Abstract
The X-ray structure of the bacterial voltage-gated sodium channel NavAb has been reported in a conformation with a closed conduction pore. Comparison between this structure and the activated-open and resting-closed structures of the voltage-gated Kv1.2 potassium channel suggests that the voltage-sensor domains (VSDs) of the reported structure are not fully activated. Using the aforementioned structures of Kv1.2 as templates, molecular dynamics simulations are used to identify analogous functional conformations of NavAb. Specifically, starting from the NavAb crystal structure, conformations of the membrane-bound channel are sampled along likely pathways for activation of the VSD and opening of the pore domain. Gating charge computations suggest that a structural rearrangement comparable to that occurring between activated-open and resting-closed states is required to explain experimental values of the gating charge, thereby confirming that the reported VSD structure is likely an intermediate along the channel activation pathway. Our observation that the X-ray structure exhibits a low pore domain-opening propensity further supports this notion. The present molecular dynamics study also identifies conformations of NavAb that are seemingly related to the resting-closed and activated-open states. Our findings are consistent with recent structural and functional studies of the orthologous channels NavRh, NaChBac, and NavMs and offer possible structures for the functionally relevant conformations of NavAb.
Keywords: voltage-gated cation channels, ion channel activation mechanism
Voltage-gated cation channels (VGCCs) are membrane-embedded protein pores that allow the selective flow of specific ions across the cell membrane in response to an external voltage stimulus. This remarkable property, which relies on a pore-gating mechanism, enables these channels to perform as critical devices in a number of electrically mediated biological processes including cellular secretion, hormone regulation, electric signaling in neurons, and contraction in excitable muscle cells (1). The superfamily of VGCCs includes the Na+, Ca2+, K+, and nonselective cationic channels, denoted Nav, Cav, Kv, and HCN channels, respectively. VGCCs are either tetrameric or pseudotetrameric with each subunit consisting of six transmembrane (TM) helices, conventionally denoted S1–S6. The helix bundle formed by segments S1–S4 constitutes the voltage-sensor domain (VSD) that triggers the closed–open transition of the conduction pore in response to TM voltage variations. The pore domain (PD) is composed of helices S5 and S6. Charge displacement across the membrane entailed by the conformational transition of the VSD, known as the gating charge (Q), results essentially from the motion of the S4 helix, which is positively charged and contains four to seven basic amino acids, mostly arginines. Gating charges have been measured for a variety of channels, as discussed extensively in recent reviews (2, 3).
In 2005, the MacKinnon group reported an X-ray crystal structure of the mammalian Kv1.2 channel in an “activated-open state” (KA), thereby providing the first structural template to decipher key aspects of VGCC function at the atomic level (4). Following early molecular dynamics (MD) simulations aimed at characterizing the structural details of the channel embedded in a lipid bilayer (5, 6), several atomistic models for the physiologically relevant deactivated or “resting-closed state” (KR) of Kv1.2 were proposed (7–12). For all of these models, the structure-based measurements of Q [12–14 units of electron charge (e)] were found to be in excellent agreement with experimental estimates for Shaker-like channels (13). Structural analyses of these models demonstrated a remarkably consensual explanation of experimental data (14). The picture of activation emerging from these studies on Kv1.2 provides support to the sliding-helix model (15, 16), which posits that, during activation, S4 moves along the TM direction by sliding through a protein-lined pathway. The motion of S4 results in a total displacement of 8–15 Å and is assisted by the formation and breaking of conformation-dependent salt bridges between the basic amino acids of S4 and negatively charged moieties located on neighboring VSD segments and on the lipid head groups. The functional relevance of this motion relies on the resulting displacement of the S4–S5 linker, the physical connector between S4 and the PD, which surrounds the conduction pore and, in the resting state, acts as a restraining cuff holding the intracellular gate closed.
In 2011, the X-ray structure of a voltage-gated sodium channel from the bacterium Arcobacter butzleri, NavAb, was resolved at 2.7 Å (17). This structure featured a closed PD and the four VSDs seemingly activated. The latter was interpreted as representative of the pre-open state of the channel or, in other words, a metastable (intermediate) structure along the pathway connecting the resting-closed state to the activated-open state. Consistently, recent MD simulations of the channel embedded in a lipid membrane have shown structural stability on timescales of ∼100 ns (18). Importantly, the NavAb structure provides a high-resolution template for enhancing our knowledge on Na+ and related Ca2+ channels, for which much less is known. However, there remains a pressing need to relate the reported NavAb X-ray structure to specific functional states of the channel. Given the significant degree of structural similarity shared by members of the VGCC family, more extensive studies on Kv channels are a resource for providing insight into this issue.
