Abstract
129/SvEv mice with a loss-of-function mutation in the heterotrimeric G protein α-subunit gene Gnai3 have fusions of ribs and lumbar vertebrae, indicating a requirement for Gαi (the “inhibitory” class of α-subunits) in somite derivatives. Mice with mutations of Gnai1 or Gnai2 have neither defect, but loss of both Gnai3 and one of the other two genes increases the number and severity of rib fusions without affecting the lumbar fusions. No myotome defects are observed in Gnai3/Gnai1 double-mutant embryos, and crosses with a conditional allele of Gnai2 indicate that Gαi is specifically required in cartilage precursors. Penetrance and expressivity of the rib fusion phenotype is altered in mice with a mixed C57BL/6 × 129/SvEv genetic background. These phenotypes reveal a previously unknown role for G protein-coupled signaling pathways in development of the axial skeleton.
Keywords: mouse, thoracic, sternum, lateral plate mesoderm
The heterotrimeric G protein α-subunits, encoded by 16 paralogous genes in humans and mice, are cytoplasmic proteins that couple a wide variety of cell-surface receptors to intracellular effectors, such as ion channels and enzymes (1–3). The complex signal-transduction activity of these widely expressed proteins has long been studied at the biochemical and cellular level, but their role in development of whole organisms is less well understood. The “inhibitory” class of α subunits (Gαi), originally named for its ability to inhibit adenylyl cyclase activity, is encoded by the Gnai1, Gnai2, and Gnai3 genes. The three Gαi subunits share 85–95% amino acid sequence identity, and they form a subfamily with the neuronal α-subunit (Gαo/Gnao), the transducin α-subunits expressed in rod (Gαt-r/Gnat1) and cone cells (Gαt-c/Gnat2), and gustducin (Gαgust/Gnat3) expressed in taste buds. The Gαi genes are linked in pairs with the transducin and gustducin genes on mouse chromosomes 3, 5, and 9. This linkage, together with their sequence homology, suggests that these subunits evolved from an ancestral G protein gene by a tandem duplication followed by two block duplications (3). A Gαi ortholog is present in Drosophila, and identification of transducin genes in the lamprey genome indicates that the initial duplication to form an ancestral Gαi and Gαt gene predates the evolution of gnathostomes (4).
Targeted loss-of-function mutations of all three Gαi genes have been generated in mice, and the resulting phenotypes indicate that Gnai1 and Gnai2 have gene-specific functions in a wide variety of tissues: loss of Gnai1 affects long-term memory (5), and Gnai2 knockout mice spontaneously develop an inflammatory bowel disease resembling ulcerative colitis (6) and have altered heart rate dynamics (7). Initial analyses of Gnai3 knockout mice did not reveal an associated phenotype (8, 9), but more recently Gnai3 has been shown to be required for insulin-mediated regulation of autophagy in hepatocytes (10). Comparison of Gnai2 knockouts and Gnai3/Gnai1 double-knockouts suggests that the three Gαi proteins may also have both overlapping and gene-specific roles in the response of macrophages and splenocytes to bacterial infection (11). Here, we demonstrate that Gnai3 expression in sclerotomal derivatives is required for normal patterning of the axial skeleton. Gnai1 and Gnai2 partially compensate for loss of Gnai3, and the phenotype is dependent on genetic background.
Results
Skeletal Defects in Gnai3−/− Mice.
Inbred 129/SvEv mice that are homozygous for a targeted loss-of-function mutation in the Gnai3 gene (12) are viable and fertile, but staining of skeletons revealed an unexpected phenotype: 95% of 129/SvEv-Gnai3−/− mice have fusions of the cartilaginous portion of the distal ribs (Fig. 1B and Table 1, first row). These fusions involve any of the true ribs (those ribs that articulate with the sternum) but usually do not affect the false ribs, the ends of which are free in the body wall. The proximal bony portions of the ribs appear normal, and in all cases the normal complement of ribs is present. The single animal that lacked rib fusions had small triangular outgrowths of cartilage at the distal end of the second rib pair where they join the sternum. One animal had an eighth rib (first false rib) with an ectopic connection to the sternum, but other than that, contacts between ribs and sternum appeared normal in all of the mice. In fetuses stained with Alcian blue, the rib fusions are visible as early as embryonic day (E) 14.5, suggesting that they occur as the ribs develop and are not caused by later overgrowth of cartilage.
