Abstract
During the course of evolution genes undergo both fusion and fission by which ORFs are joined or separated. These processes can amend gene function and represent an important factor in the evolution of protein interaction networks. Gene fusions have been suggested to be useful characters for identifying evolutionary relationships because they constitute synapomorphies or cladistic characters. To investigate the fidelity of gene-fusion characters, we developed an approach for identifying differentially distributed gene fusions among whole-genome datasets: fdfBLAST. Applying this tool to the Fungi, we identified 63 gene fusions present in two or more genomes. Using a combination of phylogenetic and comparative genomic analyses, we then investigated the evolution of these genes across 115 fungal genomes, testing each gene fusion for evidence of homoplasy, including gene fission, convergence, and horizontal gene transfer. These analyses demonstrated 110 gene-fission events. We then identified a minimum of three mechanisms that drive gene fission: separation, degeneration, and duplication. These data suggest that gene fission plays an important and hitherto underestimated role in gene evolution. Gene fusions therefore are highly labile characters, and their use for polarizing evolutionary relationships, without reference to gene and species phylogenies, is limited. Accounting for these considerable sources of homoplasy, we identified fusion characters that provide support for multiple nodes in the phylogeny of the Fungi, including relationships within the deeply derived flagellum-forming fungi (i.e., the chytrids).
Keywords: clade, neofunctionalization, monophyly, systematics, Blastocladiomycota
Gene fusions are a hybrid of two or more previously separate ORFs. They occur as a result of: chromosomal translocation, chromosomal inversion, or interstitial deletions. Gene fission involves the opposite process, i.e., separation of an ORF. Both processes have the potential to generate gene diversity and produce variant protein functions (i.e., neofunctionalization) (1, 2). It has been argued that gene-fission events occur at a low frequency because the process requires multiple simultaneous evolutionary occurrences at selectively viable positions within an ORF (3): (i) gain of a stop codon, (ii) gain of a promoter region, and (iii) appropriation of a start codon (Fig. 1A, mechanism 1). Gene fusions therefore have been suggested to represent useful tools for polarizing evolutionary relationships (3–5). This approach follows the logic that taxa possessing a gene fusion are monophyletic to the exclusion of taxa that possess unfused orthologs. Importantly, if the gene fusion is shown to be stable and monophyletic, the root of a tree can be excluded from the clade defined by the gene fusion, allowing phylogenetic relationships to be polarized. This feature can be useful for identifying ancient relationships in the tree of life, where standard sequence-based phylogenetic methods can be limited and inconsistent (3–7). However, this scenario assumes the Dollo parsimony rule applies for the gene fusion; in reality, patterns of homoplasy, including convergent evolution of the gene fusion, multiple reversions (fission), loss, or indeed horizontal gene transfer (HGT), may be present also.
Fig. 1.
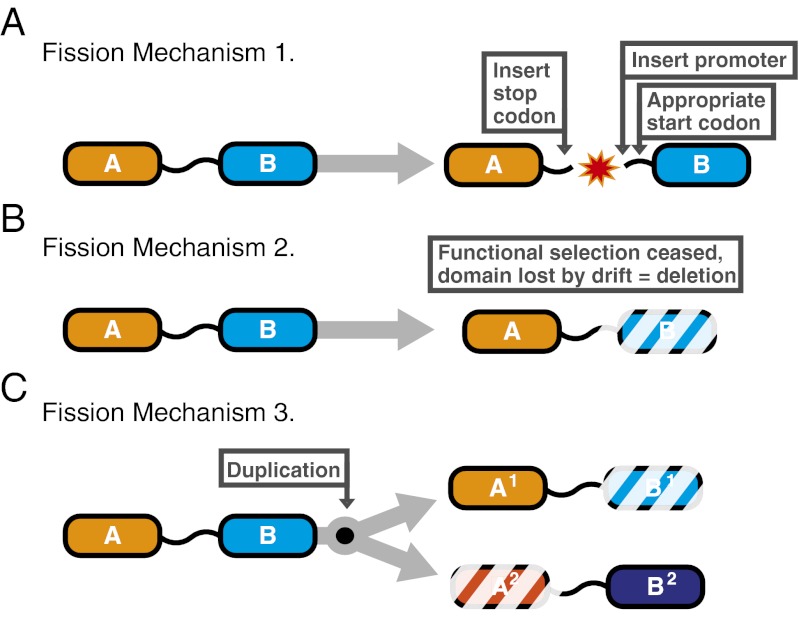
Mechanisms of gene fission. (A–C) Three mutational processes that theoretically can lead to gene fission either separately or in combination. Hypothetical conserved domains (i.e., PFAM domains) are labeled “A” and “B.”
