Abstract
Contact-dependent growth inhibition (CDI) systems encode polymorphic toxin/immunity proteins that mediate competition between neighboring bacterial cells. We present crystal structures of CDI toxin/immunity complexes from Escherichia coli EC869 and Burkholderia pseudomallei 1026b. Despite sharing little sequence identity, the toxin domains are structurally similar and have homology to endonucleases. The EC869 toxin is a Zn2+-dependent DNase capable of completely degrading the genomes of target cells, whereas the Bp1026b toxin cleaves the aminoacyl acceptor stems of tRNA molecules. Each immunity protein binds and inactivates its cognate toxin in a unique manner. The EC869 toxin/immunity complex is stabilized through an unusual β-augmentation interaction. In contrast, the Bp1026b immunity protein exploits shape and charge complementarity to occlude the toxin active site. These structures represent the initial glimpse into the CDI toxin/immunity network, illustrating how sequence-diverse toxins adopt convergent folds yet retain distinct binding interactions with cognate immunity proteins. Moreover, we present visual demonstration of CDI toxin delivery into a target cell.
Keywords: structural biology, bacterial competition, β-complementation, tRNase activity
Bacteria use a variety of strategies to compete and communicate with one another in the environment. Contact-dependent growth inhibition (CDI) is a mechanism that allows some Gram-negative bacteria to block the growth of neighboring cells (1, 2). CDI is mediated by the CdiB/CdiA family of two-partner secretion proteins. CdiB is a predicted outer membrane β-barrel protein required for secretion of the CdiA effector protein (2). CdiA exoproteins are very large (250–650 kDa) and composed of an N-terminal transport domain followed by a variable number of hemagglutinin repeats (1). The hemagglutinin-repeat region is predicted to form an extended β-helical filament capable of projecting several hundred angstroms from the inhibitor cell surface (3). The current model of CDI postulates that CdiA binds to receptors on the surface of susceptible bacteria, initiating delivery of a CdiA-derived toxin into the target cell (Fig. 1). The CDI toxin activity is contained within the C-terminal 250–300 residues of CdiA proteins—a region collectively termed “CdiA-CT” (1). CdiA-CT sequences are highly variable between CDI systems, but these toxin regions are typically demarcated by a conserved peptide motif: (Q/E)LYN in Burkholderia species (5) and VENN in most other bacteria (1). There are more than 60 CdiA-CT families based on sequence homology, suggesting that CDI+ bacteria deploy a wide variety of toxins. CdiA-CTs can dissipate the proton motive force (6), degrade DNA (1), and cleave tRNA molecules (5, 7), and each activity is sufficient to inhibit cell growth. CDI is active against bacteria, and therefore CDI+ cells must produce a CdiI immunity protein to protect themselves from autoinhibition (Fig. 1). CdiI proteins are also highly variable and bind their cognate CdiA-CTs to block toxin activity. Because CdiA-CT/CdiI binding interactions are highly specific, immunity proteins provide no protection from the toxins deployed by other CDI systems (1, 5, 8). Thus, intercellular competition is thought to drive the diversification of CDI toxin/immunity pairs. Here, we describe the crystal structures of two different CdiA-CT/CdiI complexes, which provide insights into CDI diversity and mechanisms of toxicity and immunity.
Fig. 1.
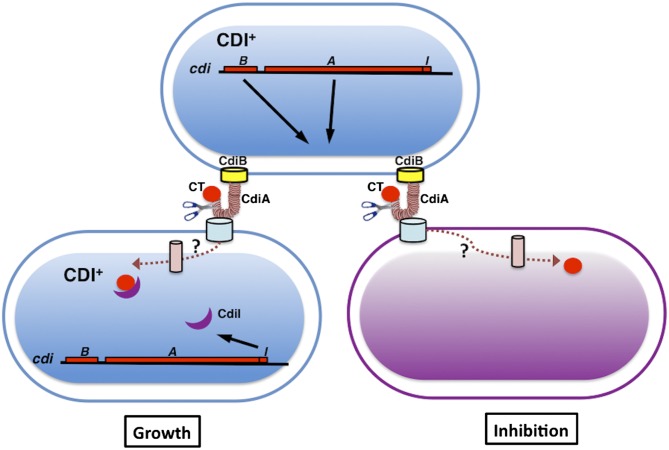
The CDI pathway. CDI+ cells containing the cdiBAI gene cluster express CdiB and CdiA at the cell surface. Contact between CdiA and the BamA receptor on the surface of target cells results in delivery of the CdiA-CT toxin into the target cell. The mechanisms of toxin translocation are not understood, but BamA (4) and unknown inner membrane components are hypothesized to mediate transport. Cells carrying the identical CDI system (depicted in blue cells) are protected from growth inhibition by the CdiI immunity protein, which specifically binds and inactivates the CdiA-CT toxin. Nonimmune cells are inhibited by CdiA-CT (depicted in purple cells).
Results
CdiA-CT/CdiI Crystallization and Structure Determination.
To explore the structural diversity of CDI toxin/immunity proteins, we focused on CdiA-CT/CdiI pairs from Burkholderia pseudomallei 1026b (Bp1026b) and Escherichia coli O157:H7 strain EC869 (EC869). The CdiA-CTIIBp1026b/CdiIIIBp1026b proteins are derived from the CDI locus on chromosome II of Bp1026b (5), and the CdiA-CTo11EC869/CdiIo11EC869 complex is encoded by the 11th “orphan” (o11) module of E. coli EC869. Orphan cdiA-CT/cdiI modules are toxin/immunity gene pairs that have been displaced from full-length cdiA genes (8). Tandem arrays of these modules are often associated with CDI systems and are thought to represent reservoirs of toxin/immunity diversity. We coexpressed each CdiA-CT together with a His6-tagged version of its immunity protein, and the resulting CdiA-CT/CdiI-His6 complexes were purified to near homogeneity (Fig. S1). The CdiA-CTo11EC869/CdiIo11EC869 complex was stable; however, the N-terminus of the CdiA-CTIIBp1026b showed significant degradation after purification, suggesting that this region is sensitive to proteolysis. Therefore, we generated a truncated version of CdiA-CTIIBp1026b beginning at residue Gly123 (numbered from Glu1 of the ELYN motif), which still binds to the CdiIIIBp1026b immunity protein and retains full toxin activity (5).