Fig. 1.
Rib and sternum defects in mice with mutations of Gαi genes. The images show the sternum with true ribs attached. False ribs that are not involved in any fusions have been removed. (A) Gnai3+/−, 19-wk-old, showing normal morphology of sternum and true ribs. (B) Gnai3-/−, 19-wk-old, with fusions of the cartilaginous portion of four rib pairs. The fusions involve true ribs only. Although there is the normal number of symmetric contacts of ribs and sternum, the sternum itself is distorted. (C) Gnai3−/− Gnai1+/−, 22-wk-old, with fusion of nine rib pairs. On the right side, the first false rib is fused to the seventh true rib (blue arrowhead). (D) Gnai3−/− Gnai1−/−, 41-wk-old, with fusions involving all but one of the true ribs and one false rib (blue arrowhead). Rib-sternum contacts are asymmetric (black arrowhead), and the second and third sternebrae are fused (red arrowhead). (E) Gnai3−/− Gnai2+/−, 4-wk-old, with fusions of ten rib pairs, including fusion of one false rib (blue arrowhead). The eighth rib on the right side is connected to the sternum (gray arrowhead). (F) Gnai1−/− Gnai2−/−, 5-wk-old, with an eighth rib connected to the sternum (gray arrowhead) but no rib fusions.
Table 1.
Thoracic skeletal defects in Gαi mutant mice
| Genetic background | Genotype | No. examined* | Skeletal fusions† |
Asymmetric sternal contacts (%) | ||
| True ribs (%) | False ribs (%) | Sternebrae (%) | ||||
| 129/SvEv | Gnai3−/− | 19 | 95 | 16 | 6‡ | 5 |
| Gnai3−/− Gnai1+/− | 10 | 100 | 80 | 10 | 0 | |
| Gnai3−/− Gnai1−/− | 21 | 100 | 100 | 67 | 29 | |
| Gnai3−/− Gnai2+/− | 6 | 100 | 100 | 17 | 33 | |
| Gnai3+/− | 15 | 0 | 0 | 0 | 0 | |
| Gnai3+/− Gnai1+/− | 5 | 0 | 0 | 0 | 0 | |
| Gnai3+/− Gnai1−/− | 7 | 0 | 0 | 0 | 0 | |
| Gnai1−/− | 10 | 0 | 0 | 0 | 0 | |
| Gnai2−/− | 2 | 0 | 0 | 0 | 0 | |
| Gnai1−/− Gnai2−/− | 6 | 0 | 0 | 0 | 17 | |
| Wild-type | 10 | 0 | 0 | 0 | 0 | |
| B6129 F2 | Gnai3−/− | 61 | 38 | 0 | 0 | 3 |
*Analysis includes adults, neonates, and E14.5 fetuses.
†Percentages are for number of animals exhibiting the trait, not number of ribs or sternebrae affected.
‡Three E14.5 fetuses not included in this calculation because the sternum was not ossified.
In the lumbar region, we observed deformation or partial fusion of one or more vertebral bodies in 9 of 10 Gnai3−/− pups (Fig. 2). In 7 of 10 pups, lumbar abnormalities consisted of a small “bridge” of bone connecting the bodies of two or three adjacent vertebrae (Fig. 2B, arrow), and in 2 of 10 pups the deformed vertebrae have pointed outgrowths that do not actually fuse. Bony lumbar fusions were not observed in wild-type and heterozygous pups, but one of eight wild-type and three of nine heterozygotes had deformed vertebrae. The frequency of bony lumbar fusions in Gnai3−/− mice is statistically significant compared with heterozygotes (P < 0.01, Fisher’s exact test).