Analyses of the evolutionary ancestry of gene fusions have demonstrated that similar domain combinations can occur by convergent evolution (8, 9) and that domains within gene fusions can have divergent ancestries (10). Yanai et al. (11) demonstrated 31 cases of HGT from 51 gene fusions. These results confirm that gene fusions can be subject to multiple sources of homoplasy. However, comparative analysis of 131 genomes showed that fusion events are approximately four times more common than fission events (12), suggesting that fissions occur at a low relative frequency and fusions therefore may be viewed as stable characters subject to transfer and convergent evolution.
Identification of gene-fission events relies on adequate genome sampling to polarize the point of fusion and to test for the number and type of fission events. To investigate evolutionary patterns of gene fusion and fission in eukaryotic genomes, we focused on the comparative analysis of the Fungi. Fungi are among the best-sampled eukaryotic higher taxonomic groups in terms of whole-genome sequence datasets (e.g., 13, 14), and there has been significant progress in identifying a resolved fungal species phylogeny (13, 15–18).
To investigate the pattern of gene fusion and fission across genomes, we developed an analysis pipeline, fdfBLAST [for “find differential fusions - Basic Local Alignment Search Tool” (19)], to identify differentially distributed gene fusions (Fig. S1). Using this tool, we identified 63 gene-fusion events. Applying phylogenetic reconstruction of constituent conserved domains, we then identified gene-fission events and, where possible, the mechanism of gene separation. This work demonstrates that gene fissions occur at a relatively high rate in the Fungi and represent an important source for both gene variation and artifact when using gene fusions as cladistic characters. Using these data, we identify multiple mechanisms that drive gene fission and that do not require complex simultaneous evolutionary events, suggesting that gene fissions play a hitherto underestimated role in gene evolution.
Results and Discussion
Detection of Gene Fusions Across the Fungi.
Using a custom-built pipeline, fdfBLAST (Fig. S1 and SI Materials and Methods), we compared nine fungal genomes to identify differentially distributed gene fusions. This process recovered 3,050 fdfBLAST hits, of which 2,885 were discarded for one or more of the following reasons: (i) the sequence had no identifiable PFAM domains (20), so it was unclear, given current PFAM sampling, if the gene identified was a combination of two or more discrete domains typical of a bona-fide gene fusion; (ii) the differential hit identified was the product of gene duplication and so was an example of a differentially distributed paralog; (iii) the gene-fusion candidate was misannotated in the genome assemblies; and (iv) the differential hit consisted of a variant number of repeat domains and therefore was likely to be the product of tandem exon duplication (21) and unlikely to be the product of a gene fusion.
We initially ran the fdfBLAST analyses with two additional Microsporidia genomes, Antonospora locustae and Encephalitozoon cuniculi, i.e, 11 genomes in total. These data identified gene fusions present in only a single Microsporidia genome, so these taxa were excluded from further analyses to avoid long-branch attraction problems in our phylogenetic analyses (22) and because these single-genome gene-fusion data points were uninformative for phylogeny or fission analyses.
Phylogenetic analysis was conducted for each domain in the remaining 165 putative gene fusions, sampling similar sequences across 138 opisthokont genome datasets (Table S1). Of these gene fusions, 105 were present in only a single genome and therefore were not useful for comparative analysis. In 30 of the remaining 60 cases, the individual phylogenies for both domain components of the gene fusion demonstrated low resolution, so it was not possible to investigate the evolutionary ancestry of these gene fusions. These datasets were excluded from further analysis, leaving 30 gene fusions for further analysis. Investigation of these 30 fusions identified 33 additional gene fusions that involved one of the domains previously analyzed but were present in an alternative fusion arrangement. These 33 extra fusions were differentially distributed between fungal genomes not previously analyzed as part of the nine fungal genomes used for fdfBLAST analysis.
We then performed phylogenetic analysis with manually corrected alignments for each of the domains present in the 63 gene fusions (SI Appendix). In nine cases (SI Appendix, fusions 10, 16, 17, 32, 33, 34, 35, 39, and 62), the phylogeny of one of the constituent domains did not resolve a phylogeny because the domain was too short, lacked resolution, or had a limited taxonomic distribution. In these cases, only a single domain phylogeny was calculated for the gene fusion. These phylogenies allowed us to map gene architectures onto individual domain phylogenies. All ORF predictions and annotations for genes branching in and around the fusion/fission events then were confirmed by searching the genome assemblies (see SI Materials and Methods for further details). This approach was used to confirm that, if a gene is annotated as unfused, it is separated on the genome assembly. These analyses led to many corrections, because several genes that were annotated as separate when identified on the genome contig resembled the gene fusion found in closely related species. Therefore, in the absence of additional data, we putatively annotate these genes as fusions (these corrections are noted on the phylogenies shown in SI Appendix, and supporting data are listed in Dataset S1).