The CdiA-CTo11EC869/CdiIo11EC869 crystal structure was solved to 2.35 Å resolution by Se-SAD (single anomalous dispersion) phasing. The crystal space group was C2221 with one complex per asymmetric unit. The structural model contains CdiA-CTo11EC869 residues Val85 – Lys297 (numbered from Val1 of the VENN motif) and Ala2 – Arg164 of CdiIo11EC869. In addition, 55 water molecules, three Y3+ ions, and one Zn2+ ion were included in the final model, resulting in an Rwork/Rfree of 18.0/22.9 (Table S1). The Bp1026b toxin/immunity complex contains no internal methionine residues for Se-Met incorporation, so crystals were soaked with bromide and the structure was solved to 2.65 Å resolution by Br-SAD phasing. The CdiA-CTIIBp1026b/CdiIIIBp1026b complex crystallized in space group F222 with four complexes per asymmetric unit. The structural model contains CdiA-CTIIBp1026b residues Gly163 – Pro294 and residues Ala2 – Arg101 of CdiIIIBp1026b. In addition, 33 water molecules were included in the final model to yield an Rwork/Rfree of 20.4/24.5 (Table S1).
Structure of the CdiA-CTo11EC869/CdiIo11EC869 Complex.
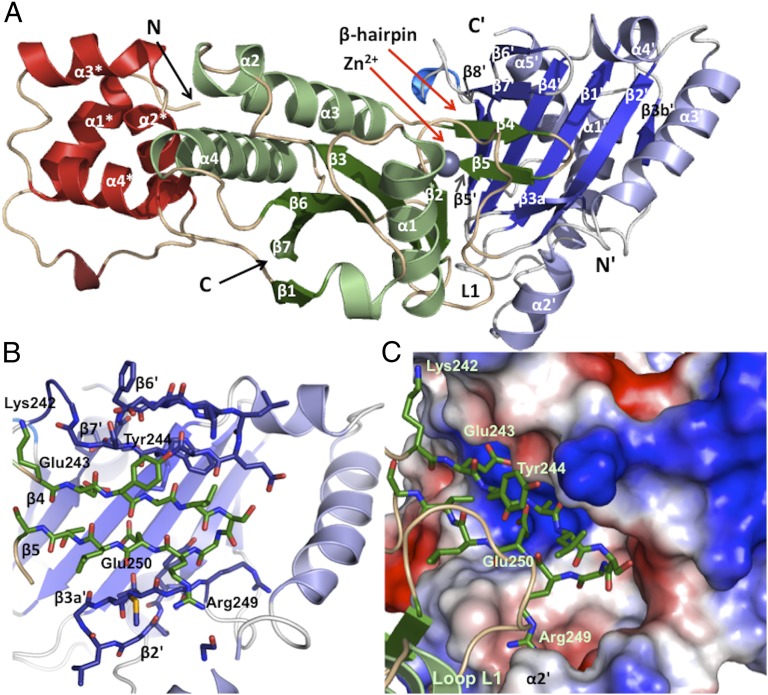
The CdiA-CTo11EC869 is composed of two domains. Residues Val85 – Arg149 form an N-terminal four α-helical bundle (α1*-α4*), and residues Thr153 – Lys297 form a C-terminal ellipsoidal α/β domain containing one 310-helix, four α-helices (α1-α4), and seven β-strands (Fig. 2A and Fig. S2B). The central mixed β-sheet (β2, β3, β6, β7, β1) of the C-terminal domain forms a half-β-barrel–like structure. Two helices (α3, α4) are located on the outside of this half-barrel, and the C-terminal end of α1 interacts with its central core. A β-hairpin (β4, β5) protrudes from the C-terminal domain near β2 and the extended loop region (L1). The CdiIo11EC869 immunity protein consists of five α-helices and eight β-strands that form two β-sheets (Fig. 2A and Fig. S2B). The central six-stranded antiparallel β-sheet (β3a′and b′, β2′, β1′, β4′, β5′, β8′) is decorated with four α-helices (α1′, α2′, α3′, α4′) inserted between strands β3′ and β4′. A fifth C-terminal helix (α5′) runs parallel to the central β-sheet, and a short two-stranded β-sheet (β6′, β7′) connects β5′ and β8′ of the central β-sheet.
Fig. 2.
Structure of the EC869 CdiA-CTo11/CdiIo11 complex. (A) Ribbon representation of the CdiA-CTo11EC869/CdiI o11EC869 complex. CdiA-CTo11EC869 contains two domains, an N-terminal α-helical bundle (red) and a C-terminal α/β nuclease domain (green). The four helices marked with asterisks (*) form the N-terminal helical bundle of CdiA-CTo11EC869. The CdiIo11EC869 immunity protein (blue) is composed of a single α/β domain. The secondary structure elements of each protein are identified and their N and C termini are indicated. All immunity protein elements are denoted with a prime symbol (′) to differentiate them from the toxin secondary structure elements. The active site Zn2+ ion is depicted as a purple sphere. (B) The CdiA-CTo11EC869 and CdiIo11EC869 proteins interact through β-augmentation. The β4,β5-hairpin of CdiA-CTo11EC869 (carbon atoms, green) inserts into the CdiI o11EC869 immunity protein (carbon atoms, blue) to form a six-stranded antiparallel β-sheet. β-hairpin residues and ion pairs are represented as sticks (where the oxygen, nitrogen, and sulfur atoms are colored red, dark blue, and yellow, respectively). (C) CdiA-CTo11EC869 β-hairpin (green sticks) along with the extended loop region L1 fits snugly into the molecular surface representation of CdiI o11EC869. White surfaces represent hydrophobic regions, and the red and blue surfaces indicate negative and positive electrostatic potential, respectively.