Fig. 2.
Fusion of lumbar vertebrae in Gnai3−/− mice. (A) B6129F2-Gnai3+/+, 5-d-old, showing wild-type morphology of the second through fifth lumbar vertebrae. (B) 129/SvEv-Gnai3−/−, 4-d-old, with partial fusion of the third and fourth lumbar vertebrae. (C) B6129F2-Gnai3−/−, 5-d-old, with fusion of the second, third, fourth, and fifth lumbar vertebrae. The fusion of the third and fourth vertebrae is more extensive than that seen in any 129/SvEv-Gnai3−/− mouse. (Magnification: 10×.)
Because skeletal abonormalities have not been reported for a different Gnai3 knockout allele that was generated on a C57BL/6 background (8), we investigated whether genetic background can modify the rib fusion phenotype. In the F2 generation of a C57BL/6J × 129/SvEv intercross we observed reduction in penetrance and expressivity of the rib fusion phenotype. Less than 40% of B6129F2-Gnai3−/− mice had rib fusions (Table 1), and in the animals with fusions, the average number was reduced from 2.7 to 1.3 (P < 0.001, unpaired t test). We observed lumbar defects in only 13 of 61 mice, a significant reduction relative to the inbred 129/SvEv background (P < 0.01, Fisher’s exact test). However, the fusions in several B6129F2-Gnai3−/− mice were more extensive than those observed in any of the 129/SvEv-Gnai3−/− mice (Fig. 2C). Whole-genome SNP genotyping of 35 B6129F2-Gnai3−/− mice (18 with rib fusions, 17 without) did not reveal a major locus associated with presence or absence of rib fusions, suggesting that these axial defects are modified by multiple loci acting additively.
Effects of Gnai1 and Gnai2 Mutations.
The amino acid sequences encoded by the mouse Gnai1 and Gnai2 genes are, respectively, 94% and 85% identical to Gnai3. Given this degree of similarity, it would not be surprising if the three proteins have some overlapping function. We intercrossed Gnai1, Gnai2, and Gnai3 knockout mice to investigate whether the other Gαi genes also contribute to skeletal development (Table 1).
In Gnai3−/− mice with either heterozygous or homozygous loss of Gnai1, the rib fusions are significantly more severe, frequently involving false ribs as well as true ribs (Figs. 1 C–D and 3). In addition to rib fusions, we observed asymmetric contacts with the sternum and fusions of sternebrae in the double mutants (Fig. 1D and Table 1). Lumbar fusions were not noticeably more severe in Gnai3−/− Gnai1−/− compared with Gnai3−/− mice.
Fig. 3.
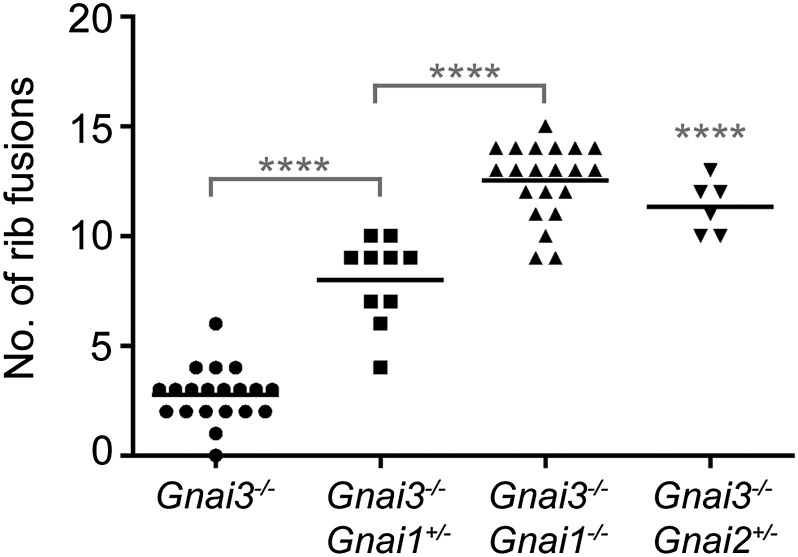
Mutation of Gnai1 or Gnai2 increases the number of rib fusions in 129/SvEv-Gnai3−/− mice. Each datapoint on the scatter plot represents an individual mouse. Black horizontal lines indicate mean number of rib fusions for a genotype. ****P < 0.0001, unpaired t test.
