RGG-repeats. The N terminus is 158 aa long and has no structural homologs (Fig. 1A). Hrp1 interacts with the AU-rich enhancer element region of the RNA, as reported by NMR (15).
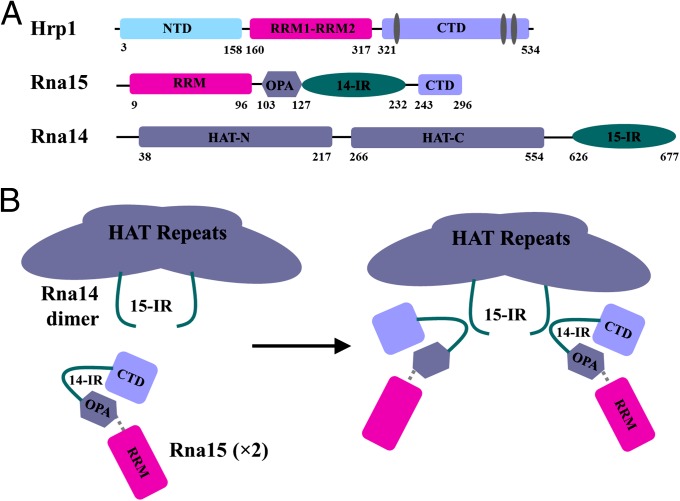
Fig. 1.
Schematic representation of CF I. (A) Hrp1, Rna15, and Rna14 are shown with functional and structural domains explicitly represented. Hrp1 contains an N-terminal domain (NTD), an RNA binding domain (RBD) in the middle with two RNA recognition motives (RRM) and a C-terminal domain. Rna15 has an RRM at the N terminus followed by the Rna14 interacting region (127–232) and the C-terminal domain (CTD), which interacts with Pcf11 (243–296). Rna14 has HAT-repeats followed by the Rna15 interacting region. (B) Proposed schematic model of Rna14–Rna15 tetramerization, which was used as a starting point for our studies. 14-IR is Rna14-interacting region and 15-IR is Rna15-interacting region.
Rna15 has 43% identity with its human counterpart CstF-64, but conservation is higher within regions with attributed function. It is a 296-aa protein containing an RNA recognition motif (RRM) at the N terminus followed by the putative Rna14 interacting domain (127–232; 14IR) and the Pcf11 interacting C-terminal domain (CTD) (16) (Fig. 1A). The RRM of Rna15 achieves sequence specificity for RNA only in the presence of other CF I proteins, specifically Rna14 and Hrp1. This property may be distinct from CstF-64, although this protein also binds GU-rich elements with only modest selectivity, and in humans there is no protein with role homologous to Hrp1.
Rna14 is the central scaffolding component of CF IA. Its homologous human protein, CstF-77, contains 12 HAT motifs at the N terminus (17, 18), which can be divided into a HAT-N and a HAT-C domain. CstF-77 forms an elongated dimer about 45 Å wide and 165 Å long through the HAT-C domain (17, 18). The HAT domain in Rna14 is followed by the Rna15 interacting region (15IR) at the C terminus (19), through which Rna14 and Rna15 form a stable dimer of heterodimers (17, 18, 20) (Fig. 1B).
The interaction of Hrp1 and Rna15 with the bridging protein Rna14 enhances 3′-end processing efficiency. Thus, it is multiple interactions between proteins and RNA, rather than a few highly specific sequence recognition events, that anchor the cleavage and polyadenynation complex and direct it to cognate processing sites. Although increasing structural information is available for individual components within CF I, only partial information is available concerning the arrangement and topology of the individual components within the complex. To understand how the individual protein and RNA components of this complex are organized during 3′-end processing, we used 1H-13C methyl transverse relaxation optimized NMR (methyl-TROSY) spectroscopy, pull-down experiments, site-directed mutagenesis, yeast genetics, and computational modeling to obtain insight into the architecture of the CF I complex. Methyl-TROSY spectroscopy identified a protein–protein interface involved in CF I complex formation and processing, which was validated using biochemical and genetics studies. We conclude that Rna14 interacts with the back of the RRM surface within Hrp1 through hydrophobic and charged residues. Mutation of these protein–protein contacts perturbs or destroys cleavage and polyadenylation efficiency, suggesting that these interactions are important for accurate recognition of 3′-end processing signals. We generated a model for CF I based on these data to generate hypotheses to test how the different protein factors are arranged within the CF I complex.
Results
We have recently reported the structure of the ternary complex of Hrp1, Rna15, and the pre-mRNA upstream processing signal (21). This structure completed earlier studies of Hrp1 with the enhancer EE element (15) and Rna15 with GU-rich RNA sequences (22) and of CstF-64 (23). To better understand how 3′-end processing signals are recognized to initiate 3′-end processing and transcription termination, we sought to obtain structural information on the complete CF I complex including Rna14, Rna15, Hrp1, and the pre-mRNA transcript. Unfortunately, crystallography has not been forthcoming with this complex, and EM studies still rely on low resolution approaches (24). Although the Rna14 and Rna15 complex has been studied biophysically (20) and by X-ray crystallography (25) and NMR (19), there is no complete structure with Hrp1. These considerations led us to investigate the complex of Rna14, Rna15, Hrp1, and pre-mRNA using NMR.