Our starting point is the well-understood KA and KR structures of Kv1.2 equilibrated in a lipid bilayer, which correspond to the activated-open (5) and resting-closed (10) conformations, respectively. These structural models are used here as the targets of a set of MD simulations, which begin with the NavAb X-ray structure that has been equilibrated in a lipid bilayer (18). Our aim is to sample selected conformations along the pathways for VSD activation and PD opening of NavAb. In so doing, we characterize these conformations by computing the value of Q and the PD-opening propensity. Anticipating our results, we find that structural rearrangements of the NavAb VSD comparable to those occurring between the Kv1.2 reference structures (KR and KA) are able to account for the experimental value of Q (13, 19). Furthermore, compared with the KA structure, the VSD conformation in the NavAb X-ray structure seems to restrict PD opening such that it can be only partially open, which agrees with previous findings showing that intermediate states of Shaker-like Kv channels are likely to be nonconductive (20). Importantly, our work has enabled us to identify channel conformations likely to be related to the resting-closed and activated-open states of NavAb. Notably, the VSD of the latter conformation is structurally similar to the corresponding domain of a recently published X-ray crystal structure for the orthologous channel NavRh (21), whereas our activated-open PD is strikingly similar to that very recently reported for NavMs (22).
Results
In previous MD simulation studies, we investigated atomic models of KA (5) and KR (10) built on the basis of the X-ray crystal structure of Kv1.2 (4). Details of these MD simulations are in the original papers. Briefly, each of these Kv1.2 channel conformations was embedded in a lipid bilayer hydrated by an ∼100 mM KCl solution, and an MD trajectory was carried out for ∼15 ns at constant temperature (300 K) and pressure (1 atm), assuming neutral pH and no applied TM electrostatic potential. The structures for KA and KR obtained at the end of the MD simulations are used here as the reference membrane-equilibrated structures of Kv1.2. Recently, we carried out an ∼100-ns MD trajectory on the X-ray crystal structure of NavAb (18) embedded in a lipid membrane using the same MD protocol as adopted for Kv1.2 (10). The conformation obtained at the end of the latter trajectory provides a structure of NavAb, hereafter referred to as NX. Superposition of NX onto the KA and KR structures reveals that the NX PD is similar to that of KR. However, the position of the NX S4 helix, relative to the bilayer center, is in-between that of KA and KR.
Sampling VSD Activated and Resting Conformations.
The VSD structures for KA and KR were used as reference points to parametrically describe the activation pathway of NavAb in terms of a generalized coordinate, R (NavAb, VSD) = R, that reports the conformational distance of the NavAb VSD from the corresponding Kv reference points. Thus, R = 0 and R = 1 are end points on the VSD activation path corresponding to KR and KA, respectively. Several findings, for both Kv and Nav channels, support the notion that the network of salt bridges provides an unambiguous descriptor of VSD conformations along the activation path (23–25). Thus, we characterize a given VSD conformation via a distance matrix, M (NavAb, VSD), mapping the domain electrostatic interactions, specifically the matrix of distances between the VSD-charged S4 residues (R1 to R4) and VSD acidic residues plus lipid phosphate groups (B1 to B6). Table S1 gives the identity and numbering of these residues/groups for VGCCs considered in this study.
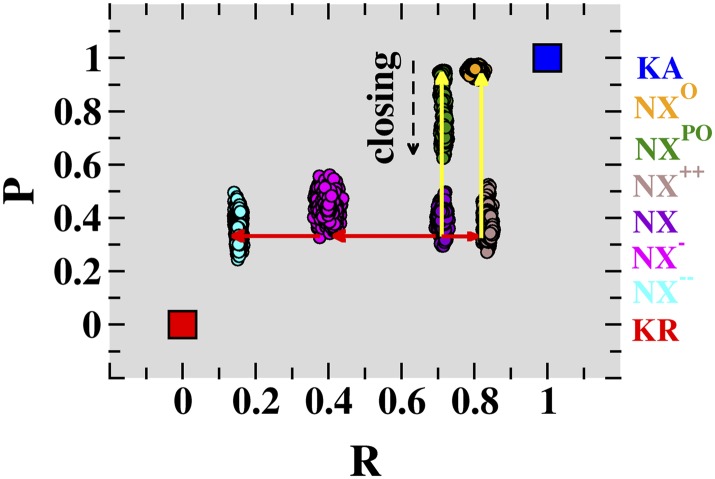
In this framework, the conformational distance of structure NX averaged over the four VSD subunits is R = 0.68 ± 0.03, which quantifies the intermediate nature of VSD with respect to KA and KR. Starting from NX, we applied biased MD simulations to sample NavAb conformations with differing values of R, seeking possible activated and resting conformations of this channel (Fig. 1). The procedure consisted of combining a biased and an equilibrium MD simulation to first drive the NX VSD toward a given value of the generalized coordinate, R, and then relaxing the resulting structure at that R value without constraints (Fig. S1A). Importantly, the pore domain was unconstrained in all phases of the MD protocol. In total, three independent trajectories were generated, sampling channel conformations at R = 0.85 ± 0.02, 0.38 ± 0.06, and 0.14 ± 0.01 (averaged over four VSD subunits), hereafter referred to as conformations NX++, NX−, and NX−−, respectively. These specific R values, above and below the value for NX (R = 0.68 ± 0.03), were chosen aiming to sample channel conformations distinct from the NavAb crystal structure in both directions of the activation path. The R(t) profiles for these MD runs and for the equilibrium trajectory of NX indicate that within the timescale of these simulations (∼30 ns), the structural relaxation of the conformations NX++, NX−, and NX−− is similar to that of the crystal structure (Fig. S1A). Together, these findings suggest that NX++, NX−, and NX−−, along with NX, can be envisaged as possible milestones marking the activation pathway of NavAb. Notably, in all of these MD simulations the PD remained closed, in a similar conformation to that in the crystal structure.