Complete loss of both Gnai3 and Gnai2 is lethal before E10 (10), but mice with the genotype Gnai3−/− Gnai2+/− are viable and have rib fusions equivalent in severity to Gnai3−/− Gnai1−/− (Figs. 1E and 3). Animals with the genotype Gnai1−/− Gnai2−/− had no rib fusions, but one had an eighth rib ectopically connected to the sternum (Fig. 1F). This phenotype was also observed in one Gnai3−/− Gnai2+/− mouse and one Gnai3−/− mouse. Taken together, these data indicate that all three Gαi genes participate in skeletal development, but Gnai3 is the most important; rib fusions were never observed in any animal that was not homozygous for loss of Gnai3 (Table 1).
Gαi Is Required in Rib Precursors.
During vertebrate development, somites differentiate into dermotome, myotome (which gives rise to the intercostal muscles), and sclerotome (which gives rise to the ribs and spinal column) (13). Growth of ribs is controlled by a signaling cascade initiated by Hox gene expression in myotome of the thoracic region and transmitted to the sclerotome by PDGF and FGF signaling (14–16). In mice with mutations that disrupt expression of myotome-specific genes required in this signaling pathway, rib fusions are preceded by disorganization and fusion of developing intercostal muscles, with initial myotome defects visible by E10.5–E11.5 (17, 18). Therefore, the rib fusions in Gnai3 mutant mice could reflect a requirement for Gαi in either developing musculature or skeleton.
We used a muscle-specific Myog (myogenin) probe to reveal morphology of developing intercostal muscles in wild-type and Gnai3−/− Gnai1−/− embryos at E11.5 and E12.5. The staining pattern in wild-type and mutant embryos was indistinguishable at both developmental stages (Fig. 4 A–D). At E12.5, a Sox9 probe revealed what may be the initial stages of fusion of the rib primordia; in three of five mutant embryos, the distal ends of the Sox9-expressing domains appeared broader than the equivalent regions of four wild-type embryos, and in several places adjacent domains were in contact (Fig. 4F, arrowhead). The lack of obvious myotome defects during this period suggested that the skeletal defects in Gnai3−/− mice are not secondary to a requirement for Gαi in muscle.
Fig. 4.
Rib fusions in Gnai3−/− Gnai1−/− mice are not associated with morphological defects of myotome. (A) Wild-type embryo, E11.5, side view of trunk between the limb buds. In situ hybridization with a Myog probe reveals morphology of the developing intercostal muscles. (B) Gnai3−/− Gnai1−/− embryo, E11.5, Myog probe. Intercostal muscle morphology is indistinguishable from that of the wild-type embryo. (C) Wild-type, E12.5, Myog probe. (D) Gnai3−/− Gnai1−/−, E12.5, Myog probe. (E) Wild-type, E12.5. In situ hybridization with a Sox9 probe reveals rib primordia. (F) Gnai3−/− Gnai1−/−, E12.5, Sox9 probe. Arrowhead indicates possible fusion of rib primordia. (Magnification: A and B, 11×; C–F, 10×.)