The CdiA-CTo11EC869/CdiIo11EC869 binding interaction is mediated by β-augmentation, in which the toxin donates its β-hairpin (β4, β5) to the immunity protein to produce a six-stranded antiparallel β-sheet. The augmented sheet consists of CdiIo11EC869 β6′ and β7′, followed by the β4-β5 hairpin from CdiA-CTo11EC869, and completed by CdiIo11EC869 β3a′ and β2′ (Fig. 2B). The interface is stabilized by ion pairs between the toxin β-hairpin and the immunity central β-sheet and α2′ (Fig. S2C and Table S2). Additionally, there are contributions by the toxin L1 loop region interacting with α2′ of CdiIo11EC869 facilitated by ion pairs and hydrophobic contacts (Fig. 2C and Fig. S2C). The toxin/immunity interface buries 1,996 Å2 of the surface area, ∼12% and 10% of the solvent-accessible surface area of CdiA-CTo11EC869 and CdiIo11EC869, respectively. The EC869 toxin/immunity proteins have high affinity for one another (Kd = 17.8 ± 7 nM), and the complex has greater thermal stability [melting temperature (Tm) 65.1 ± 0.9 °C] than isolated CdiA-CTo11EC869 (Tm 53.8 ± 1.4 °C) and CdiIo11EC869 (Tm 50.1 ± 0.9 °C) (Fig. S2D).
Structure of the CdiA-CTIIBp1026b/CdiIIIBp1026b Complex.
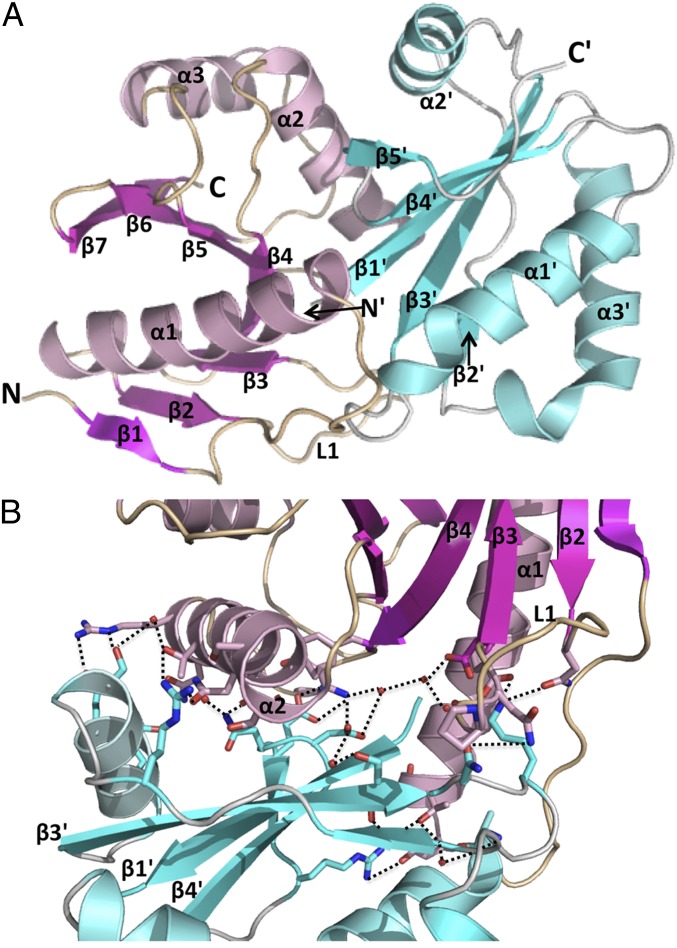
The CdiA-CTIIBp1026b toxin consists of a seven-stranded mixed β-sheet and three α-helices. Like the C-terminal domain of the EC869 toxin, the central β-sheet of CdiA-CTIIBp1026b forms a half-β-barrel–like structure with the C-terminal end of a long α-helix (α1) running through its central cavity. The remaining α-helices (α2, α3) decorate the outside of the half-barrel (Fig. 3A and Fig. S3A). The CdiIIIBp1026b immunity protein has a simple topology with a five-stranded antiparallel β-sheet decorated with three α-helices (Fig. 3A and Fig. S3B). The CdiA-CTIIBp1026b/CdiIIIBp1026b complex interface is dominated by electrostatic interactions via residue side-chains (Fig. 3B and Fig. S3C). Toxin residues within the N-terminal half of long-helix α1, α2, and extended loop L1 interact with immunity protein residues at the end of the β-sheet and in helix α2′. The interaction network is extensive, with at least 20 ion pairs and direct hydrogen bonds between the toxin and immunity proteins (Table S2). In addition, a network of water-mediated hydrogen bonds also contributes to the CdiA-CTIIBp1026b/CdiIIIBp1026b interface. The CdiA-CTIIBp1026b/CdiIIIBp1026b interface buries 2,044 Å2, which corresponds to 17% and 22% of CdiA-CTIIBp1026b and CdiIIIBp1026b total surface area (respectively). The Bp1026b complex has a dissociation constant of 21.1 ± 9 nM and a Tm of 70.4 ± 0.7 °C (Fig. S3D). The Tm of CdiA-CTIIBp1026b is 52.3 ± 0.7 °C and CdiIIIBp1026b is 60.9 ±1.2 °C (Fig. S3D), again demonstrating that the complex is more stable than the isolated toxin and immunity proteins.
Fig. 3.
Structure of the Bp1026b CdiA-CTII/CdiIII complex. (A) The CdiA-CTllBp1026b toxin (pink) and CdiIllBp1026b immunity protein (cyan) are depicted in ribbon representation with secondary structure elements indicated. All immunity protein elements are denoted with a prime symbol (′) to differentiate them from the toxin secondary structure elements. (B) The interface between CdiA-CTllBp1026b (pink) and CdiIllBp1026b (cyan) is formed by an extensive network of ion pairs and hydrogen bonds. Within the network, interacting residue side chains are represented as sticks (oxygen and nitrogen atoms are colored red and blue, respectively), water molecules as red spheres, and interacting bonds as black dotted lines.
Comparison of CdiA-CT/CdiI Complex Structures.