Fig. 1.
Conformations associated with voltage-sensor domain activation and pore-domain opening of NavAb. The present unconstrained MD trajectories for membrane-equilibrated NavAb conformations NX−−, NX−, NX, NX++, NXPO, and NXO are shown as projections onto a conformational distance space, defined by the generalized coordinates R (NavAb, VSD; t) and P (NavAb, PD; t). These six conformations correspond to distinct positions along the VSD-activation (red arrows) and PD-opening (yellow arrows) pathways of NavAb. The conformational distance positions corresponding to the activated-open (blue square) and resting-closed (red square) conformations of Kv1.2 are also indicated. The time series of R (NavAb, VSD; t) and P (NavAb, PD; t), including the biased and equilibrium phases of the simulations, are presented in Fig. S1.
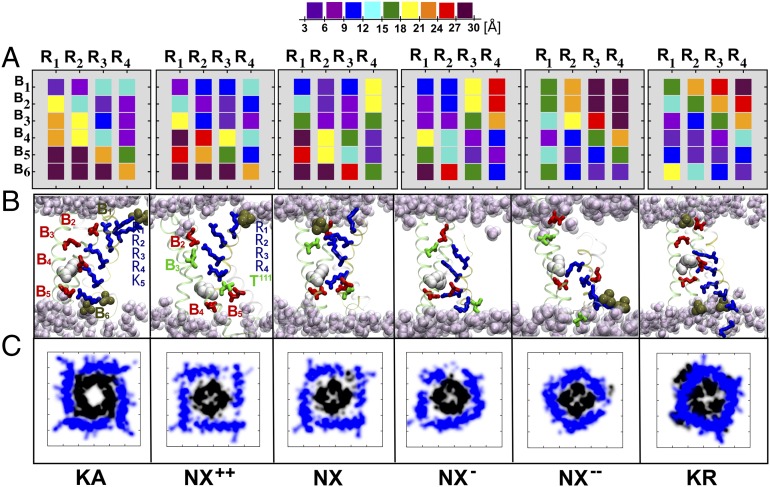
Each of the above-mentioned simulations of NavAb features a distinct set of electrostatic interactions in the VSD, as easily visualized by dissection of their salt-bridge interactions (Fig. 2). Here, a given VSD ion pair arginine (R)–B was considered to form a salt bridge for distances <9 Å. In the particular case of site B3 of NavAb, hydrogen bonds rather than salt bridges are considered, as this site involves an asparagine instead of the glutamate observed in Kv1.2. In NX++, R1 to R4 are exposed to the extracellular milieu, forming salt bridges or hydrogen bonds with binding sites B1, B2, and B3, in an arrangement that closely resembles the KA structure (Fig. 2). By contrast, in NX−−, the S4 residues are paired with sites B4, B5, and B6 (Fig. 2 and Table S1) near the intracellular entrance of the domain, which resembles the structure of KR. The VSD structures of NX and NX− present intermediate salt-bridge arrangements such that the sequence NX−−, NX−, NX, and NX++ indeed suggests an activation pathway, with the so-called catalytic center occupied sequentially by the S4 residues R2, R3, R4, and T111, respectively (Fig. 2). Recall that this center, which is located at an occluded site in the domain formed by B4, B5, and F56 (F233 in Kv1.2), has been suggested to catalyze the sequential transfer of the S4 basic residues across the membrane during activation (26). In this respect, NX−− and NX++ are similar to KR and KA, with R2 and K306 from Kv1.2 (corresponding to T111 in NavAb) occupying the catalytic site in KR and KA, respectively. Furthermore, aside from the salt-bridge arrangement, other measures of structural distance, for example the rmsd, also highlight the structural similarity between NX−− and KR and NX++ and KA (Table S2).
Fig. 2.
Voltage-sensor conformations of NavAb. (A) Distance matrices Mij (NavAb, VSD) mapping the domain electrostatic interactions between the S4 basic residues (numbered R1 to R4) and their binding sites [numbered B1 to B6 for (Upper) PO4−, E32, N49, E59, and D80 and (Lower) PO4−, respectively] in each of the VSD conformations NX++, NX, NX−, and NX−−. For the purpose of comparison, the reference, distance matrices Mij (Kv1.2, VSD) for Kv1.2 in the KA and KR conformations are also depicted; Kv1.2 binding sites are numbered B1 to B6 for (Upper) PO4−, E183, E226, E236, and D259 and (Lower) PO4−, respectively. Distance matrices are computed by averaging over the four independent subunits. (B) Snapshots of the VSD for the NavAb and Kv1.2 conformations, highlighting the position of the S4 basic residues (blue sticks) and the salt bridges/hydrogen bonds they form with the acidic/polar residues (red/green sticks) of other VSD segments or with lipid phosphate moieties (ochre). Note that the NavAb “catalytic center,” formed by B4, B5, and F56 (white; F233 in Kv1.2), is occupied by the S4 residues R2, R3, R4, and T111. (C) Atomic-density plots for the selected NavAb and Kv1.2 conformations, depicting the positions of the S4–S5 linkers (blue) and S6 helices (black) at the membrane plane placed at the intracellular entrance of the channel. The S4–S5 linker constrains the bundle-crossing region of S6 in the conformations NX− and NX−−, thereby prohibiting pore opening in these structures.