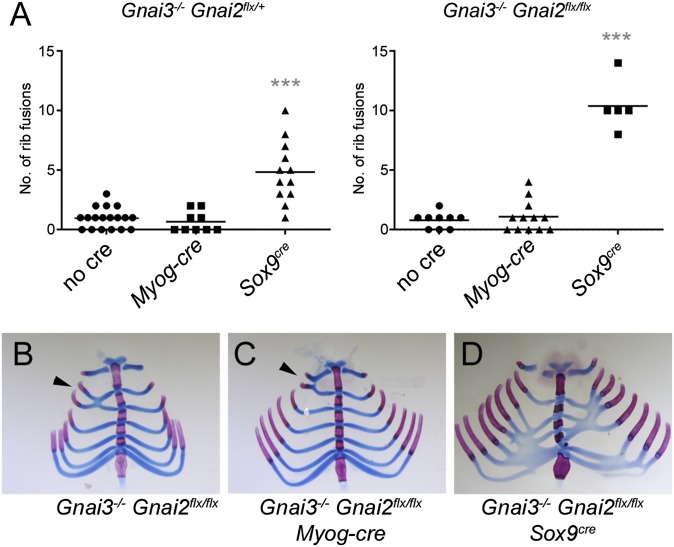
Because mutation of Gnai1 or Gnai2 increases the severity of rib fusions in Gnai3−/− mice, we reasoned that a conditional allele of Gnai2 could be used as an independent test of the tissue-specific origin of the rib fusion phenotype. We made a conditional allele of Gnai2 (Fig. 5) in which loxP sites flank exons 2, 3, and 4, and we used a Myog-cre transgene (19) to drive Cre expression in skeletal muscle precursors and a Sox9cre knock-in allele (20) to drive Cre expression in cartilage precursors, including the sclerotomal cells that give rise to the ribs. In mice that have the genotype Gnai3−/− Gnai2flx/+ or Gnai3−/− Gnai2flx/flx, severity of the rib phenotypes should be increased by expression of Cre recombinase in the critical tissue. The Myog-cre transgene had no effect, but the Sox9cre knock-in significantly increased the severity of rib fusions in the double-mutant mice (Fig. 6).
Fig. 5.
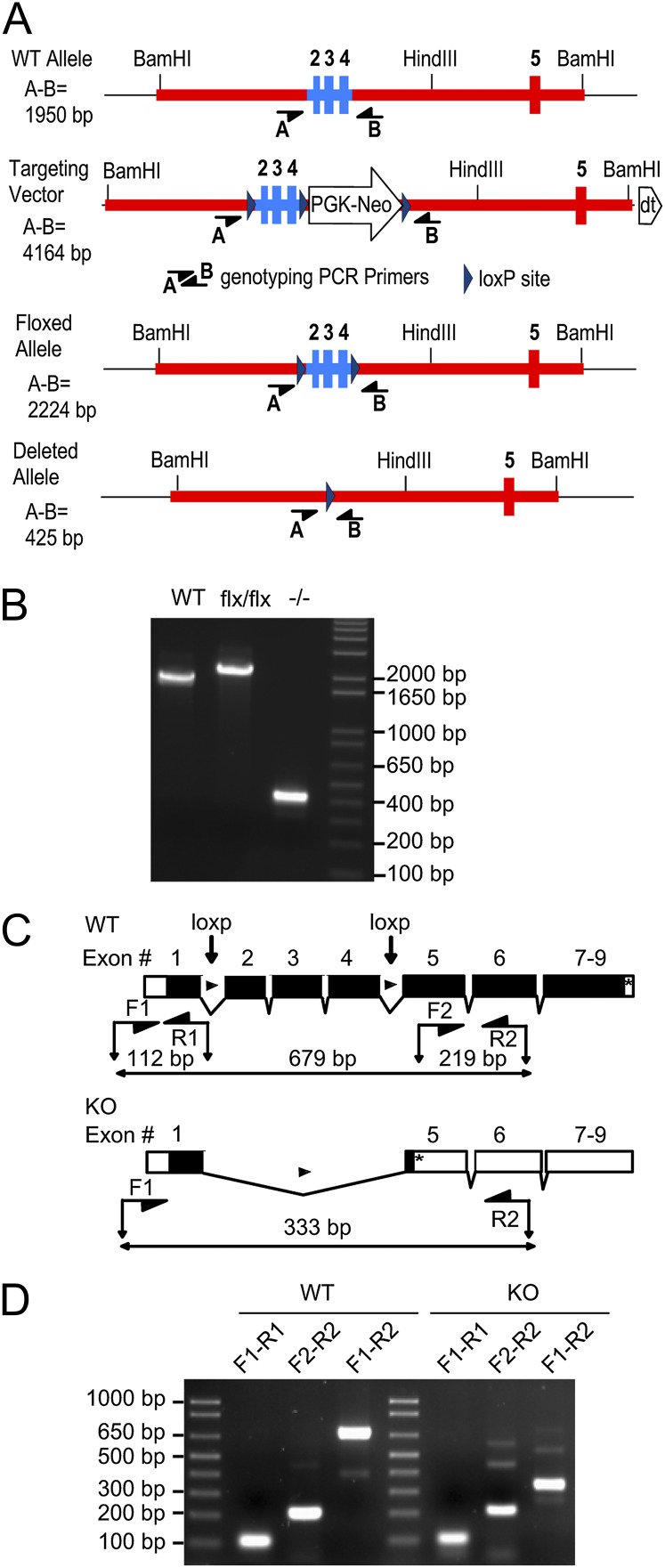
Conditional allele of Gnai2. (A) Targeting strategy. WT allele: depicts the region of the wild-type Gnai2 gene containing the targeted exons 2, 3, and 4 (blue) used to construct the targeting vector. Targeting vector: depicts the portion of the targeting vector used to target the Gnai2 locus. Floxed allele: depicts the structure of the targeted allele after Cre-mediated excision of the PGK-Neo cassette. Deleted allele: depicts the structure of the disrupted allele from which exons 2, 3, and 4 have been removed by the action of Cre recombinase. The position of key restriction endonuclease sites and the location of genotyping primers A and B are indicated. Rectangles, exons included in targeting vector; heavy red line, intronic sequence included in the targeting vector; PGK-Neo, neomycin