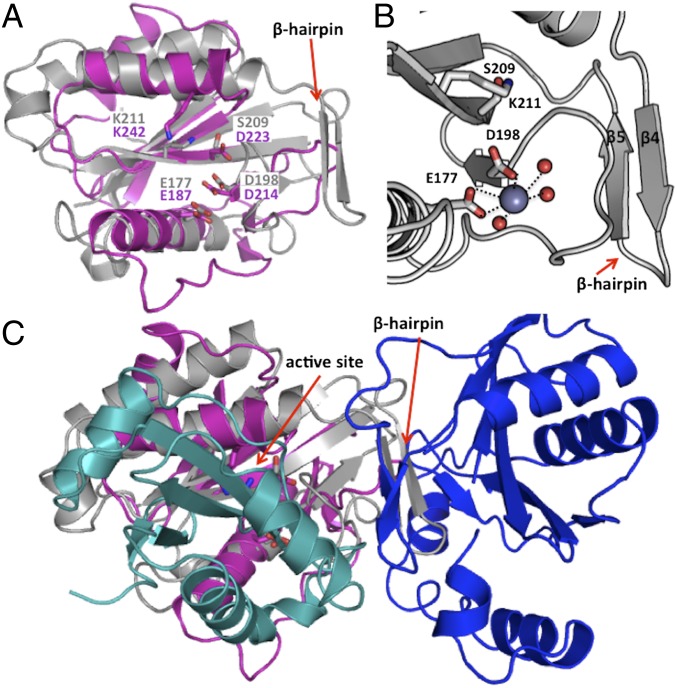
Although the two toxin domains share only ∼15% sequence identity and have distinct topologies, their 3D structures superimpose with an rmsd of 3.9 Å and Z-score of 5.8 (9) (Fig. 4A and Fig. S4A). Notably, CdiA-CTIIBp1026b lacks the β-hairpin element found in the CdiA-CTo11EC869 toxin. Both CdiA-CT C-terminal domains are structurally similar (9) to type IIS restriction endonucleases (10) (Table S3), suggesting that the toxins have metal-dependent DNase activity. Furthermore, metal K-edge absorption analysis revealed that native CdiA-CTo11EC869/CdiIo11EC869 crystals have significant zinc content. Based on structural homology (9) to the BspD6I endonuclease, CdiA-CTo11EC869 residues Glu177, Asp198, Ser209, and Lys211 are predicted to form the nuclease active site. Additionally, extra electron density within the active site vicinity was modeled as a Zn2+ ion, which is coordinated by Glu177, Asp198, and three water molecules in a ββα–metal motif (β2, β3, α1) (Fig. 4B and Table S4) (11). Similarly, CdiA-CTIIBp1026b residues Glu187, Asp214, Asp223, and Lys242 are predicted to form an active site and coordinate a catalytic metal ion within a ββα–metal motif (β3, β4, α1) (Fig. 4A and Table S4). However, there is no density attributable to an active-site cation in the CdiA-CTIIBp1026b/CdiIIIBp1026b complex, presumably because direct hydrogen bonds between the immunity protein and active site residues preclude metal binding. These predictions are supported by our previous work showing that CdiA-CTIIBp1026b is a Mg2+-dependent tRNase and that its nuclease activity is ablated by the Asp214Ala mutation (5).
Fig. 4.
Structural superimposition of EC869 and Bp1026b CdiA-CT/CdiI protein complexes. (A) Predicted active site residues of the EC869 and Bp1026b toxin domains. The two toxin domains are superimposed and active site residues are rendered as stick representations. EC869 and Bp1026b carbon atoms are colored gray and pink, respectively; oxygen and nitrogen atoms are colored red and blue, respectively. (B) Coordination of Zn2+ within the CdiA-CTo11EC869 active site. The Zn2+ ion is depicted as a purple sphere, ordered waters as smaller red spheres, and interacting bonds with Zn2+ are depicted as black dotted lines. (C) Superimposition of the EC869 and Bp1026b CdiA-CT/CdiI protein complexes. Ribbon representations of CdiA-CTllBp1026b and CdiIllBp1026b are colored pink and cyan, respectively, and the C-terminal domains of CdiA-CTo11EC869 and CdiI o11EC869 are colored gray and blue, respectively. The C-terminal toxin domains superimpose upon one another, whereas the immunity proteins do not. The N-terminal α-helical domain of CdiA-CTo11EC869 has been omitted for clarity.
Despite their common α/β fold, the CdiIo11EC869 and CdiIIIBp1026b immunity proteins share little sequence identity (∼12%) or structural homology (Z-score of 0.2) (9) (Fig. S4B). Moreover, each CdiI protein binds its cognate toxin at a completely different location (Fig. 4C), consistent with the specificity of CDI immunity. The CdiIIIBp1026b protein binds directly over the central core of CdiA-CTIIBp1026b to produce a “closed clam” structure. This structure provides a mechanism for immunity because CdiIIIBp1026b occludes the predicted nuclease active site (Fig. 4C), and presumably prevents the toxin from binding substrate. In contrast, the CdiIo11EC869 immunity protein binds to the C-terminal side of the toxin domain in a “lock-and-key” type of manner, producing an elongated complex that buries little of the toxin’s central core (Fig. 4C). Because the CdiA-CTo11EC869 active site is solvent-exposed in the complex, it is not immediately clear how CdiIo11EC869 neutralizes the toxin. It is possible that CdiIo11EC869 prevents nucleic acid binding, or alternatively, the conformation of the toxin could be altered upon binding the immunity protein.
CdiA-CT Toxins Have Distinct Nuclease Activities.