Measurement of the Gating Charge.
Following the direct measurement protocol (27) (SI Materials and Methods and Fig. S2), the generated NavAb conformations with distinct R values were then subjected to structure-based Q measurements to identify a pair of VSD structures most likely corresponding to the active and resting states, that is, with a Q value comparable to QA, the experimental gating charge. Typically, estimates of QA range from 12 to 15 e (13, 19), as measured for a variety of VGCCs. Using NX++ as the reference conformation, the computed Q for the structure pairs NX++/NX++, NX++/NX, NX++/NX−, and NX++/NX−− are, respectively, 0.0, −2.14, −8.32, and −12.08 e (Table S3); the negative sign implies that, relative to NX++, the set of VSD conformations transfers charge inwardly (−) across the membrane capacitance. This analysis indicates unambiguously that only the conformation pair NX++/NX−− results in a value of Q (∼12 e) comparable to the experimental, QA. Therefore, NX++ and NX−− are potential active and resting conformations, respectively. As a consequence, NX and NX− are putative intermediates on the activation path, as already suggested by their R values. Importantly, given a certain displacement of the basic residues of S4 along the TM direction, the value of Q depends strongly on the local electrostatic field. In light of this, it is remarkable that, despite differences in primary structure between the VSDs of NavAb and Kv1.2 (e.g., number of charged S4 residues and binding sites), the computed Q associated with the activated–resting transition is about the same (∼12 e) for both channels. This observation raises the intriguing hypothesis that sensitivity to variations in the TM potential, being directly subject to evolutionary pressure, may be more conserved than the VSD sequence itself. As a consequence, typical VSDs are expected to transfer approximately three elementary units of charge, and thus the activation mechanism may be insensitive to minor structural variations.
Sampling Open-Pore Conformations.
The membrane-equilibrated structures NX++, NX, NX−, and NX−− all feature a closed pore. As expected from their distinct VSD conformations, major structural differences among them relate to the positions of S4 and the S4–S5 linker. In NX−−, S4 is positioned near the intracellular face and the S4–S5 linker is in close contact with the pore. Relative to NX−−, the S4 helix in NX−, NX, and NX++ is, respectively, displaced by 6.5, 13.7, and 15.4 Å in the outward direction along the normal to the membrane, in such a way that the linker is gradually moved apart (detached) from the PD (Fig. 2); in the NX++ structure, the linker is displaced laterally by 3.45 Å from the reference position in NX−−. Seemingly, the S4–S5 linkers constrain the motion of the S6 bundle to a different extent in each conformation, thereby implying that the PD-opening propensity can be finely modulated by the VSD conformation; in other words, the PD-opening propensity increases as the constraint on the closed pore imposed by the linker is released. In this respect, compared with NX++, NX−− and NX− show a lower PD-opening propensity, but this is less clear for NX. To test this hypothesis, we performed additional MD simulations to probe the PD-opening propensity for different VSD conformations. Following the previous framework, the PD-opening route was described in terms of the generalized coordinate, R (NavAb, PD) = P, which reports the conformational distance of the NavAb PD from the reference conformations in KA and KR, featuring, respectively, an open and closed PD. Here, the PD conformation was described by means of a matrix of distances between the Cα atoms of three PD-lining S6-bundle residues (F1 to F3), highly conserved over the entire family of VGCCs and forming the hydrophobic gate along the ionic permeation pathway (Table S1). Here, P = 0 and P = 1 correspond to PD conformations of KR and KA, respectively.
Starting from the membrane-equilibrated NX structure (P = 0.33), we have generated two independent MD trajectories in which first the protein is driven simultaneously toward a conformation with the VSD activated and the PD open (P ∼ 0.95), and second, the starting structure is guided only to an open-PD conformation. During the procedure where only the PD is biased, the VSD and the S4–S5 linker are left unconstrained. At the end of both independent transformations, the structures were then subjected to an unbiased MD run without any constraints (Materials and Methods). For the simulation biased toward NX++, the R value of 0.84 agrees with the previous VSD transformation (Fig. S1B). In addition, the distribution of P values is centered at 0.95, indicating PD stability throughout the trajectory (Fig. 1). It is worth noting that both R and P values were computed from the equilibrium part of the MD trajectory. Consistent with its P value, the final equilibrated structure obtained from this simulation, hereafter referred to as NXO, exhibits an open PD, as demonstrated by analysis of the PD volume (28).