selection cassette; dt, diptheria toxin selection cassette. (B) PCR analysis of mouse genomic DNA using primers A and B. All Gnai2 genotypes produce PCR products of the expected sizes indicated in Fig 4A. WT, DNA from wild-type mouse; flx/flx, DNA from mouse homozygous for the floxed allele; −/−, DNA from mouse homozygous for the deleted allele which exons 2–4 have been excised by a Cre transgene driven by the ubiquitous Sox2 promoter. (C) (Upper) Diagram of the wild-type intron/exon organization of the Gnai2 gene. Locations of LoxP sites in the floxed allele and RT-PCR primers are indicated. (Lower) Diagram of the deleted allele. Deletion of exons 2–4 by Cre recombinase is predicted to result in a frame-shift and premature stop codon in exon 5. The lengths of the depicted amplicons include the primers. Black boxes, coding sequence; open boxes, untranslated exon sequence; *, stop codon. (D) RT-PCR analysis of brain RNA from a wild-type and a Gnai2−/− mouse. Sequencing of the RT-PCR products confirmed splicing from exon 1 to exon 5 and the presence of a premature stop codon in mRNA transcribed from the deleted allele.
Fig. 6.
Rib fusion phenotype of Gnai3−/− mice is enhanced by loss of Gnai2 in cartilage. (A) Scatter plot showing the distribution of rib fusions in mice of different genotypes. Statistically significant increase in the number of fusions is seen in Gnai3−/− Gnai2flx/+ Sox9cre/+ and Gnai3−/− Gnai2flx/flx Sox9cre/+ mice. Horizontal lines indicate mean number of rib fusions. ***P < 0.001, unpaired t test. (B) Gnai3−/− Gnai2flx/flx neonate with one rib fusion (black arrowhead). (C) Gnai3−/− Gnai2flx/flx Tg(Myog-Cre) neonate with one rib fusion (black arrowhead). (D) Gnai3−/− Gnai2flx/flx Sox9cre/+ neonate with 10 rib fusions.