Assignments of Methyl ILV Groups in Hrp1.
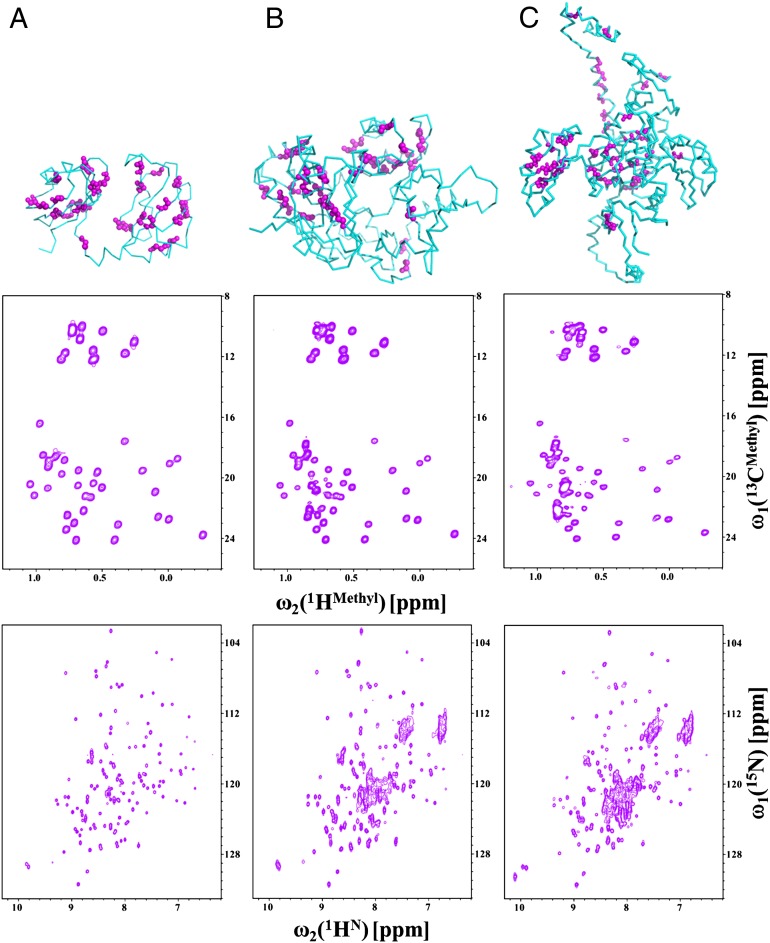
The CF I complex including Hrp1, Rna14, Rna15, and RNA adds up to ∼300 kDa and exceeds the limit of standard NMR spectroscopy. This limitation led us to use methyl-TROSY experiments (26, 27) to investigate the architecture of CF I and the arrangement of individual components in the complex. Full-length Hrp1 protein is challenging to study due to the presence of unstructured domains at both the N and C termini, whereas the RRMs in the center are well structured. Because Ile, Leu, and Val residues are very well distributed over the Hrp1 RRMs, we specifically 13C- and 1H-labeled the δ-methyl groups of Ile side chains, δ-methyl groups of all Leu and γ-methyl groups of all Val side chains in deuterated protein to observe these signals with high resolution. Thus, we recorded high-resolution methyl-TROSY spectra of methyl Ile/Leu/Val-labeled Hrp1 RRMs (RBD: 19.1 kDa), Hrp1 RBD+C-terminal domain (RBDCTD; 42.9 kDa), and full-length Hrp1 (59.7 kDa) (Fig. 2). The side-chain assignments for these constructs were completed using a suite of 3D HCCH-COSY and 13C-NOESY spectra, and ambiguous assignments were cross-validated using site-directed mutagenesis; for example, I452L in the RBDCTD construct was used to verify the identification of Ile452 from the disappearance of the weak Ile452 signal in the 1H-13C spectrum of the I452L mutant. Furthermore, fractionally labeled (10%) 13C-labeled and ILV-labeled samples were used to stereospecifically assign the methyl groups in the Hrp1–RBD (27, 28). Altogether, we assigned the 10 δ-methyl groups in the Ile side chains out of 12 total Ile side chains, 14 methyl groups for all Leu, and 19 methyl groups out of 20 Val side chains present in the Hrp1 RBD. It was also possible to transfer assignments from Hrp1–RBD to Hrp1–RBDCTD and Hrp1–FL proteins because the middle region of Hrp1, the RRMs, contains a large fraction of the methyl containing residues. The N- and C-terminal domains do not have many methyl containing residues and structure prediction algorithms predict highly flexible structures. Interestingly, the methyl groups originating from the terminal regions of Hrp1 have weaker intensities, suggesting residual disorder or conformational averaging (Fig. 2).
Fig. 2.
Methyl TROSY experiments on Hrp1 protein. Results obtained for Hrp1–RBD (A), Hrp1–RBDCTD (B), and the Hrp1-full length proteins (C) are shown. (Top) The structure of each segment with Ile, Leu, and Val residues highlighted with pink spheres. The Hrp1–RBD structure was taken from the 2CJK and 2KM8 structures, whereas Hrp1–RBDCTD and Hrp1-full length were modeled using I-TASSER. (Middle) 1H-13C HMQCs of Hrp1–RBD, Hrp1–RBDCTD, and Hrp1-full length proteins. (Bottom) 1H-15N HSQCs for the same constructs.