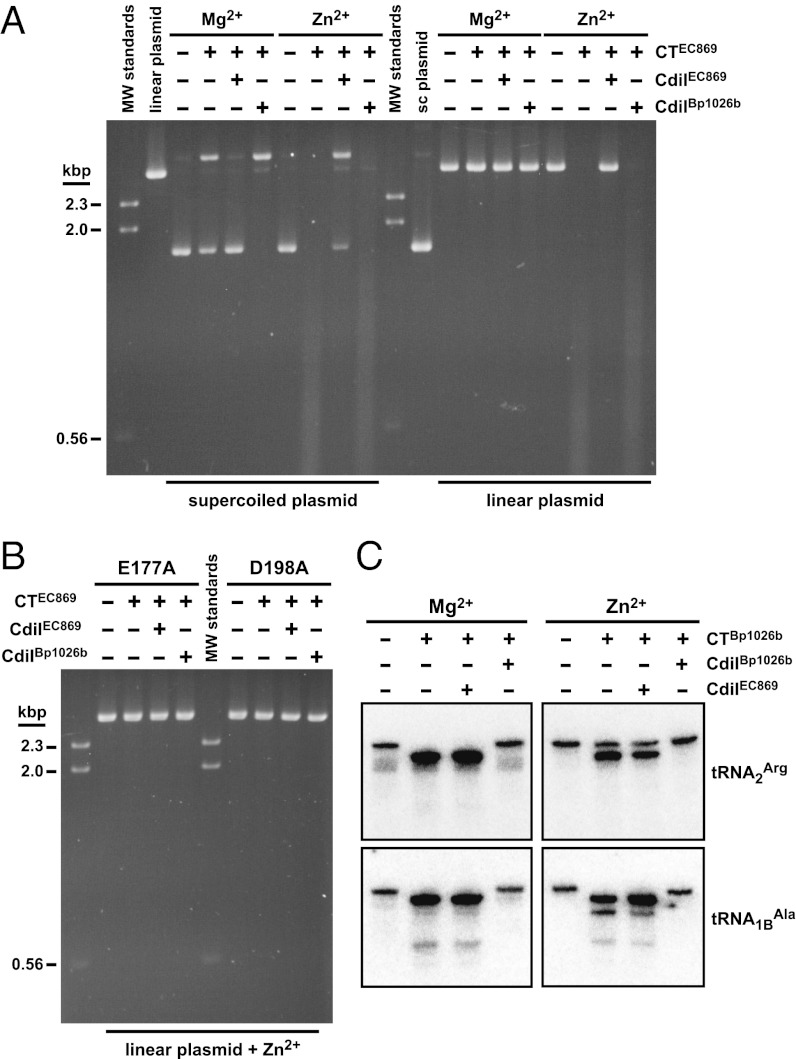
To test whether CdiA-CTo11EC869 is a DNase, we isolated the domain from its immunity protein and assayed the purified toxin for nuclease activity in vitro. CdiA-CTo11EC869 converted supercoiled plasmid DNA into an open-circular form in the presence of Mg2+ (Fig. 5A), consistent with “nickase” activity in which only one strand of double-stranded DNA is cleaved (10). Because Zn2+ is coordinated in the predicted CdiA-CTo11EC869 active site, we also tested nuclease activity with this cation. Remarkably, Zn2+ greatly stimulated DNase activity, leading to the complete degradation of both supercoiled and linear plasmid substrates (Fig. 5A). CdiA-CTo11EC869 activity was completely blocked by CdiIo11EC869 in reactions supplemented with Mg2+, but nickase activity was still apparent in the presence of Zn2+ (Fig. 5A). In contrast, noncognate CdiIIIBp1026b immunity protein had no effect on DNase activity (Fig. 5A). We also mutated two of the predicted active site residues in CdiA-CTo11EC869 to test their role in catalysis. Both Glu177Ala and Asp198Ala mutants of CdiA-CTo11EC869 copurified with His6-tagged CdiIo11EC869, indicating that the toxin variants retain their native fold, but neither mutant exhibited DNase activity in vitro (Fig. 5B).
Fig. 5.
CdiA-CT toxins have distinct nuclease activities. (A) DNase activity of the CdiA-CTo11EC869 toxin on supercoiled and linear plasmid substrates. Plasmid DNA was incubated with purified CdiA-CTo11EC869 in the presence of either Mg2+ or Zn2+ and reactions were analyzed by agarose gel electrophoresis and ethidium bromide staining. Reactions also included either purified CdiIo11EC869 or CdiIIIBp1026b immunity proteins where indicated. Untreated supercoiled linear plasmid substrates were included as controls for the migration of undigested DNA. The migration positions of linear molecular weight (MW) DNA standards are indicated in kilobase pairs (kbp). (B) Mutation of predicted active site residues ablates CdiA-CTo11EC869 DNase activity. Linear plasmid DNA was incubated with purified CdiA-CTo11EC869 containing the Glu177Ala (E177A) and Asp198Ala (D198A) mutations in buffer supplemented with Zn2+. Reactions also contained CdiIo11EC869 or CdiIIIBp1026b immunity proteins where indicated. (C) The CdiA-CTIIBp1026b toxin has tRNase activity. Purified E. coli tRNA was treated with CdiA-CTIIBp1026b toxin in reactions supplemented with Mg2+ or Zn2+. Reactions contained CdiIo11EC869 or CdiIIIBp1026b immunity proteins where indicated and were run on denaturing polyacrylamide gels and analyzed by Northern blot hybridization using radiolabeled probes to tRNA2Arg and tRNA1BAla.
We previously reported that CdiA-CTIIBp1026b is a tRNase (5), but its structural resemblance to CdiA-CTo11EC869 suggests that it may also possess Zn2+-dependent DNase activity. However, purified CdiA-CTIIBp1026b showed no nuclease activity on plasmid DNA in the presence of either Zn2+ or Mg2+ (Fig. S5A) but readily cleaved tRNA under the same conditions (Fig. 5C). Similarly, the CdiA-CTo11EC869 toxin is specific for DNA, with no nuclease activity detected on tRNA substrates (Fig. S5B). We next sought to determine where the CdiA-CTIIBp1026b toxin cleaves its tRNA target. Digested tRNA appears to be nearly the same size as full-length tRNA (Fig. 5C), suggesting the toxin cleaves near either the 5′ or 3′ ends of the molecules. S1 nuclease protection analysis revealed that CdiA-CTIIBp1026b cleaves E. coli tRNA2Arg after residues A70 and U71 in the aminoacyl acceptor stem (Fig. S5 C–E). These sites suggest that the toxin binds double-stranded RNA but cuts only one strand of the duplex to inactivate tRNA. Together, these observations demonstrate that each CdiA-CT has a distinct nuclease activity and metal requirement despite sharing a common fold.