Before analysis, we knew of two differentially distributed gene fusions across the nine taxa compared in the fdfBLAST analysis: the fusion of an aldose-1-epimerase domain with an UDP-galactose-4-epimerase domain (GenBank GAA21569) present in Saccharomycotina and Schizosaccharomycetes (23) and a trifusion in the pterin branch of the de novo folate biosynthesis pathway between dihydroneopterin aldolase, 2-amino-4-hydroxymethyldihydropteridine diphosphokinase, and dihydropteroate synthase domains (GenBank EGA60749). We note that the fdfBLAST analysis recovered both exemplar gene fusions (SI Appendix, fusions 2 and 17).
To investigate the putative functional annotation of the 63 gene fusions identified, we took an example of all of the individual domains from the gene fusions and ran a BLAST2GO analysis (24). These data suggest that the 63 fusions identified are distributed across a diversity of functional annotation categories with no functional bias evident (Fig. S2).
Sources of Character Instability in 63 Fungal Gene Fusions.
HGT involves the transmission of genetic material across species boundaries (25). Our comparative analysis of gene fusions identified one case of HGT, the previously described transfer of the aldose-1-epimerase and UDP-galactose-4-epimerase gene fusion (e.g., GenBank GAA21569), from the Saccharomycotina to Schizosaccharomycetes (SI Appendix, fusion 2) (23). This result confirms that HGT of gene fusions can be an issue, as identified in other work (11, 26). However, of the 63 gene fusions analyzed, we identified only one clear example of HGT of a gene fusion. This pattern contrasts that observed by Yanai et al. (11), who report 31 transfers of gene fusions from a cohort of 51 gene fusions analyzed. The most likely explanation for this discrepancy is that Yanai et al. largely identified transfers between or into prokaryotic genomes, where HGT is thought to occur at high frequency (25, 27–29); HGT is thought to occur at a lower frequency into fungal genomes (30, 31).
The most significant source of homoplasy identified in the 63 gene fusions analyzed is gene-fission events leading to revision of the fusion state in derived taxa/genomes. It has been argued that fission events are unlikely because the process requires multiple and simultaneous evolutionary changes in the correct order (3). Analysis of an evolutionarily diverse collection of 131 genomes demonstrated that fusion events are approximately four times more common than fission events (12), as is consistent with the idea that gene fissions occur at a lower frequency. However, in contrast, comparisons of