Hrp1 Interacts with Rna14–Rna15 in Vitro.
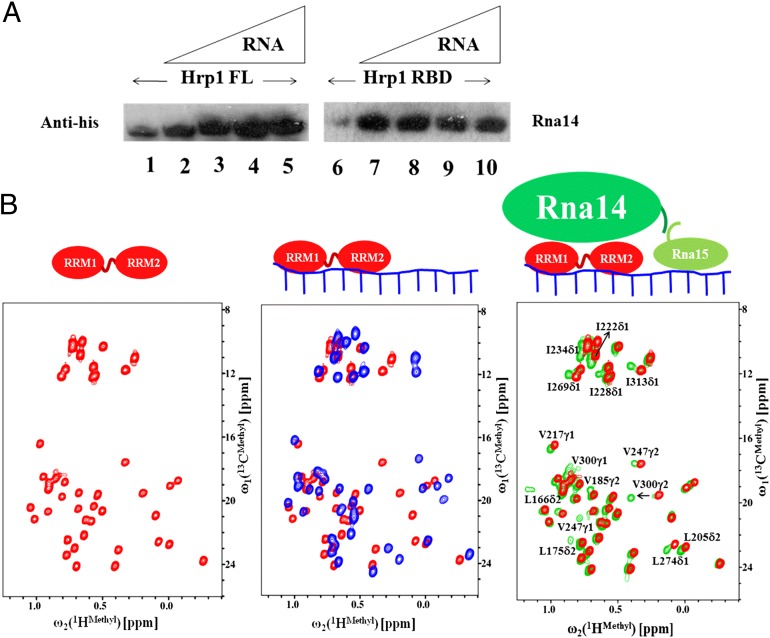
To identify the regions of Hrp1 that interact with Rna14, we first used different fragments of Hrp1 (RBD, RBDCTD, and FL) to pull down Rna14 in the presence or absence of RNA. As shown in Fig. 3A, His-tagged Rna14 is pulled down by either Hrp1–RBD or Hrp1–FL (as GST-fused proteins). There is some difference in the band intensities for pulled-down proteins in the presence and absence of RNA. The bands in the presence of RNA are slightly more intense than in the absence of RNA, suggesting that the presence of RNA stabilizes the protein complex.
Fig. 3.
Interaction of Hrp1 with the Rna14–Rna15 tetramer by pull-down experiments and NMR chemical shift perturbation analysis. (A) GST Pull-down of Hrp1 (full length; FL) and Hrp1 (RBD) with His-tagged Rna14–Rna15. Lanes 1 and 6 are Hrp1 FL and Hrp1 RBD with Rna14–Rna15 in the absence of RNA. Lanes 2–5 are Hrp1 FL incubated with Rna14–Rna15 with different amounts of RNA, 7–10 are Hrp1 RBD and Rna14–Rna15 with different amounts of RNA. (B) Chemical shift perturbation of Hrp1 upon addition of RNA and of the Rna14–Rna15 complex. Hrp1 RBD (free) is represented in red, the RNA-bound complex is shown in blue, and the Rna14–Rna15 bound complex is shown in green. A cartoon model summarizing the chemical shift perturbation results is shown at the top.
Hrp1 Interacts with Rna14 Through the Back Surface of the RRMs.
We used chemical shift perturbation analysis to map the binding interface between Hrp1 and Rna14. Labeled Hrp1 was titrated with unlabeled RNA, followed by the addition of the unlabeled preformed Rna14–Rna15 complex. We also conducted the experiment by adding unlabeled Rna14–Rna15 to labeled Hrp1 without RNA, and the final spectra were similar to those with Rna14-Rna15 and RNA, except that the peaks were much more intense and sharper in the presence of RNA.
The structure of Hrp1, RNA, and Rna15 has already been reported (21): Hrp1 binds to the 5′-portion of the anchoring RNA, and Rna15 binds to the 3′-part of the same element (Fig. 3B). The spectra of Hrp1 recorded in the presence of RNA are consistent with the known structure of the Hrp1–RNA complex. Upon addition of the Rna14–Rna15 complex to Hrp1, methyl signals corresponding to L175δ2, L205δ2, I228δ1, L274δ1, V300γ2, and I313δ1 of Hrp1–RBD showed large chemical shift perturbations (Fig. 3B), whereas L166δ2, V217γ1, I222δ1, I234δ1, V247γ2, and I269δ1 of the RBD showed moderate chemical shift changes. Other methyl peaks were not perturbed at all upon addition of the Rna14–Rna15 complex. The proximity of these residues to Rna14–Rna15 could be verified using site-directed mutagenesis and in vivo experiments (as discussed later). Interestingly, all these methyl groups are on the back of the RRM surface which binds to RNA and form a contiguous hydrophobic patch. The results with Hrp1–RBDCTD and Hrp1 full-length proteins are shown in Figs. S1 and S2, respectively, where we see very similar changes occurring for the same residues, demonstrating that the interactions with Rna14 resides on the back of the RRMs.
Site-Directed Mutagenesis.