CdiA-CTo11EC869 Toxin Degrades Target Cell DNA During CDI.
We next asked whether the CdiA-CTo11EC869 toxin displays DNase activity when expressed inside cells. We also examined a truncated CdiA-CTo11EC869 protein (residues Ala142 – Lys297) in these experiments to determine whether the N-terminal α-helical bundle (Fig. 2A) is required for DNase activity. This toxin-encoding sequence could not be cloned in the absence of the cognate immunity gene; therefore, we used controlled proteolysis of CdiIo11EC869 to activate the CdiA-CTo11EC869 toxin inside E. coli cells (8, 12). Briefly, the C terminus of CdiIo11EC869 was tagged with the ssrA(DAS) peptide, which targets the immunity protein for degradation by the ClpXP protease, thereby liberating the CdiA-CT to exert its toxic activity. Visualization of DAPI-stained cells showed that chromosomal DNA was lost after 3 h of toxin activation (Fig. S6). This avid DNase activity is consistent with the Zn2+-dependent activity observed in vitro and strongly suggests that Zn2+ is the relevant cation for in vivo activity. In contrast, CdiA-CTo11EC869 carrying the Asp198Ala active site mutation had no effect on cellular DAPI staining (Fig. S6). These results demonstrate that the C-terminal α/β domain of CdiA-CTo11EC869 is sufficient for DNase activity. Presumably, the α-helical bundle domain and the remainder of the N-terminal region perform another function during CDI.
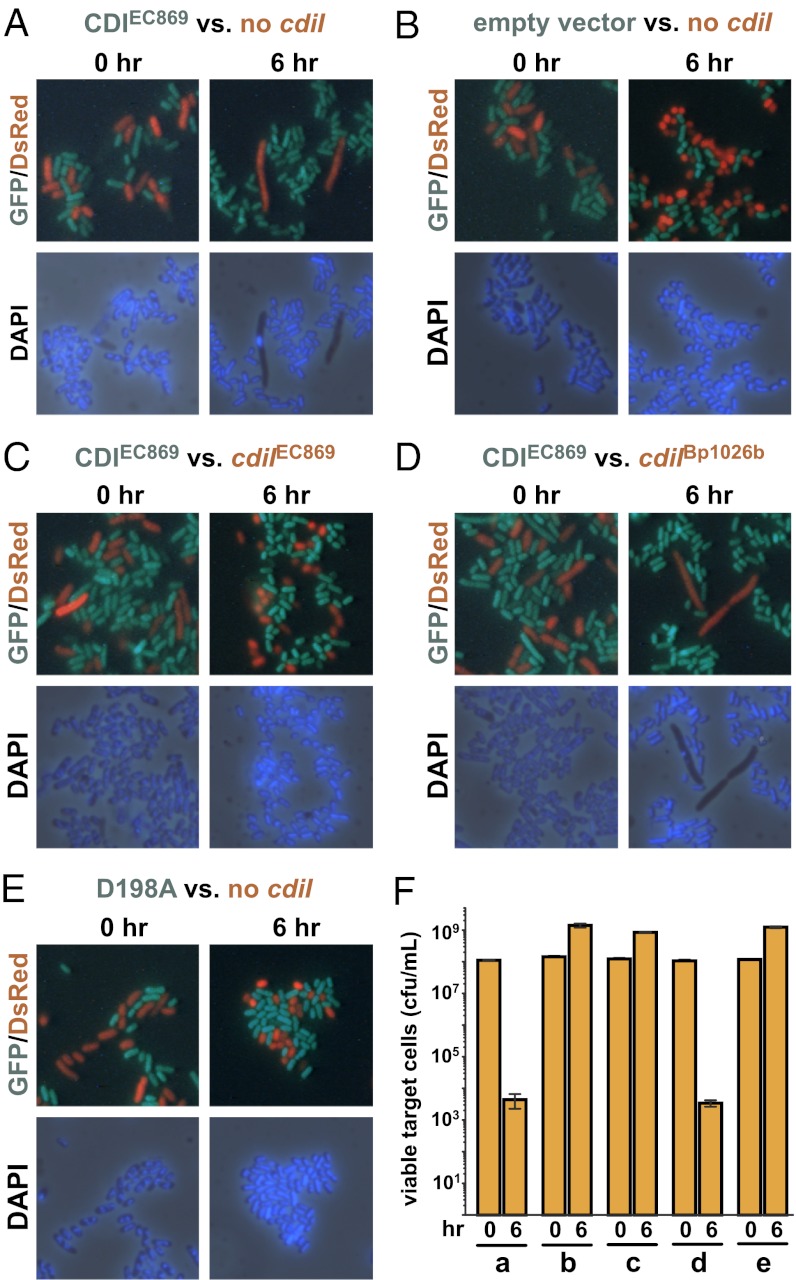
Finally, we tested whether CdiA-CTo11EC869 DNase activity is responsible for growth inhibition during cell-mediated CDI. We generated a cosmid-borne chimeric CDI system, in which the cdiA-CTo11EC869/cdiIo11EC869 coding sequences are fused to the E. coli EC93 cdiA gene at the VENN-encoding sequence. The resulting EC93-EC869o11 chimeric system was introduced into GFP-labeled E. coli to produce green fluorescent inhibitor cells. The inhibitor cells were then cocultured with DsRed-labeled target cells, allowing the two cell populations to be distinguished by fluorescence microscopy. Upon initial mixing, the green inhibitor and red target cells both have the same nucleoid morphology as assessed by DAPI staining (Fig. 6). However, target cells underwent substantial changes in morphology and lost DAPI staining after 6 h of coculture with inhibitor cells (Fig. 6A). These changes were paralleled by a dramatic loss of target cell viability during coculture (Fig. 6F). Target cells expressing the CdiIo11EC869 immunity protein retained genomic DNA during coculture with inhibitor cells and suffered no loss of viability (Fig. 6C), but the noncognate CdiIIIBp1026b immunity protein provided no protection (Fig. 6D). Moreover, introduction of the CdiA-CTo11EC869 Asp198Ala active site mutation into the EC93-EC869o11 chimera system resulted in a loss of growth inhibition and DNase activity (Fig. 6E). Together, these results indicate that DNase activity is responsible for growth inhibition and that the CdiA-CTo11EC869 toxin domain is translocated into the target cell cytoplasm during CDI (Fig. 1).