These results confirm that the rib fusion phenotype is caused by loss of Gαi in cartilage. The reduced penetrance of the rib phenotype in Gnai3−/− Gnai2flx/flx mice relative to 129/SvEv-Gnai3−/−, and the similarity of the Gnai3−/− Gnai2flx/flx Sox9cre/+ mice to 129SvEv-Gnai3−/− Gnai2+/− mice, is likely because of C57BL/6 alleles of modifier genes contributed by the Sox9cre and Tg(Myog-cre) mice. Number and severity of the lumbar vertebral fusions was not increased in Gnai3−/− Gnai2flx/flx Sox9cre/+, again indicating that this phenotype is dependent primarily on loss of Gnai3.
Discussion
The restriction of rib fusions in Gnai3−/− mice to the cartilaginous, distal portion of the ribs and the apparently normal development of the bony proximal ribs suggest that the primaxial/abaxial classification system for somitic development (21, 22) may provide insight into Gαi function. Unlike the epaxial/hypaxial classification, which originally defined somite-derived muscle on the basis of innervation by dorsal or ventral branches of the spinal nerves and was later extended to embryonic position relative to the notochord (23), the primaxial/abaxial classification is based on interaction between migrating somitic cells and the lateral plate mesoderm (LPM). The primaxial domain, close to the body axis, consists solely of somitic cells, but in the abaxial domain somite derivatives migrate away from the axis and differentiate in close proximity to tissue derived from the LPM. The LPM forms the connective tissue that surrounds and penetrates the somite-derived bone and muscle of the abaxial domain. Analysis of chick development indicates that the spinal column and vertebral ribs are primaxial, but the sternal ribs are abaxial and dependent on bone morphogenetic protein (BMP) signaling from the LPM (24). One part of the axial skeleton, the sternum, is derived directly from the LPM, not somites (25, 26). Mammals do not have distinct, ossified sternal ribs, but the distal, cartilaginous portion of the ribs appears to be analogous to the sternal ribs of birds. Analysis of the Tg(Prrx1-cre)1Cjt transgenic mouse, which labels the LPM, suggests that the distal, cartilaginous portion of the first rib is abaxial, but the remaining ribs are surrounded by somite-derived periosteum, technically making them primaxial (26). The distal portions of those ribs are nevertheless in close proximity to LPM-derived connective tissue, the sternum, and to the distal portion of intercostal muscles, which are labeled by the Prrx1-cre transgene. Although the vertebrae are definitively primaxial, the lateral surfaces of the lumbar vertebrae are attached to an abaxial muscle, the psoas major (26). Thus, all three of the regions in which we observe axial skeletal fusions in the Gαi mutant mice (ribs, sternum, and lumbar vertebrae) are either LPM-derived or develop close to LPM-derived tissue.
Consistent with a hypothesis that Gαi is required for transduction of signals from LPM to rib primordia, fusions of the distal rib cartilage almost identical to those of Gnai3−/− mice are seen in Bmp7−/− knockout mice (27) and in Bmp4+/− Bmp7+/− compound heterozygotes (28). Both Bmp4 and Bmp7 are expressed by the LPM (29, 30), and in the chick, Bmp4 is required for growth of somitic cells into the LPM domain (24). There is little evidence that BMP signaling requires heterotrimeric G proteins, but sonic hedgehog (Shh) signaling appears to alter the response of sclerotome cells to BMP and is required for chondrogenesis (31). Hedgehog signaling in Drosophila has been linked to heterotrimeric G proteins through Smoothened (Smo), which couples to Gαi (32, 33), and Shh-induced proliferation of rat cerebellar granule cell precursors is reduced by knock-down of Gnai2 and Gnai3 expression (34). If the rib fusions in Gnai3−/− mice result from defects in an interaction between hedgehog and BMP signaling pathways, disruption of hedgehog signaling in chondrocytes might be expected to resemble loss of Gαi. However, neither chondrocyte-specific overexpression of Shh nor conditional knockout of Smo or the hedgehog receptor Ptch1 in chondrocytes closely resemble the Gnai3 knockout phenotype (35–37).