Based on these results, we prepared thirteen single and seven multiple mutants of Hrp1 to confirm that the interacting interface of Hrp1 and Rna14 coincide with what was identified by NMR. These mutants were used in pull-down and/or NMR titration experiments to evaluate their interaction with Rna14–Rna15. The list of mutants is given in Table S1. All mutants bind RNA normally, as judged by chemical shift changes, suggesting strongly that the protein structure was not affected by the changes, but either show attenuated or no binding to the Rna14–Rna15 complex. The effect of L205S (Fig. S3 A1–C1), I228N (Fig. S3 A2–C2), and I313N (Fig. S4) on Rna14–Rna15 binding under NMR titration experiments are shown. These residues show maximum perturbation and their mutation abolish the interaction with Rna14. Additionally, reduced NMR perturbation for I222S confirms the involvement of this residue in the binding interface (Fig. S5).
Structural Model for the CF I Complex.
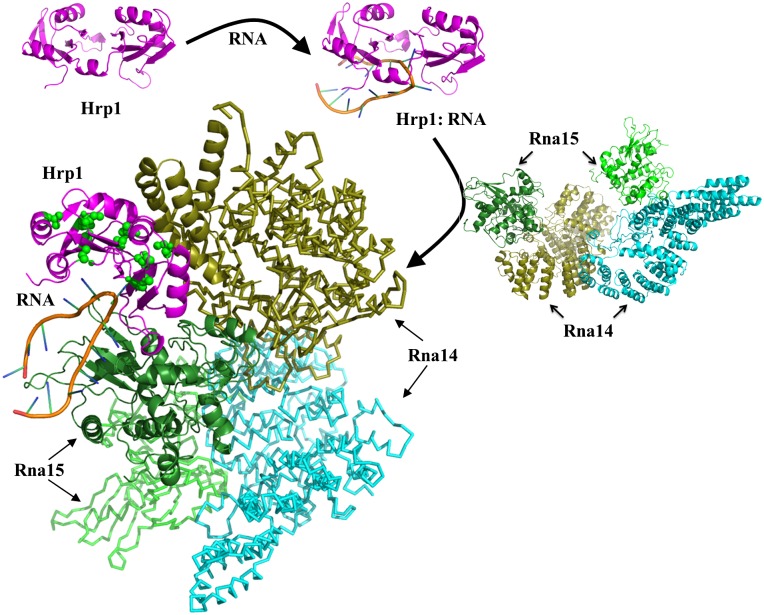
The CF I complex contains five proteins (Rna14, Rna15, Clp1, Pcf11, and Hrp1). In this study, we have investigated the interaction of three proteins (Hrp1, Rna14, and Rna15) with themselves and the anchoring RNA. Using the information derived from the methyl-NMR spectroscopy, site-directed mutagenesis, and pull-down experiments, we generated a model of CF I with HADDOCK (29).
The modeling was done in two steps. In the first step, all individual components were modeled or taken, when available, from the Protein Data Bank (PDB). I-TASSER (30) was used to model Rna14 (1–677), Rna15 (1–296), and Hrp1 (1–534) based on existing structures deposited in the PDB. Rna14 was modeled based on structures 2OND, 2OOE, and 2L9B, whereas Rna15 was modeled based on structures 2X1A, 2KM8, 2J8P, 1P1T, and 2L9B. Hrp1 (1–534) was modeled based on structures 2CJK and 2KM8, whereas Hrp1 (156–322) and the 16-nt RNA were taken from structures 2CJK and 2KM8.
In the second step, the HADDOCK online server was used to model the CF I complex including Hrp1, RNA, Rna14, and Rna15 (http://haddock.science.uu.nl/services/HADDOCK/haddockserver-multi.html). The GenTBL software web portal of HADDOCK was used to generate restraints for multicomponent docking based on active and passive residues. The active residues are experimentally proven to be directly involved in the interaction between two molecules and have >40–50% solvent accessibility, whereas passive residues are solvent accessible surface neighbors of active residues. Residues used as active and passive residues are given in Table S2. Active and passive residues for Rna14/5p to Rna15/4p were taken from structure 2L9B, whereas active and passive sites for Rna15 to RNA, and vice versa, were taken from 2KM8 and 2X1F. The 2KM8 and 2CJK structures were used to obtain information about active and passive residues of Hrp1 and RNA. Rna14 forms its own dimer through residues 516–585 of one unit interacting with residues 317–480 of the other (18). Rna14 also interacts with Rna15 through residues 626–677, whereas Rna15 interacts with Rna14 through residues 127–232 (16, 19). Results from the current study were used to define active and passive residues for the Hrp1 and Rna14 pair. At this moment, we do not have any information about the residues of Rna14 interacting with Hrp1. However, because residues 317–480 and 516–585 of Rna14 are involved in homodimerization and residues 626–677 are involved in the Rna15 interaction, they were excluded during the modeling of CF I complex of Hrp1, Rna14, Rna15, and RNA, whereas the remaining residues (1–316) of Rna14 were used for docking to Hrp1 as passive residues (Fig. 4).
Fig. 4.