Fig. 6.
The CdiA-CTo11EC869 toxin degrades DNA during contact-dependent growth inhibition (CDI). GFP-labeled E. coli inhibitor cells (green) were mixed with DsRed-labeled target cells (red) and grown in shaking broth cultures. Cocultures were sampled at 0 and 6 h and stained with DAPI to visualize cellular DNA by fluorescence microscopy. (A) EC93-EC869o11 inhibitor cells versus targets that lack an immunity gene. (B) Mock inhibitor cells (carrying an empty vector cosmid) versus targets that lack an immunity gene. (C) EC93-EC869o11 inhibitors versus target cells that carry the cognate cdiIo11EC869 gene. (D) EC93-EC869o11 inhibitors versus target cells that carry the noncognate cdiIIIBp1026b immunity gene. (E) EC93-EC869o11 inhibitors carrying the Asp198Ala (D198A) missense mutation versus target cells that lack an immunity gene. (F) Quantification of viable target cells during CDI. The number of viable target cells at 0 and 6 h were determined as colony forming units (cfu) per milliliter. The data from competitions corresponding to panels A–E are indicated. Values are the mean ± SEM for three independent experiments.
Discussion
The CdiA-CT/CdiI structures presented here provide a glimpse into the CDI toxin/immunity protein network. These complexes bear no resemblance to the well-studied toxin/antitoxin “addiction module” proteins (13) and are only distantly related to the colicin toxin/immunity family (14). Although structurally distinct, the CDI complexes share some general features with colicin nuclease domains and their immunity proteins. The extensive shape and charge complementarity of the Bp1026b complex interface is reminiscent of the interactions between colicins E5 and D and their cognate immunity proteins (15, 16). Moreover, CdiIo11EC869 appears to inactivate its toxin in a manner analogous to a number of colicin systems (e.g., E3, E7/E8/E9), in which the immunity protein binds an “exosite” adjacent to the toxin active site (17–19). However, the elegant β-augmentation interaction between CdiA-CTo11EC869 and CdiIo11EC869 has not been reported for any other toxin/immunity complex. Homotypic β-augmentation has been observed in viral capsid assembly (20) and appears to be the underlying mechanism of β-sheet expansion in amyloid diseases (21). Additionally, some signal transduction pathways exploit β-augmentation to mediate heterodimeric interactions. For example, the PDZ domain of neuronal nitric oxide synthase (nNOS) extends a β-hairpin that docks into the peptide-binding groove of syntrophin to produce an expanded β-sheet (22). Although the nNOS-syntrophin interface resembles the EC869 complex, we note that β-augmentation interactions during signal transduction are dynamic and transient. In contrast, the CdiA-CTo11EC869/CdiIo11EC869 complex appears to be a unique example of a stable heterodimeric interface mediated by β-augmentation.
Comparative sequence analysis suggests that many CdiA-CTs are composites built from two variable domains (5, 7). The structures also indicate that each CdiA-CT is composed of at least two domains, with the C-terminal nuclease domain forming a stable complex with its cognate immunity protein. The C-terminal domains are also sufficient to inhibit growth when expressed in E. coli cells (5), suggesting that they constitute the functional CDI toxin. In contrast, the CdiA-CT N-terminal regions are not fully resolved in the structures and their functional significance remains unclear. The N-terminal regions are exceptionally labile to proteolysis, suggesting these domains are flexible and perhaps partially disordered. Intrinsically flexible domains are critical for colicin toxin import (23, 24), so perhaps the N-terminal region mediates CdiA-CT transport across the target cell envelope. The N-terminal α-helical bundle of CdiA-CTo11EC869 has weak structural homology to diverse membrane-associated proteins, consistent with the translocation hypothesis, but the function of these domains in CDI remains to be determined. Our results together with previous predictions (25) also suggest that many other CdiA-CT toxin domains may have similar structures despite sharing very little sequence identity. However, we note that some CDI toxin family members must possess other folds because the E. coli EC93 toxin forms pores in target cell membranes (6), and CdiA-CTs from B. pseudomallei K96243 and Erwinia chrysanthemi EC16 share significant sequence identity with colicins E5 and E3, respectively (5, 26).
Methods
CdiA-CT/CdiI expression constructs and toxin/immunity protein complex purification have been described previously (5, 8). Site-directed mutagenesis and construction of the chimeric EC93-EC869 CDI system are outlined in SI Methods. Protein crystallization was as described (27). Briefly, crystals were grown by hanging-drop, vapor-diffusion method at room temperature against a reservoir containing 0.1 M sodium acetate (pH 5.5), 0.2 M NaCl, 18% (wt/vol) PEG-6000, and 10 mM YCl3 for the EC869 complex and 0.49 M sodium phosphate monobasic, 0.96 M potassium phosphate dibasic, and trace amounts of chymotrypsin for the Bp1026b complex. Structural models were determined as described (28, 29). Isolation of toxin and immunity proteins is described in SI Methods. Determination of binding affinities for the complexes and thermal stabilities of isolated toxins, immunity proteins, and complexes were determined by differential scanning fluorimetry (30) as outlined in SI Methods. Nuclease activity assays were performed essentially as described (5) with modifications outlined in SI Methods. Growth competitions were carried as described previously (1) except CDI+ inhibitor and target cells were mixed at a 1:1 ratio and incubated at 37 °C with shaking for 6 h. Cells from the CDI competition experiments were visualized by fluorescence microscopy as described in SI Methods.
Supplementary Material
Acknowledgments
We thank Angelina Iniguez for technical assistance, Stephanie Aoki for providing cosmid pDAL930, Bruce Braaten for strain DL4259, and Drs. Tom Poulos and Nicholas Chim for critical reading of the manuscript. We thank the Advanced Light Source at Berkeley National Laboratories and the Stanford Synchrotron Radiation Lightsource for their invaluable help in data collection. This work was supported by Grants AI099687 (to C.W.G and C.S.H.), GM078634 (to C.S.H.), and GM102318 (to C.W.G., C.S.H., and D.A.L.) from the National Institutes of Health and University of California, Irvine Council on Research, Computing and Libraries funds (to C.W.G.). J.L.E.W. is supported by National Science Foundation Graduate Research Fellowship DGE-1144085.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.K.B. is a guest editor invited by the Editorial Board.