relatively closely related eukaryotic genomes: (Oryza sativa and Arabidopsis thaliana) identified six polarized fusion events and eight polarized fission events (32), suggesting that fissions may occur at a higher relative frequency than previously observed. Following on from the examples set out by Nakamura et al. (32), we therefore argue that, to investigate the relative rate of fusions and fissions, it is necessary to compare closely related genomes across an established species phylogeny.
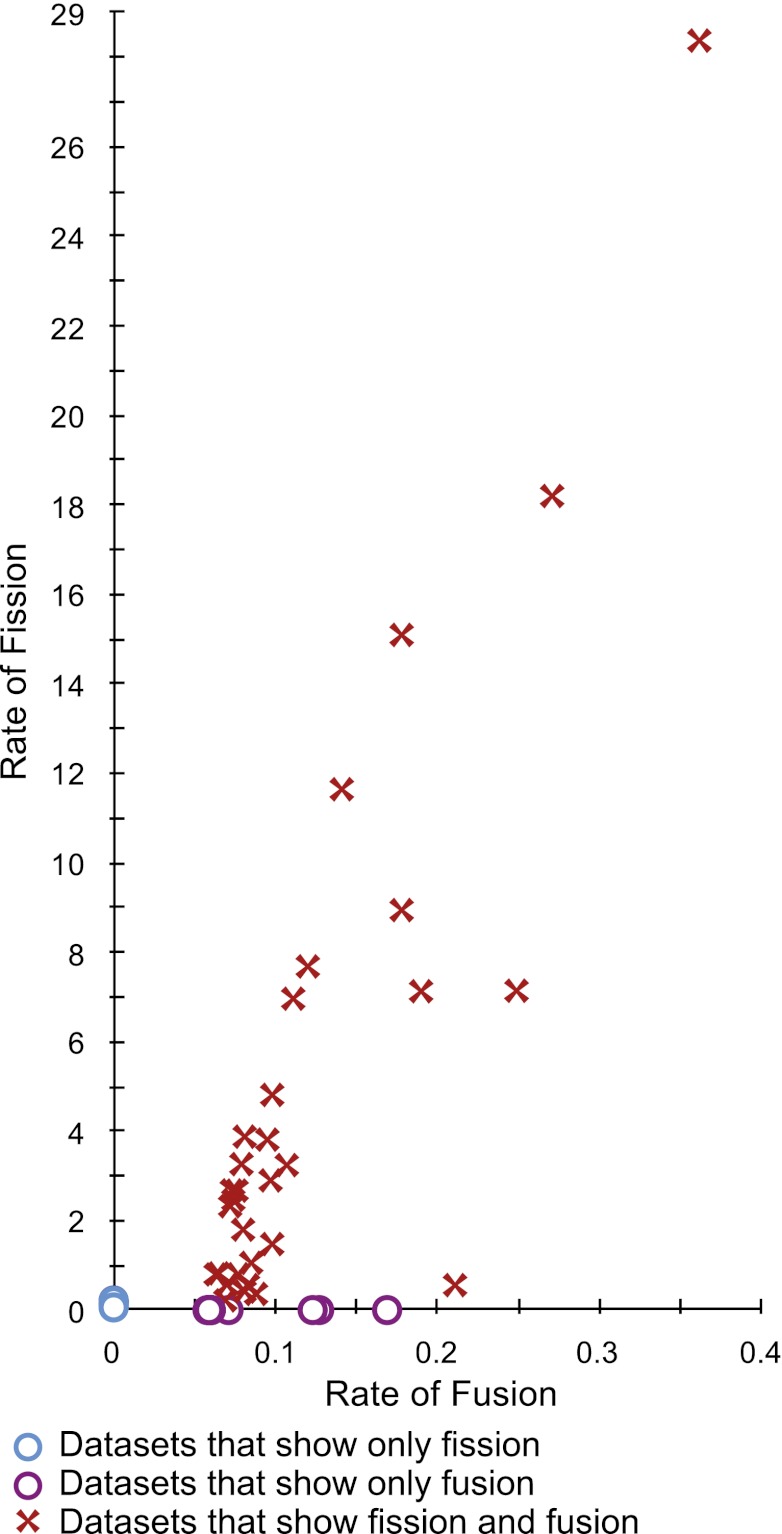
To investigate patterns of gene fusion and fission, we mapped these events onto a concatenated 67-protein phylogeny (Fig. 2) calculated from the 115 fungal genomes used in this study. This tree is generally consistent with previously published fungal phylogenies (13, 15–17) but represents the genomes used in the comparative analysis reported here (Table S1). Using proportional likelihood character state reconstruction analysis in Mesquite (33) and correcting for possible gene annotation errors (described in SI Materials and Methods) and paralog problems, we identified 110 gene-fission events. This result represents a high ratio of gene fissions relative to gene fusions (1:1.746). Using the output data of Mesquite, we also plotted the relative rate of fusion verses fission for the 63 datasets, demonstrating that, among the datasets that show both fusion and fission, the relative rate of fission is much higher than the relative rate of fusion (Fig. 3). Taken together these data suggest that gene fission is a hitherto underestimated force in genome evolution in the Fungi, and potentially in other groups, although this observation needs to be tested on a group-by-group basis.
Fig. 2.
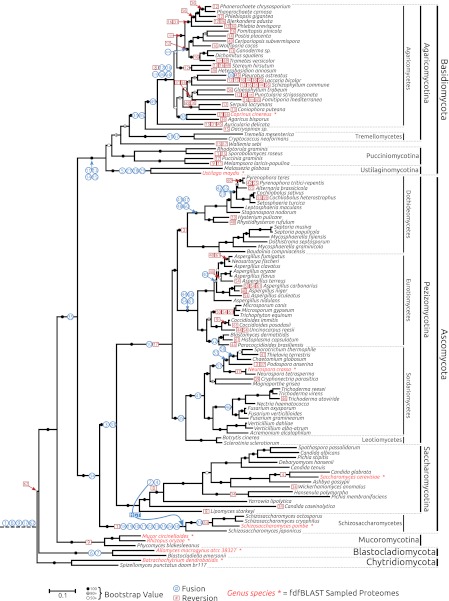
Phylogeny of fungi with complete genomes demonstrating 63 gene fusions and 110 nested gene fissions. A 67-gene concatenated phylogeny of 115 fungal taxa sampling 19,858 amino acid alignment characters. Genes sampled in the alignment are listed in Table S4. Topology support was calculated using 1,000 bootstrap replicates; see the key for guide to visual representation of the bootstrap values. Taxa marked with an asterisk and written in red text are the genomes used for the initial fdfBLAST search. Gene-fusion events are represented as blue circles. The number corresponds to the fusion number used in SI Appendix. Reversion/fission events are shown by red squares, using the same number convention. Note that one case of HGT is identified (SI Appendix, fusion 2), which is represented on the tree as a blue line illustrating the transfer.