Model of the CF I complex. The Hrp1, 3′-end processing substrate RNA and Rna14–Rna15 complexes were used in HADDOCK to model the organization and interaction of components of the complete CF I complex. Starting from the complex of Hrp1 with RNA, the Rna14–Rna15 complex was added to generate a model of the complete CF I complex. Here, Hrp1 is shown in magenta, Rna15 proteins are shown in green-chartreuse and green-forest colors, whereas Rna14 monomers are shown in cyan and olive colors and the RNA is depicted by its secondary structure. The residues of Hrp1 interacting with Rna14 are shown with green spheres. Images were generated with Pymol (www.pymol.org).
Functional Analysis of Hrp1 Mutants.
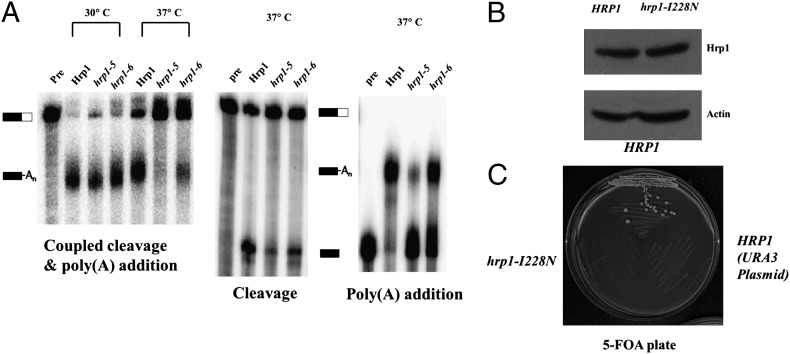
To test whether the interaction between the Hrp1 RRMs and Rna14-Rna15 contributes to the function of these proteins, we took advantage of two hrp1 mutants that had been previously isolated (14). These mutations, L205S (hrp1-5) and I313N (hrp1-6), are viable but fail to grow at 36 °C. Based on our NMR analysis presented earlier, they also abolish the interaction of Hrp1 and Rna14-Rna15.
Extracts were prepared from the wild-type and mutant hrp1 strains grown at the permissive temperature of 30 °C and were tested for mRNA 3′-end processing activity using in vitro cleavage and polyadenylation assays. The two mutants were normal for processing of RNA substrates containing the GAL7 poly(A) site (Fig. 5 Left). In these reactions, which are done in the presence of ATP and at 30 °C, cleavage is immediately followed by poly(A) addition such that all of the cleaved RNA is adenylated. However, if the reaction temperature is raised to 37 °C, processing was greatly decreased in the two hrp1 mutants (Fig. 5 Left). To examine the cleavage step alone, ATP was replaced with dATP, which prevents elongation of the poly(A) tail. The two hrp1 mutants were also defective for cleavage in reactions performed at 37 °C (Fig. 5 Center). To analyze polyadenylation separately from cleavage, RNA ending at the GAL7 poly(A) site was used as substrate. Poly(A) addition was also impaired in extract from the hrp1 mutants compared with wild-type (Fig. 5 Right). The L205S (hrp1-5) mutant was always more defective than I313N (hrp1-6). Thus, the mutant hrp1 proteins are competent for 3′-end processing at 30 °C, but deficient at the higher temperature, which is likely to destabilize the weakened interaction between Hrp1 and Rna14-Rna15. From these results, we conclude that this contact is important for the function of these proteins in mRNA processing.
Fig. 5.
Functional analysis of Hrp1 mutants. (A) In vitro 3′ end processing reactions. For coupled cleavage-polyadenylation assays (Left), extracts were prepared from the strains expressing the wild-type Hrp1 or the hrp1-5 or hrp1-6 mutants grown at 30 °C. These extracts were incubated with ATP and 32P-labeled full-length GAL7-1 RNA (Pre, unreacted precursor) for 20 min at 30 °C. The same conditions were used for cleavage reactions (Center) except that ATP was replaced with dATP. Poly(A) addition assays (Right) included ATP and precleaved GAL7-9 RNA as precursor. Products were resolved on a denaturing 5% polyacrylamide gel and visualized with a PhosphorImager. Positions of substrate and product are indicated. (B) The I228N hrp1 mutant expresses at normal levels. Western blot of extracts from the PSY818 strain expressing Myc-tagged versions of wild-type Hrp1 or hrp1–I228N mutant on a LEU2-marked plasmid and untagged Hrp1 on a URA3 plasmid. Anti-Myc antibody was used to detect the tagged Hrp1 species, and an actin blot serves as the loading control. (C) The I228N hrp1 mutant is not viable. The strains used in B were streaked on 5-FOA medium to counter select the pRS316–HRP1(URA3) covering plasmid. Cells carrying only the URA3 plasmid, or the hrp1–I228N mutant plasmid plus the URA3 plasmid, could not grow on this plate, whereas those expressing Myc–Hrp1 grew well. Plates were photographed following incubation for 4 d at 24 °C.
We also created an Ile to Asn replacement at position 228 of Hrp1, another residue involved in the interaction of Hrp1 and Rna14-Rna15. The I228N mutant is expressed well (Fig. 5B), but the mutant was not viable when it is the only form of Hrp1 in the cell (Fig. 5C), whereas hrp1-I222S supports wild-type growth.