Database deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4G6V and 4G6U).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216238110/-/DCSupplemental.
References
- 1.Aoki SK, et al. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468(7322):439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki SK, et al. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309(5738):1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- 3.Kajava AV, et al. Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol Microbiol. 2001;42(2):279–292. doi: 10.1046/j.1365-2958.2001.02598.x. [DOI] [PubMed] [Google Scholar]
- 4.Aoki SK, et al. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol. 2008;70(2):323–340. doi: 10.1111/j.1365-2958.2008.06404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikolakakis K, et al. The toxin/immunity network of Burkholderia pseudomallei contact-dependent growth inhibition (CDI) systems. Mol Microbiol. 2012;84(3):516–529. doi: 10.1111/j.1365-2958.2012.08039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoki SK, Webb JS, Braaten BA, Low DA. Contact-dependent growth inhibition causes reversible metabolic downregulation in Escherichia coli. J Bacteriol. 2009;191(6):1777–1786. doi: 10.1128/JB.01437-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diner EJ, Beck CM, Webb JS, Low DA, Hayes CS. Identification of a target cell permissive factor required for contact-dependent growth inhibition (CDI) Genes Dev. 2012;26(5):515–525. doi: 10.1101/gad.182345.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poole SJ, et al. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet. 2011;7(8):e1002217. doi: 10.1371/journal.pgen.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24(23):2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kachalova GS, et al. Structural analysis of the heterodimeric type IIS restriction endonuclease R.BspD6I acting as a complex between a monomeric site-specific nickase and a catalytic subunit. J Mol Biol. 2008;384(2):489–502. doi: 10.1016/j.jmb.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 11.Sokolowska M, Czapinska H, Bochtler M. Crystal structure of the β β α-Me type II restriction endonuclease Hpy99I with target DNA. Nucleic Acids Res. 2009;37(11):3799–3810. doi: 10.1093/nar/gkp228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGinness KE, Baker TA, Sauer RT. Engineering controllable protein degradation. Mol Cell. 2006;22(5):701–707. doi: 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi Y, Park JH, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet. 2011;45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- 14.Cascales E, et al. Colicin biology. Microbiol Mol Biol Rev. 2007;71(1):158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luna-Chávez C, Lin YL, Huang RH. Molecular basis of inhibition of the ribonuclease activity in colicin E5 by its cognate immunity protein. J Mol Biol. 2006;358(2):571–579. doi: 10.1016/j.jmb.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 16.Graille M, Mora L, Buckingham RH, van Tilbeurgh H, de Zamaroczy M. Structural inhibition of the colicin D tRNase by the tRNA-mimicking immunity protein. EMBO J. 2004;23(7):1474–1482. doi: 10.1038/sj.emboj.7600162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleanthous C, et al. Structural and mechanistic basis of immunity toward endonuclease colicins. Nat Struct Biol. 1999;6(3):243–252. doi: 10.1038/6683. [DOI] [PubMed] [Google Scholar]
- 18.Maté MJ, Kleanthous C. Structure-based analysis of the metal-dependent mechanism of H-N-H endonucleases. J Biol Chem. 2004;279(33):34763–34769. doi: 10.1074/jbc.M403719200. [DOI] [PubMed] [Google Scholar]
- 19.Ko TP, Liao CC, Ku WY, Chak KF, Yuan HS. The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure. 1999;7(1):91–102. doi: 10.1016/s0969-2126(99)80012-4. [DOI] [PubMed] [Google Scholar]
- 20.Gronenborn AM. Protein acrobatics in pairs: Dimerization via domain swapping. Curr Opin Struct Biol. 2009;19(1):39–49. doi: 10.1016/j.sbi.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamasaki M, Li W, Johnson DJ, Huntington JA. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature. 2008;455(7217):1255–1258. doi: 10.1038/nature07394. [DOI] [PubMed] [Google Scholar]
- 22.Hillier BJ, Christopherson KS, Prehoda KE, Bredt DS, Lim WA. Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science. 1999;284(5415):812–815. [PubMed] [Google Scholar]
- 23.Housden NG, Loftus SR, Moore GR, James R, Kleanthous C. Cell entry mechanism of enzymatic bacterial colicins: Porin recruitment and the thermodynamics of receptor binding. Proc Natl Acad Sci USA. 2005;102(39):13849–13854. doi: 10.1073/pnas.0503567102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loftus SR, et al. Competitive recruitment of the periplasmic translocation portal TolB by a natively disordered domain of colicin E9. Proc Natl Acad Sci USA. 2006;103(33):12353–12358. doi: 10.1073/pnas.0603433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang D, Iyer LM, Aravind L. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 2011;39(11):4532–4552. doi: 10.1093/nar/gkr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker D, Lancaster L, James R, Kleanthous C. Identification of the catalytic motif of the microbial ribosome inactivating cytotoxin colicin E3. Protein Sci. 2004;13(6):1603–1611. doi: 10.1110/ps.04658504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goulding CW, Perry LJ. Protein production in Escherichia coli for structural studies by X-ray crystallography. J Struct Biol. 2003;142(1):133–143. doi: 10.1016/s1047-8477(03)00044-3. [DOI] [PubMed] [Google Scholar]
- 28.Goulding CW, et al. Crystal structure of a major secreted protein of Mycobacterium tuberculosis-MPT63 at 1.5-A resolution. Protein Sci. 2002;11(12):2887–2893. doi: 10.1110/ps.0219002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goulding CW, et al. Thiol-disulfide exchange in an immunoglobulin-like fold: structure of the N-terminal domain of DsbD. Biochemistry. 2002;41(22):6920–6927. doi: 10.1021/bi016038l. [DOI] [PubMed] [Google Scholar]
- 30.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2(9):2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.