Fig. 3.
Relative rates of gene fusions and gene fission. Data points were identified using Mesquite. See Table S2 and SI Appendix for results of the Mesquite analysis.
Identifying gene fission depends on the number of genomes sampled that branch within the fusion clade. Therefore, the number of observable gene-fission events depends on the phylogenetic depth of the fusion event. By plotting phylogenetic depth against the number of fissions, we observe a correlation between phylogenetic depth of the fusion and number of fission events (Fig. S3). This analysis also identifies a subset of five gene fusions that, given nine or more derived genomes (eight or more derived nodes), appear not to undergo gene fission. This observation suggests there are two categories of gene fusions: gene fusions that are reversible (undergo fission), and a subset that are fixed. Identifying examples of the second category will be especially useful for identifying phylogenetic markers.
Fig. S3 suggests that, as node depth increases, the number of observed gene fissions tends to increase, however, the low r2 values (0.083 and 0.052) indicate that factors other than node depth are at play. To investigate this possibility further, we plotted node depth against fission rates identified from the Mesquite analyses. This analysis showed that the gene fissions grouped into two types: recent fusions with a fast rate of fission and old fusions with a slow rate of fission. To investigate if these groupings were the result of the predicted function of these gene-fusion domains, we ran BLAST2GO annotation for 13 recent/fast and 13 old/slow gene fusion domains and found no clear pattern of gene function associated with either type (Fig. S4).
Mechanisms of Gene Fission.
These data identified a high relative rate of gene fission (Figs. 2 and 3), in contradiction to previous data (12) that, together with theoretical conjecture regarding the mechanisms driving gene fission (3), have been suggested to demonstrate that gene fissions occur at a low relative rate. Because our data strongly contradict the idea that fissions occur at a low relative rate, we were interested in investigating the possibility that alternative mechanisms may drive gene fission. We identified three theoretical mechanisms that, individually or collectively, could result in gene fission: (i) splitting of the ORF, whereby a stop codon, promoter region, and start codon are inserted into the ORF at selectively viable positions, resulting in two separate genes (3) (we name this mechanism “fission by separation”) (Fig. 1A); (ii) gene fission by loss of function and degeneration of the sequence encoding one domain (we name this mechanism “fission by degeneration”) (Fig. 1B); and (iii) duplication of a gene fusion and differential loss of constituent domains by either the first or the second mechanism (we name this mechanism ”fission by duplication”) (Fig. 1C). Fission by separation and fission by degeneration does not require multiple, concurrent, complex sequence changes. Additional unidentified mechanisms, for example, transposon insertion, may play a role also.
Incomplete sampling, patterns of gene loss, and phylogenetic uncertainty make it difficult to identify which mechanism drove fission in each of the 110 events. However, we identified nine cases in which gene duplication within a gene-fusion clade has led to multiple paralogs, one with a fused version and one with an unfused version (SI Appendix, fusions 4, 8, 13, 14, 16, and 34). These nine cases are consistent with fission by duplication (Fig. 1C, mechanism 3). Taken together these data demonstrate that a diversity of mechanisms can lead to gene fission, and in some cases these mechanisms do not require complex simultaneous mutational events as previously argued (3), consistent with the idea that fissions can occur at a high relative rate.
Gene Fusions Provide Additional Support for Multiple Clades in the Fungal Tree of Life.
Allowing for both gene fissions and gene losses, the 63 gene fusions were mapped to the last common ancestor of the taxa possessing the gene fusion, using Mesquite to evaluate the proportional likelihood for the position of each character change, i.e., each fusion and each fission (SI Appendix and Table S2). By comparing the distribution of gene-fusion characters with the fungal phylogeny, we were able to identify gene-fusion characters consistent with nodes in the multigene phylogeny that add further support to the fungal phylogeny. It is important to note that further genome sampling may amend these results and demonstrate that these fusions support different and deeper cladistic relationships; however, the fusions identified represent characters consistent with a significant proportion of the backbone of the fungal phylogeny and several relationships within the Ascomycota and Basidiomycota (13, 15–17).