Discussion
We have investigated, by NMR and then biochemical and yeast molecular genetics methods, the interaction between Hrp1 and the Rna14–Rna15 dimer of dimers during 3′-end processing in yeast. The NMR analysis, GST pull-down, site-directed mutagenesis and in vivo recruitment experiments reveal that the RRM of Hrp1 interacts with Rna14 via the latter protein’s N-terminal residues (Asn201 to Gln240) and through the back surface of the two RRMs that are also used to bind to RNA. Here, residues such as L175δ2, L205δ2, I228δ1, L274δ1, V300γ2, and I313δ1 of Hrp1–RBD experience very large chemical shift perturbations upon binding to Rna14–Rna15 (Fig. 3B), consistent with their closeness to Rna14, whereas L166δ2, V217γ1, I222δ1, I234δ1, V247γ2, and I269δ1 of the RBD exhibit only moderate chemical shift changes upon binding to Rna14–Rna15, suggesting that they are further away from Rna14 in the CF I complex.
The identity of the binding residues identified by the methyl-TROSY experiments was confirmed to be functionally relevant by in vivo studies of the Hrp1 mutants. We found that mutations in the Hrp1 interaction surface perturb mRNA 3′-end processing in vitro and are nonviable at 37 °C, a condition expected to destabilize protein–protein interactions. In agreement with this conclusion, the L205S (hrp1-5) and I313N (hrp1-6) alleles were found to be synthetically lethal with the rna14-1 and rna15-1 mutants, respectively (14). These defects in the rna14/15 mutants can be attributed to the disruption of the interface between Hrp1 and Rna14, an interaction known to be important to make a functional CF I complex (11, 20, 21, 31).
The use of the back of the RRM to interact with Rna14 provides a mechanism to explain why we observe enhanced interaction between Hrp1 and Rna14–Rna15 in the presence of RNA. As in many other RRM proteins, the two Hrp1 RRMs are unrelated structurally in the absence of RNA but form a defined conformation when bound to RNA, bringing the two RRMs in a well defined physical orientation with respect to the RNA and each other. Because both RRMs interact with Rna14–Rna15, this conformation is favored when the Hrp1 protein is bound to RNA, leading to a more effective interaction. This result also suggests that recognition of RNA processing sequences may initiate assembly of a processing complex or its reorganization by favoring protein–protein interactions which are weaker in the absence of specific RNA recognition.
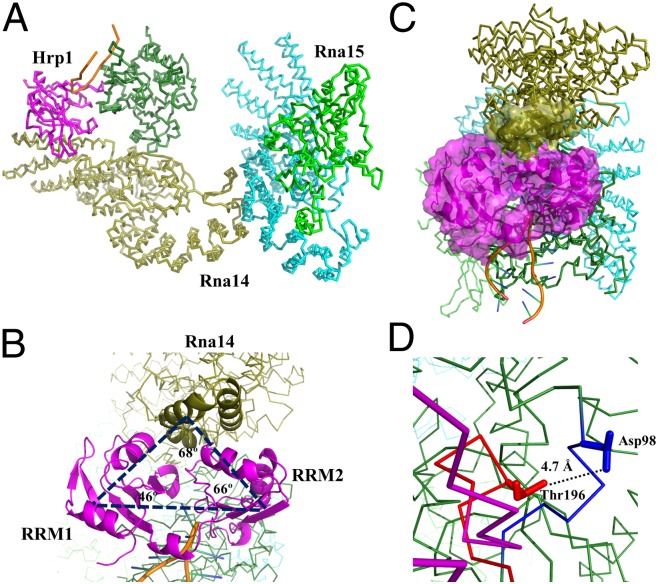
Based on the NMR titration experiments, pull-down assay, and in vivo recruitment experiments, it was possible to generate a model of CF I using the multidock approach of HADDOCK. The model of CF I complex contains protein–protein contacts between Hrp1 and Rna14, Rna14 and Rna14, Rna14 and Rna15, and Hrp1 and Rna15, as well as interactions of both Hrp1 and Rna15 with RNA (21). The modeled structure has the architecture of an inverted-U with Rna14 in the middle of the U, and Hrp1 and Rna15 providing the end arms (Fig. 6A). The distance between the two end arms is ∼120 Å, whereas the bottom of the U has a depth of ∼80 Å. The RRMs (S158 to A233 and K244 to A318) of Hrp1 form two corners of the triangle, whereas N201–Q240 of Rna14 provide the third corner to the triangle (Fig. 6B) and fill the void between the RRMs of Hrp1, as seen in Fig. 6C. The model includes interactions between loop 3 of Hrp1 and loop 5 of Rna15, where loop 3 (residues D193–R198) from Hrp1 makes contact with loop 5 (residues Y93–D98) of Rna15 (Fig. 6D), as reported in the earlier study (21). In the model, loop 3 of Hrp1 is also positioned in close proximity to residues N111–N121 from the OPA region of Rna15. The RNA is situated at the edge of the complex interacting with Rna15 and Hrp1 and might stabilize the whole complex as suggested by NMR and pull down experiments. Other important interactions in the complex are between residues Asn536-Glu583 of Rna14 with residues Ser365–Asp395 and Leu429–Ile445 of another Rna14 molecule, and those between Rna14 and Rna15, as described in earlier studies (Fig. S6) (15, 19, 21). These features reported in a piecemeal fashion are now integrated in a single model of the CF I complex (Fig. S7).