In NXO, Ile217 constitutes the major constraining element along the permeation pathway, delineating a hydrophobic constriction with a PD radius of ∼3.0 Å which, compared with KA (29), is large enough to allow full hydration of the permeation pathway and ion conduction. Indeed, in a short independent MD simulation performed in the presence of a TM voltage potential, we observe permeation of Na+ ions through the lumen of NXO (Movie S1). In contrast, in an MD simulation with the VSD in the NX conformation, the target PD structure was not stable, and relaxed into an intermediate configuration, as shown by the drift from the initial biased P value (dashed black arrow in Fig. 1). The final equilibrated structure resulting from this simulation, NXPO, exhibits a partially open PD conformation, in which the Met221 constrains the PD radius at ∼2.0 Å. Although large enough to allow partial hydration, this conformation does not allow ion conduction, as indicated by a recent investigation on the orthologous channel, NaChBac (30). The equilibrated PD structures are significantly different, suggesting a larger PD-opening propensity for NX++ compared with NX. Moreover, as seen in Fig. S3, the selectivity filter radius in the NXO conformation is slightly larger than in NXPO and NX++, suggesting that the selectivity filter in the crystal structure may be different from that of an active conformation of the channel. Additional studies are required to clarify whether these structural differences have functional relevance or originate from an incomplete sampling of the structural fluctuations of the channel or possibly an experimental artifact (e.g., low temperature or bicelle environment). Finally, note that, in Kv1.2, the existence of two hinge points along S6—the first corresponding to a conserved glycine residue at the N terminus of the segment and the second to the well-known Pro-Val-Pro motif—favor a kinked helical structure for the segment in the open conformation of the channel (KA). In contrast, for NavAb, the S6 flexibility seems to rely primarily on Pro200 at the N terminus, and thus S6 adopts a tilted (nonkinked) helical structure in the open conformation of NavAb (NXO). This difference is reflected in the atomic-density plots, which show a distinct pattern of occupancy at the membrane plane on the intracellular side of the channel (Fig. 2 and Fig. S3).
Discussion
Owing to the structural similarity among members of the large family of VGCCs, both the voltage-sensing process and the gating mechanism are expected to be common features, analogous to that unveiled for Kv1.2. Indeed, results from site-directed spin labeling and EPR spectroscopy support the notion that the VSD of NaChBac, and particularly its voltage-sensing pathway, may be similar to that of several K+ channels (31). Here, under this working hypothesis, structural models (KA and KR) of Kv1.2 were used to drive biased and equilibrium MD simulations that explored the conformational space of NavAb. Importantly, although in our strategy the biasing phase depends strongly on the chosen templates, relaxation potentially allows discovery of independent configurations. Gating-charge measurements then suggest the VSD structure in NX++ and NX−− as conformations potentially related to the activated and resting states of this channel.
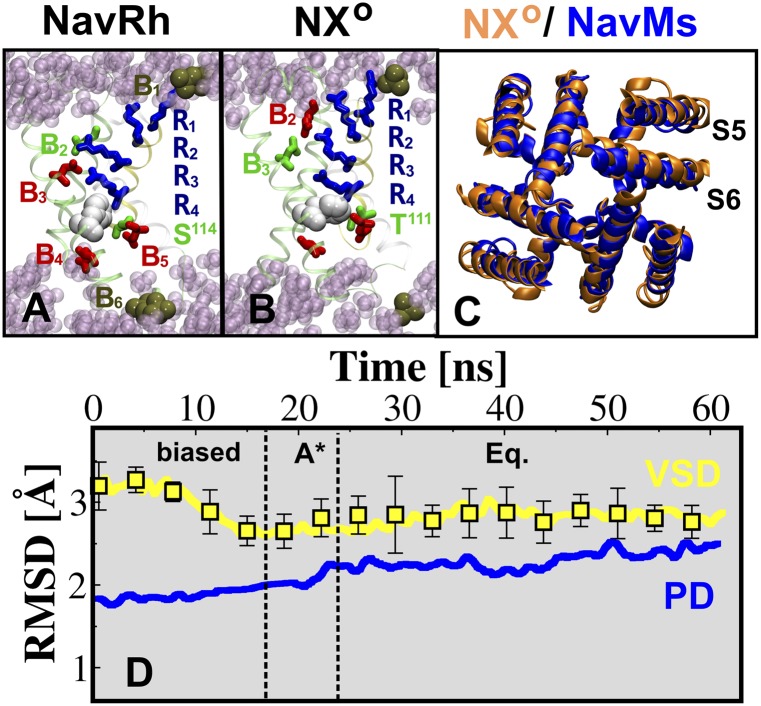
While this manuscript was in preparation, the X-ray structure of NavRh, an NaChBac ortholog from the marine Alphaproteobacterium HIMB114, was solved at 3.05-Å resolution showing the VSD in a depolarized (active) conformation but with a closed PD structure (21). To compare the NavAb conformations against NavRh, we performed an additional MD simulation to relax the NavRh in a membrane. Although moderate to substantially different from NX, NX−, and NX−−, the VSD conformation in the relaxed NavRh structure bears strong similarity to the domain in NX++ (Fig. 3). Specifically, the S4 gating charges occupy the aqueous cleft lying above the occluded site on the extracellular side in the VSD of both structures, forming a similar salt-bridge arrangement as indicated by their conformational distances, namely RAb = 0.85 ± 0.02 and RRh = 0.86 ± 0.01. Rmsd values of 2.47 ± 0.99 Å and 2.55 ± 0.29, computed for the main-chain atoms of S4 and VSD, respectively, further indicate their structural similarity (Table S2). Moreover, a recent investigation of the orthologous bacterial channel, NaChBac (25), has provided an extensive set of state-dependent interactions between residues in S1 and S2, and the gating residues of S4, based on analysis of disulfide locking of cysteine double mutants. The latter study shows that the NaChBac residues R122 (R4) and D60 (B3) interact in the active conformation of the VSD, providing unambiguous structural information. Among the NavAb VSD conformations, this constraint is best satisfied by NX++, in which the average distance between the Cβ atoms of R4 and N49 (B3) is 10.70 ± 0.33 Å, consistent with their probed propensity of interacting in the active state of the channel (Fig. S4 and Table S4). Taken together, these findings give strong support to the notion that the VSD conformation in NX++ is fully activated.