These clades (Fig. 2) included monophyly of the Dikarya (fusion 35), Ascomycota (fusion 34), and Basidiomycota (fusions 11, 13, 14, and 15). The gene fusions analysis also identified several characters consistent with phylogenetic relationships within the Dikarya. These include, for example, within the Basidiomycota, fusion characters that support the monophyly of the: Agaricomycetes (fusion 49), Tremellomycetes (fusions 51 and 57), Ustilaginomycotina (fusions 5, 27, 28, and 29), placement of Auricularia as the primary branch among the Agaricomycetes (fusions 12, 16, 32B, 36, and 56) and, placement of Tremellomycetes as the primary branch among the Agaricomycotina (fusions 31 and 45). Within the Ascomycota, we found gene fusions consistent with monophyly of Pezizomycotina (fusion 10), Eurotiomycetes (fusions 44, 50, 54, and 61), Sordariomycetes (fusion 41), Schizosaccharomycetes (fusions 18, 19, 20, 21, 22, 23, 25, 26, and 30), and the monophyletic grouping of the Pezizomycotina and the Saccharomycotina (fusions 3 and 33), while fusion gene data provided support for sisterhood of the Sordariomycetes and Leotiomycetes (fusion 39).
Making use of new genome data from the chytrid fungus Blastocladiella emersonii, we identified two gene-fusion characters consistent with the monophyly of the recently proposed phylum Blastocladiomycota (15) (fusions 6 and 7).
Using gene-annotation data, we could not see any clear link between patterns in gene fusion and specific fungal clades or taxonomic groups. Interestingly, however, by far the highest numbers of gene-fusion events were identified in the Schizosaccharomycetes clade, with 10 fusion and two fission events. The Schizosaccharomycetes have some of the smallest genomes among the Ascomycota (34), suggesting that gene fusions may correlate with genome size or an ancestral pattern of genome reduction. However, we note that a similar pattern was not evident in the Saccharomycotina, which have comparatively small genomes (34).
Conclusion
These results demonstrate that gene fusions often are affected by homoplasy in the form of reversion (gene fission) and that in many cases this is a considerable factor in the evolution of a gene family. Taken together, these results suggest that gene-fusion characters are much more rarely fixed or immutable than previously hypothesized and require considerable analytical attention on a case-by-case basis before being adopted as phylogenetic markers. We then show that gene fission acts by a combination of multiple mechanisms that do not require complex patterns of simultaneous sequence evolution, supporting the hypothesis that fission can occur at a high frequency. These results also are consistent with the conclusion that gene fission represents a significant factor for neofunctionalization within gene families. The use of gene fusions as evolutionary markers therefore is dependant on adequate genome sampling that allows (i) accurate identification of fission events, or (ii) demonstration that the fusion event has remained stable. In both cases reference to a resolved species phylogeny and phylogenetic analysis of the fusion gene are important to investigate the stability of the fusion state. Therefore, we recommend that gene fusions be used only as additional lines of support in combination with other phylogenetic data rather than as an alternative. Using this approach, we identified support for many branching relationships in the fungal tree of life.
Materials and Methods
Finding Differentially Distributed Gene Fusions.
We developed a custom-built five-step bioinformatics pipeline, fdfBLAST, to identify differentially distributed gene fusions between genomes (described in SI Materials and Methods and Fig. S1; all pipeline scripts are available at https://github.com/guyleonard/fdfBLAST). We used this pipeline to compare nine fungal genomes. This analysis used the following seed genomes: Allomyces macrogynus, Batrachochytrium dendrobatidis, Coprinus cinereus, Mucor circinelloides, Neurospora crassa, Rhizopus oryzae, Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Ustilago maydis. The fdfBLAST pipeline was run with the following parameters: e-value upper limit 1e-10, lower limit 0, and hit number limit 250 with the comparison of right and left hit ratios set to a lower ratio value of 0.1 and a higher ratio value of 1.0. This process identified 3,050 candidate gene fusions that were refined by manual curation using the graphical output data from steps 4 and 5 of the fdfBLAST pipeline (described in SI Materials and Methods and illustrated in Fig. S1). This process, combined with phylogenetic analysis and manual inspection of genome annotation, identified 63 gene fusions as described in Results and Discussion.
Phylogenetic Analysis of the Individual Domains Within the Gene Fusions.
For all candidate gene fusions we calculated a phylogeny for each individual domain component of the gene fusion using a custom-built pipeline (35, 36). We followed this pipeline analysis by a series of manual steps: BLAST checks of taxon sampling, multiple sequence alignment, alignment masking (37), amino acid substitution model selection (38), and tree calculation using PHYML with 100 bootstrap replicates (39). See Table S3 for details of models selected and SI Materials and Methods for details of the analysis pipeline used for phylogenetic analysis.