Fig. 6.
Architecture of the CF I complex. (A) The CF I complex composed of Rna14, Rna15, Hrp1 and RNA is shown to have an inverted U-like structure where Rna14 provides the base of inverted U, whereas Hrp1 and Rna15 form the end arms. (B) The top view of CF I complex provides a different impression of the shape of the complex. The two RRMs of Hrp1 (colored in magenta) form the two corners of the triangle, whereas Rna14 (colored in olive) forms the third corner. (C) Surface view of the Hrp1 and Rna14 interface. Hrp1 is shown in magenta, whereas Rna14 is in olive. Residues N201 to Q240 of Rna14 are shown in a surface representation. (D) The interactions between loop 3 of Hrp1 and loop 5 of Rna15 are highlighted; the distance between Thr196 (Cγ2) of Hrp1 and Asp98 (Cγ) of Rna15 is shown explicitly.
In conclusion, we have successfully investigated components of the ~300-kDa CF I complex by NMR spectroscopy using methyl-specific labeling techniques combined with pull-down and site-directed mutagenesis experiments, and identified a protein–protein interface related to an RNA recognition surface. The identification of interactions between different protein factors of CF I and their spatial juxtaposition will help us better understand the cleavage and polyadenylation reaction during yeast 3′-end mRNA processing. In this complex, Hrp1 is sandwiched between other protein factors and RNA, and communicates between specific RNA sequences recognized by Hrp1 and Rna15 and the assembly of the processing complex, thereby providing a physical link between recognition of processing signals and complex assembly.
Experimental Procedures
Cloning, Overexpression, and Purification.
We prepared full length Hrp1 (1–535 aa), the RNA binding domain (RBD) containing two RNA recognition motifs (RRMs; 156–324 aa), and RBD plus C-terminal region (RBDCTR; 156–535). All Hrp1 constructs were expressed in Rosetta cells grown in minimal M9 media. For the production of U-[2H], Ile-d1-[13CH3] and Val, Leu-[13CH3, 12CD3] samples, 64 mg/L of α-ketobutyric acid (methyl-13CH3), and 105 mg/L of α-ketoisovaleric acid (dimethyl-13CH3, 12CD3) were added to the culture 1 h before induction. Proteins were purified using nickel-affinity chromatography. The His6-tag was removed by tobacco etch virus (TEV) protease, followed by size-exclusion chromatography.
Rna14 and Rna15 were coexpressed and copurified from a pET-Duet vector as described (20). This complex was concentrated up to 16 mg/mL in 200 mM NaCl/20 mM Tris (pH 8.0) buffer and stored at −80 °C in small aliquots.
NMR.
Backbone and side chain assignments for Hrp1 (RBD) were verified using standard suites of NMR experiments, including HNCACB, CBCACONH, HNCO and HCCH/CCH-TOCSY. Protein concentrations ranged from 80 μM to 1.2 mM. All spectra were processed with NMRPipe (32) or Topspin (Bruker). Spectral analysis was performed using CCPNMR (33) or CARA (www.cara.nmr.ch).
Stereospecific Assignment of Ile, Leu, and Val.
Stereospecific resonance assignments were done using fractionally labeled samples and CT-HSQC experiments, as described (34, 35). We also used 13C-NOESY-HSQC and HCCH-COSY experiments to confirm side-chain assignments.
Haddock Modeling.
All protein components (Hrp1, Rna15, and Rna14) of the CF I complex were modeled using I-TASSER (30) based on homology with other proteins or known structures of part of any CF I component, as available in the Protein Data Bank (PDB). The nucleic acid component was taken from the earlier published structure 2KM8. After generating the protein components, these structures were docked as a complex using the Haddock multidock server (http://haddock.science.uu.nl/services/HADDOCK/haddockserver-multi.html) with appropriate binding interface definition. The final protocol for docking and refinement was executed using the default parameter sets on the server using automated semiflexible simulated annealing with 200 lowest energy solutions, followed by water shell refinement.
In Vitro 3′-End Processing Assays.
Preparation of yeast cell extracts, transcription of [α-32P] UTP-labeled GAL7-1 RNA or precleaved GAL7-9, and processing assays were performed as described (36).
Supplementary Material
Acknowledgments
We thank Drs. A Bonvin and Marc van Dijk for their valuable help during HADDOCK docking, Dr. A Bohm for Rna14–Rna15 construct, and other members of G.V. group for many helpful discussions and suggestions. This work was supported National Institutes of Health Grants GM064440 (to G.V.) and GM41752 (to C.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214102110/-/DCSupplemental.