Fig. 3.
(A and B) Snapshots of the VSD of NavAb NXO and NavRh, highlighting the position of the S4 basic residues (blue sticks) and the salt bridges/hydrogen bonds they form with the acidic/polar residues (red/green sticks) of other VSD segments or with the lipid phosphate moieties (ocre). The highly conserved Phe residue (white), forming the catalytic center, is shown for both structures. (C) Structural superposition of the PD of NXO (orange) and NavMs (blue). (D) Rmsd profiles of selected segments of NavAb NXO, VSD (yellow), and PD (blue). The rms deviations of the backbone atoms for the VSD and PD are calculated with respect to the X-ray structure of NavRh and NavMs, respectively; for the VSD, the acidic/polar binding sites within the VSD were superposed and the average over the four subunits is reported, with associated error bars.
A recent study (25) suggested the existence of significant energetic coupling between T110 and D60 (B3) in the resting state of NaChBac. Among our NavAb structures, NX−, with a distance of 11.01 ± 1.71 Å between the Cβ atoms of residues E96 and N49 (B3), best explains the experimental observation. However, our analysis also supports the notion that a deeper NX−−-like conformation of the VSD is likely to exist in the resting state of the channel. In the latter, the Cβ atoms in the pair of residues E96 and N49 (B3) are 11.38 ± 0.93 Å apart. Thus, from the perspective of the present work, two conformations are consistent with the experiment (25). However, we note the nonconductive nature of NX− and NX−−, as suggested by their low PD-opening propensities. Very recent metal-ion bridge experiments have tracked the VSD cycle in the Kv channel (12). The latter work suggests that NX−−-like conformations could be distinguishable from other nonconductive structures by considering deeper sites, rather than B3, which is positioned at the top of S2, to probe the conformation-dependent interactions of S4 (Fig. S4). Indeed, only NX−− shows E96 in close proximity (8.66 ± 0.51 Å) to N25 in S1, which is inwardly positioned ∼10 Å away from the site of N49 (B3) (Table S4); the corresponding residues in the Shaker channel spontaneously form disulfide and metal bridges at hyperpolarized potentials, suggesting their close proximity in the resting state (32). All of this corroborates our finding that the VSD of NX−− is likely related to the resting conformation.
As mentioned above, presently there is an activated-open X-ray crystal structure of Kv1.2 and several proposed molecular models for the resting-closed state of the channel. Specifically, in the model by Pathak et al. (7), S4 is ∼8 Å inwardly positioned relative to the activated state. The models by Delemotte et al. (10) and Jensen et al. (11), although agreeing with the former model, show a larger vertical inward displacement of S4 relative to the activated state, namely ∼15 Å. Very recently, Henrion et al. (12) proposed a resting-state model (they call C3) for which the up-down S4 displacement is ∼12 Å. In the same study, another VS conformation (C4), in which S4 is more deeply located within the domain (∼17 Å), was further considered to match the experimental data; this latter conformation is, however, thought to be reached solely under enduring hyperpolarized pulses. Thus, the picture of activation emerging from these structural studies suggests that S4 moves about 8–15 Å in the TM direction. It is noteworthy that here, relative to NX++, the S4 helix in NX−− is displaced by 15.4 Å in the inward direction, which is consistent with the previous studies on Kv1.2. Furthermore, the sequential VS conformational transitions on passing from NX++ to NX−− involve a sliding motion of the salt-pairing interactions as a result of the S4 basic residues moving from external to internal binding sites along the domain; the sequential transitions are, respectively, accompanied by the motion of one of the S4 residues, T111, R4, R3, and R2, across the catalytic center (Movie S2).