Multigene Concatenated Phylogeny of the Fungi.
Sixty-seven protein sequences were selected for multigene concatenated phylogeny (Table S4). Fifty-seven of these proteins were from a selection of conserved single-copy protein domains (18) that were present in 106 or more of the fungal genomes analyzed (Table S1) using a BLAST gather of 1e-30 (Table S4). The remaining 10 genes were selected from a list of genes we favor for phylogenetic analysis (Table S4, gray shading). We then generated an alignment for each individual gene family using a modification of a custom built gene-by-gene phylogeny pipeline (35, 36) integrating the trimAL (37) alignment masking tool. The resulting alignments were concatenated together using a custom Perl script. The final concatenated data matrix encompassed 115 taxa and 19,858 characters. We then calculated a maximum likelihood phylogeny using the program RAxML (40) with our previously developed easyRAx script (https://github.com/guyleonard/easyRAx). One hundred best-known-likelihood starting trees were computed, followed by 1,000 bootstrap trees using the PROT-CAT-LG substitution model parameters.
Mapping Gene-Fusion/-Fission Characters onto the Species Phylogeny.
To map the distribution of fusion and fission characteristics onto the species phylogeny (Fig. 2), we used character-mapping functions in Mesquite (33). We compiled a character matrix using the phylogenies calculated for each individual protein domain present in the gene fusions (SI Appendix) to identify cases of gene loss or potential paralogs, so that fusions were coded as 1, unfused orthologs as 0, and lost genes or paralogs as “−”. The distribution of characters was analyzed using the likelihood approach, using the Asymm2 model if both fusion and fission events were present or the MK1 model if only one form of character transition was evident. These results are summarized in Table S2. The results of the Mesquite character mapping analyses also are shown diagrammatically in SI Appendix.
Supplementary Material
Acknowledgments
We thank Joint Genome Institute and The Broad Institute for making their genome data publicly available. We also thank Prof. Suely Gomes (Universidade de São Paulo), our consortium partner on the Blastodadiella genome, for allowing us to make use of a preliminary assembly of this genome. We thank Dr. Bill Wickstead (University of Nottingham) for letting us use his PFAM domain tree-labeling script. We thank the reviewers for helping us improve this manuscript. G.L. is supported by a Biotechnology and Biological Sciences Research Council Grant BB/G00885X/1 (to T.A.R.)
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence alignments reported in this paper are available through the authors’ Web site, http://gna-phylo.nhm.ac.uk/content/leonard_and_richards_2012. For all sequences from public access databases an accession number or ID term (DOE JGI) is provided in SI Appendix.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210909110/-/DCSupplemental.
References
- 1.Apic G, Gough J, Teichmann SA. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J Mol Biol. 2001;310(2):311–325. doi: 10.1006/jmbi.2001.4776. [DOI] [PubMed] [Google Scholar]
- 2.Doolittle RF. The multiplicity of domains in proteins. Annu Rev Biochem. 1995;64:287–314. doi: 10.1146/annurev.bi.64.070195.001443. [DOI] [PubMed] [Google Scholar]
- 3.Stechmann A, Cavalier-Smith T. Rooting the eukaryote tree by using a derived gene fusion. Science. 2002;297(5578):89–91. doi: 10.1126/science.1071196. [DOI] [PubMed] [Google Scholar]
- 4.Philippe H, et al. Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Proc Biol Sci. 2000;267(1449):1213–1221. doi: 10.1098/rspb.2000.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stechmann A, Cavalier-Smith T. The root of the eukaryote tree pinpointed. Curr Biol. 2003;13(17):R665–R666. doi: 10.1016/s0960-9822(03)00602-x. [DOI] [PubMed] [Google Scholar]
- 6.Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436(7054):1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- 7.Gribaldo S, Poole AM, Daubin V, Forterre P, Brochier-Armanet C. The origin of eukaryotes and their relationship with the Archaea: Are we at a phylogenomic impasse? Nat Rev Microbiol. 2010;8(10):743–752. doi: 10.1038/nrmicro2426. [DOI] [PubMed] [Google Scholar]
- 8.Stover NA, Cavalcanti AR, Li AJ, Richardson BC, Landweber LF. Reciprocal fusions of two genes in the formaldehyde detoxification pathway in ciliates and diatoms. Mol Biol Evol. 2005;22(7):1539–1542. doi: 10.1093/molbev/msi151. [DOI] [PubMed] [Google Scholar]
- 9.Nara T, Hshimoto T, Aoki T. Evolutionary implications of the mosaic pyrimidine-biosynthetic pathway in eukaryotes. Gene. 2000;257(2):209–222. doi: 10.1016/s0378-1119(00)00411-x. [DOI] [PubMed] [Google Scholar]
- 10.Wolf YI, Kondrashov AS, Koonin EV. Interkingdom gene fusions. Genome Biol. 2000;1(6):H0013. doi: 10.1186/gb-2000-1-6-research0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yanai I, Wolf YI, Koonin EV. 2002. Evolution of gene fusions: Horizontal transfer versus independent events. Genome Biology 3(5)::research0024.