References
- 1.Wahle E, Rüegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23(3):277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 2.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38(9):2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11(21):2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 4.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65(7-8):1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmid M, Jensen TH. Quality control of mRNP in the nucleus. Chromosoma. 2008;117(5):419–429. doi: 10.1007/s00412-008-0166-4. [DOI] [PubMed] [Google Scholar]
- 6.Iglesias N, Stutz F. Regulation of mRNP dynamics along the export pathway. FEBS Lett. 2008;582(14):1987–1996. doi: 10.1016/j.febslet.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 7.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4(7):223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63(2):405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danckwardt S, Hentze MW, Kulozik AE. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27(3):482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rondon AG, Proudfoot NJ. Nuclear roadblocks for mRNA export. Cell. 2008;135(2):207–208. doi: 10.1016/j.cell.2008.09.053. [DOI] [PubMed] [Google Scholar]
- 11.Kim Guisbert KS, Li H, Guthrie C. Alternative 3′ pre-mRNA processing in Saccharomyces cerevisiae is modulated by Nab4/Hrp1 in vivo. PLoS Biol. 2007;5(1):e6. doi: 10.1371/journal.pbio.0050006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graber JH, McAllister GD, Smith TF. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 2002;30(8):1851–1858. doi: 10.1093/nar/30.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dichtl B, Keller W. Recognition of polyadenylation sites in yeast pre-mRNAs by cleavage and polyadenylation factor. EMBO J. 2001;20(12):3197–3209. doi: 10.1093/emboj/20.12.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kessler MM, et al. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11(19):2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pérez-Cañadillas JM. Grabbing the message: Structural basis of mRNA 3’UTR recognition by Hrp1. EMBO J. 2006;25(13):3167–3178. doi: 10.1038/sj.emboj.7601190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu X, et al. The C-terminal domains of vertebrate CstF-64 and its yeast orthologue Rna15 form a new structure critical for mRNA 3′-end processing. J Biol Chem. 2007;282(3):2101–2115. doi: 10.1074/jbc.M609981200. [DOI] [PubMed] [Google Scholar]
- 17.Legrand P, Pinaud N, Minvielle-Sébastia L, Fribourg S. The structure of the CstF-77 homodimer provides insights into CstF assembly. Nucleic Acids Res. 2007;35(13):4515–4522. doi: 10.1093/nar/gkm458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai Y, et al. Crystal structure of murine CstF-77: Dimeric association and implications for polyadenylation of mRNA precursors. Mol Cell. 2007;25(6):863–875. doi: 10.1016/j.molcel.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 19.Moreno-Morcillo M, Minvielle-Sébastia L, Fribourg S, Mackereth CD. Locked tether formation by cooperative folding of Rna14p monkeytail and Rna15p hinge domains in the yeast CF IA complex. Structure. 2011;19(4):534–545. doi: 10.1016/j.str.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Noble CG, Walker PA, Calder LJ, Taylor IA. Rna14-Rna15 assembly mediates the RNA-binding capability of Saccharomyces cerevisiae cleavage factor IA. Nucleic Acids Res. 2004;32(11):3364–3375. doi: 10.1093/nar/gkh664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leeper TC, Qu X, Lu C, Moore C, Varani G. Novel protein-protein contacts facilitate mRNA 3′-processing signal recognition by Rna15 and Hrp1. J Mol Biol. 2010;401(3):334–349. doi: 10.1016/j.jmb.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pancevac C, Goldstone DC, Ramos A, Taylor IA. Structure of the Rna15 RRM-RNA complex reveals the molecular basis of GU specificity in transcriptional 3′-end processing factors. Nucleic Acids Res. 2010;38(9):3119–3132. doi: 10.1093/nar/gkq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pérez Cañadillas JM, Varani G. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 2003;22(11):2821–2830. doi: 10.1093/emboj/cdg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon JM, et al. Reconstitution of CF IA from overexpressed subunits reveals stoichiometry and provides insights into molecular topology. Biochemistry. 2011;50(47):10203–10214. doi: 10.1021/bi200964p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulson AR, Tong L. Crystal structure of the Rna14-Rna15 complex. RNA. 2012;18(6):1154–1162. doi: 10.1261/rna.032524.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiaux J, Bertelsen EB, Horwich AL, Wüthrich K. NMR analysis of a 900K GroEL GroES complex. Nature. 2002;418(6894):207–211. doi: 10.1038/nature00860. [DOI] [PubMed] [Google Scholar]
- 27.Sprangers R, Kay LE. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007;445(7128):618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- 28.Neri D, Szyperski T, Otting G, Senn H, Wüthrich K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989;28(19):7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 29.Dominguez C, Boelens R, Bonvin AM. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 30.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gross S, Moore CL. Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol Cell Biol. 2001;21(23):8045–8055. doi: 10.1128/MCB.21.23.8045-8055.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 33.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59(4):687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 34.Barnwal RP, Atreya HS, Chary KV. Chemical shift based editing of CH3 groups in fractionally 13C-labelled proteins using GFT (3, 2)D CT-HCCH-COSY: Stereospecific assignments of CH3 groups of Val and Leu residues. J Biomol NMR. 2008;42(2):149–154. doi: 10.1007/s10858-008-9273-z. [DOI] [PubMed] [Google Scholar]
- 35.Fairbrother WJ, et al. Polypeptide backbone resonance assignments and secondary structure of Bacillus subtilis enzyme IIIglc determined by two-dimensional and three-dimensional heteronuclear NMR spectroscopy. Biochemistry. 1991;30(28):6896–6907. doi: 10.1021/bi00242a013. [DOI] [PubMed] [Google Scholar]
- 36.Zhao J, Kessler M, Helmling S, O’Connor JP, Moore C. Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition of mRNA precursor. Mol Cell Biol. 1999;19(11):7733–7740. doi: 10.1128/mcb.19.11.7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.