Our findings are consistent with recent experiments demonstrating the existence of sequential S4 salt-bridge pairing during the activation process (23–25) and with previous MD simulations studies on the intermediates states of the VSD in Kv1.2 (10). Importantly, in this scenario, the conformational transition from NX++ to NX thus corresponds to an early stage of the deactivation process of NavAb (21). Indeed, although an intermediate, the net value of Q associated with NX++/NX was found to correspond to only ∼20% of QA, suggesting that the X-ray structure is functionally closer to the active than to the resting conformation. In agreement with findings of a recent computational investigation (33), we observe that some of the residues of S4 populate the 310-helix conformation in a significant fraction of the molecular configurations sampled along the trajectory (Fig. S5). The α- to 310-helix transition has been proposed to facilitate the sliding of S4 through the constrained region, and therefore different residues are expected to be in the 310 conformation in different activation states. Our analysis of the secondary structure content of S4 suggests that, despite a significant degree of heterogeneity among the four subunits, the region populating the 310 conformation moves from the N to the C terminus of S4 on passing from the resting to the activated state, thereby providing support to the picture of a moving helix transitioning from α to 310 as it crosses a constriction.
MD simulations exploring channel conformations along the PD-opening pathway reveal that the NX VSD constrains the pore up to a partially open state, as observed in the NXPO structure. Only for NXO, which exhibits four NX++ VSDs in the activated conformation, is the pore structure remarkably stable in the open conformation. Very recently, the X-ray structure of an ortholog of NavAb, namely NavMs, has been determined at 3.5 Å in a micelle-embedded open-PD conformation (22). The PDs of NXO and NavMs show significant structural similarity, as indicated by the small rmsd values and, most importantly, by the highly similar arrangements of the helices in the S6 bundle (Fig. 3 C and D). Thus, we conclude that NXO and NX−− are likely to be reasonable representations of the activated-open and resting-closed states of NavAb.
Materials and Methods
Conformational Analysis of VSD and PD.
We describe the instantaneous conformation of the VSD and PD by means of a distance matrix, Mij (NavAb, l; t) = rij (NavAb, l; t), where l is the domain considered (l ∈ {VSD, PD}) and rij is the distance between the geometrical centers of the charged moiety of a given S4 residue (j ∈ {R1, R2, ..., R4}) and the charged moiety of a given negatively charged or polar group (i ∈ {B1, B2, ..., B6}). For the PD distance matrix, we consider three PD-lining residues in the S6-helix bundle (i, j ∈ {F1, F2, F3}); rij is the Cα–Cα average distance between nonadjacent subunit (i, j) pairs. Table S1 presents the identities of the (i, j) sets for the NavAb, NavRh, and Kv1.2 channels.
By describing the conformations of KA and KR in Kv1.2 as Mij (KA, l; t) = rij (KA, l; t) and Mij (KR, l; t) = rij (KR, l; t), respectively, we define the conformational distance R (NavAb, l; t) of NavAb from the Kv-reference structures as follows:
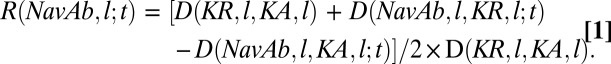 |
Here, D (KR, l, KA, l) is the rmsd between the distance matrices of domain l in KA and KR. D (NavAb, l, KR, l; t) and D (NavAb, l, KA, l; t) are the rms deviations between the instantaneous distance matrix of domain l in NavAb and the corresponding matrices of KA and KR, respectively. Note that R (NavAb, l; t) = 0.5 for NavAb conformations with equivalent deviations from the reference conformations; otherwise, R (NavAb, l; t) tends to 0 and 1 in the limits of NavAb conformations identical to KR and KA, respectively.
Sampling Selected VSD and PD Conformations.
The present procedure consisted of applying a biased and an equilibrium MD simulation to the membrane-bound NavAb structure, NX, to guide it to a given R value and then relaxing the structure at that conformational distance without constraints. Note the PD structure is also unconstrained. Three independent trajectories were generated, sampling NavAb conformations at R = 0.85 ± 0.02, 0.38 ± 0.06, and 0.14 ± 0.01 (average values over the four VSD subunits). The final membrane-equilibrated structures obtained from these simulations are referred to as NX++, NX−, and NX−−, respectively. In two additional independent MD simulations, the PD-closed structures NX and NX++ were first biased toward an open-PD conformation (P ∼ 0.9) and then submitted to relaxation without any constraints. The final membrane-equilibrated structures obtained from these MD runs are labeled NXPO and NXO, respectively. For details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grant NIGMS P-01 55876 and the Commonwealth of Pennsylvania. The computations were performed using resources from XSEDE (www.xsede.org/high-performance-computing) Grant MCA93S020. W.T. thanks the National Council of Technological and Scientific Development (CNPq) for support under Grants 470406/2011-9 and 141009/2009-8.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218087109/-/DCSupplemental.
See Commentary on page 21186.