- 12.Kummerfeld SK, Teichmann SA. Relative rates of gene fusion and fission in multi-domain proteins. Trends Genet. 2005;21(1):25–30. doi: 10.1016/j.tig.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 13.James TY, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443(7113):818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 14.Stajich JE, et al. The fungi. Curr Biol. 2009;19(18):R840–R845. doi: 10.1016/j.cub.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.James TY, et al. A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota) Mycologia. 2006;98(6):860–871. doi: 10.3852/mycologia.98.6.860. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, et al. Phylogenomic analyses predict sistergroup relationship of nucleariids and fungi and paraphyly of zygomycetes with significant support. BMC Evol Biol. 2009;9:272. doi: 10.1186/1471-2148-9-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitzpatrick DA, Logue ME, Stajich JE, Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torruella G, et al. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single-copy protein domains. Mol Biol Evol. 2012;29(2):531–544. doi: 10.1093/molbev/msr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32(Database issue):D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letunic I, Copley RR, Bork P. Common exon duplication in animals and its role in alternative splicing. Hum Mol Genet. 2002;11(13):1561–1567. doi: 10.1093/hmg/11.13.1561. [DOI] [PubMed] [Google Scholar]
- 22.Hirt RP, et al. Microsporidia are related to Fungi: Evidence from the largest subunit of RNA polymerase II and other proteins. Proc Natl Acad Sci USA. 1999;96(2):580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slot JC, Rokas A. Multiple GAL pathway gene clusters evolved independently and by different mechanisms in fungi. Proc Natl Acad Sci USA. 2010;107(22):10136–10141. doi: 10.1073/pnas.0914418107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conesa A, Götz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 2008;2008:619832. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doolittle WF. Lateral genomics. Trends Cell Biol. 1999;9(12):M5–M8. [PubMed] [Google Scholar]
- 26.Andersson JO, Roger AJ. Evolutionary analyses of the small subunit of glutamate synthase: Gene order conservation, gene fusions, and prokaryote-to-eukaryote lateral gene transfers. Eukaryot Cell. 2002;1(2):304–310. doi: 10.1128/EC.1.2.304-310.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boucher Y, et al. Lateral gene transfer and the origins of prokaryotic groups. Annu Rev Genet. 2003;37:283–328. doi: 10.1146/annurev.genet.37.050503.084247. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence JG, Ochman H. Reconciling the many faces of lateral gene transfer. Trends Microbiol. 2002;10(1):1–4. doi: 10.1016/s0966-842x(01)02282-x. [DOI] [PubMed] [Google Scholar]
- 29.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 30.Richards TA, Leonard G, Soanes DM, Talbot NJ. Gene transfer into the fungi. Fungal Biol Rev. 2011;25:98–110. [Google Scholar]
- 31.Marcet-Houben M, Gabaldón T. Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 2010;26(1):5–8. doi: 10.1016/j.tig.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura Y, Itoh T, Martin W. Rate and polarity of gene fusion and fission in Oryza sativa and Arabidopsis thaliana. Mol Biol Evol. 2007;24(1):110–121. doi: 10.1093/molbev/msl138. [DOI] [PubMed] [Google Scholar]
- 33.Maddison WP, Maddison DR. 2011. Mesquite: A modular system for evolutionary analysis. Version 2.75. Available at: http://mesquiteproject.org.
- 34.Kelkar YD, Ochman H. Causes and consequences of genome expansion in fungi. Genome Biol Evol. 2012;4(1):13–23. doi: 10.1093/gbe/evr124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards TA, et al. Phylogenomic analysis demonstrates a pattern of rare and ancient horizontal gene transfer between plants and fungi. Plant Cell. 2009;21(7):1897–1911. doi: 10.1105/tpc.109.065805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richards TA, et al. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc Natl Acad Sci USA. 2011;108(37):15258–15263. doi: 10.1073/pnas.1105100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 40.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.