References
- 1.Hille B. Ionic Channels of Excitable Membranes. 2nd Ed. Sunderland, MA: Sinauer; 1992. [Google Scholar]
- 2.Bezanilla F. The voltage-sensor structure in a voltage-gated channel. Trends Biochem Sci. 2005;30(4):166–168. doi: 10.1016/j.tibs.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Catterall WA. Ion channel voltage sensors: Structure, function, and pathophysiology. Neuron. 2010;67(6):915–928. doi: 10.1016/j.neuron.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long SB, Campbell EB, MacKinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309(5736):897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 5.Treptow W, Tarek M. Environment of the gating charges in the Kv1.2 Shaker potassium channel. Biophys J. 2006;90(9):L64–L66. doi: 10.1529/biophysj.106.080754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jogini V, Roux B. Dynamics of the Kv1.2 voltage-gated K+ channel in a membrane environment. Biophys J. 2007;93(9):3070–3082. doi: 10.1529/biophysj.107.112540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pathak MM, et al. Closing in on the resting state of the Shaker K(+) channel. Neuron. 2007;56(1):124–140. doi: 10.1016/j.neuron.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 8.Nishizawa M, Nishizawa K. Coupling of S4 helix translocation and S6 gating analyzed by molecular-dynamics simulations of mutated Kv channels. Biophys J. 2009;97(1):90–100. doi: 10.1016/j.bpj.2009.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalili-Araghi F, et al. Calculation of the gating charge for the Kv1.2 voltage-activated potassium channel. Biophys J. 2010;98(10):2189–2198. doi: 10.1016/j.bpj.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delemotte L, Tarek M, Klein ML, Amaral C, Treptow W. Intermediate states of the Kv1.2 voltage sensor from atomistic molecular dynamics simulations. Proc Natl Acad Sci USA. 2011;108(15):6109–6114. doi: 10.1073/pnas.1102724108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen MØ, et al. Mechanism of voltage gating in potassium channels. Science. 2012;336(6078):229–233. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- 12.Henrion U, et al. Tracking a complete voltage-sensor cycle with metal-ion bridges. Proc Natl Acad Sci USA. 2012;109:8552–8557. doi: 10.1073/pnas.1116938109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16(6):1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 14.Vargas E, Bezanilla F, Roux B. In search of a consensus model of the resting state of a voltage-sensing domain. Neuron. 2011;72(5):713–720. doi: 10.1016/j.neuron.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Catterall WA. Voltage-dependent gating of sodium channels: Correlating structure and function. Trends Neurosci. 1986;9:7–10. [Google Scholar]
- 16.Guy HR, Seetharamulu P. Molecular model of the action potential sodium channel. Proc Natl Acad Sci USA. 1986;83(2):508–512. doi: 10.1073/pnas.83.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475(7356):353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carnevale V, Treptow W, Klein ML. Sodium ion binding sites and hydration in the lumen of a bacterial ion channel from molecular dynamics simulations. J Phys Chem Lett. 2011;2(19):2504–2508. [Google Scholar]
- 19.Kuzmenkin A, Bezanilla F, Correa AM. Gating of the bacterial sodium channel, NaChBac: Voltage-dependent charge movement and gating currents. J Gen Physiol. 2004;124(4):349–356. doi: 10.1085/jgp.200409139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zagotta WN, Hoshi T, Aldrich RW. Shaker potassium channel gating. III: Evaluation of kinetic models for activation. J Gen Physiol. 1994;103(2):321–362. doi: 10.1085/jgp.103.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486(7401):130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCusker EC, et al. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat Commun. 2012;3 doi: 10.1038/ncomms2077. Article 1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeCaen PG, Yarov-Yarovoy V, Sharp EM, Scheuer T, Catterall WA. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc Natl Acad Sci USA. 2009;106(52):22498–22503. doi: 10.1073/pnas.0912307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu D, et al. State-dependent electrostatic interactions of S4 arginines with E1 in S2 during Kv7.1 activation. J Gen Physiol. 2010;135(6):595–606. doi: 10.1085/jgp.201010408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yarov-Yarovoy V, et al. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc Natl Acad Sci USA. 2012;109(2):E93–E102. doi: 10.1073/pnas.1118434109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328(5974):67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Treptow W, Tarek M, Klein ML. Initial response of the potassium channel voltage sensor to a transmembrane potential. J Am Chem Soc. 2009;131(6):2107–2109. doi: 10.1021/ja807330g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smart OS, Goodfellow JM, Wallace BA. The pore dimensions of gramicidin A. Biophys J. 1993;65(6):2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Treptow W, Tarek M. Molecular restraints in the permeation pathway of ion channels. Biophys J. 2006;91(3):L26–L28. doi: 10.1529/biophysj.106.087437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barber AF, et al. Hinge-bending motions in the pore domain of a bacterial voltage-gated sodium channel. Biochim Biophys Acta. 2012;1818(9):2120–2125. doi: 10.1016/j.bbamem.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakrapani S, Sompornpisut P, Intharathep P, Roux B, Perozo E. The activated state of a sodium channel voltage sensor in a membrane environment. Proc Natl Acad Sci USA. 2010;107(12):5435–5440. doi: 10.1073/pnas.0914109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campos FV, Chanda B, Roux B, Bezanilla F. Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K channel. Proc Natl Acad Sci USA. 2007;104(19):7904–7909. doi: 10.1073/pnas.0702638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwaiger CS, Bjelkmar P, Hess B, Lindahl E. 310-Helix conformation facilitates the transition of a voltage sensor S4 segment toward the down state. Biophys J. 2011;100(6):1446–1454. doi: 10.1016/j.bpj.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.