Other signaling pathways involved in skeletal development and dorsal-ventral patterning that plausibly could require Gαi activity include Wnt and PDGF. Frizzled proteins, the receptors for Wnt, are putative G protein-coupled receptors (38), although they have not been specifically linked to Gαi. Negative regulators of Wnt signaling include Axin1, which may modulate heterotrimeric G-protein activity through its regulator of G-protein signaling domain. Mutation of Axin1 in the spontaneous mouse mutant Fused results in fusions of both vertebrae and ribs, but the rib fusions tend to involve the proximal ribs close to the spine (39). As described above, PDGF is involved in signaling from myotome to sclerotome during rib development, and activation of PDGFRα, a receptor tyrosine kinase, on sclerotome cells leads to altered cell migration via a PI3 kinase-Akt pathway (40). Gαi3 is known to regulate migration of HeLa cells by an Akt-dependent pathway (41), and a growing body of data indicates that heterotrimeric G proteins can function within receptor tyrosine kinase signaling pathways (42–44).
We have identified a previously undescribed requirement for heterotrimeric G proteins in skeletal development. The specifics of the rib fusion phenotype suggest that Gαi is required at the interface between somitic and lateral plate mesoderm. Additional crosses between the Gnai3 mutants and mice with targeted mutations in some of the genes mentioned above (e.g., Smo, Axin1, or Pdgfra) may reveal the pathway in which Gαi participates. Mapping of the genes involved in modifying the rib fusion phenotype in C57BL/6 may also be informative, possibly identifying other members of the signaling cascade.
Materials and Methods
Mice.
Targeted mutations of the Gnai1, Gnai2, and Gnai3 genes were previously described (12, 45). A colony of 129/SvEv-Gnai3tm1Lbi Gnai1tm1Drs mice was maintained by intercrossing homozygotes (hereafter indicated Gnai3−/− Gnai1−/−). To generate other genotypes, a Gnai3−/− Gnai1−/− mouse was crossed to a wild-type 129/SvEv mouse, and the offspring were intercrossed. The 129/SvEv- Gnai3−/− Gnai1+/+ and 129/SvEv-Gnai3+/+ Gnai1−/− sublines were established by intercrossing homozygotes. The 129/SvEv-Gnai2tm1Uru mouse colony was maintained by intercrossing heterozygotes (Gnai2+/−).
A conditional allele of Gnai2 (Gnai2flx) with loxP sites upstream of exon 2 and downstream of exon 4 was generated by homologous recombination in 129/SvEv embryonic stem cells. Chimeric mice derived from the targeted cells were crossed with 129/SvEv mice, and offspring were intercrossed to produce an inbred 129/SvEv-Gnai2flx/flx homozygous line. DNA genotyping and RT-PCR of brain RNA from mice homozygous for the targeted allele and hemizygous for the Tg(Sox2-cre)1Amc transgene (46), which drives ubiquitous expression of Cre recombinase in the early embryo, were used to confirm recombination of the loxP sites and deletion of exons 2–4 in the presence of Cre recombinase (Fig. 4). Genotyping primers were A (5′- GTGGTAAGCCTGTGTTTGTGAGAG) and B (5′-GGAGCCTGGACTTTGCTTCTGACC). Primers for RT-PCR were F1 (5′-TGCACCGTGAGCGCCGAGGACAAG), F2 (5′-ACCTGAATGATCTGGAGCGCATTG), R1 (5′-CTAACAGAAGCAACTTCACCTCCC), and R2 (5′-TCAAGGCGACACAGAAGATGATGG). Tg(Myog-cre)1Eno, and Sox9tm3(cre)Crm mice were previously described (19, 20).
For the B6 × 129 intercross, C57BL/6J mice were purchased from the Jackson Laboratory. SNP genotyping was performed by the Mutation Mapping and Developmental Analysis Project of Brigham and Women’s Hospital, Harvard University. All animal experiments were performed with approval of the National Institute of Environmental Health Sciences Institutional Animal Care and Use Committee.
Skeleton Staining and Analysis.
Skeletons of neonatal and adult mice were stained with Alcian blue and Alizarin red (47). Mouse fetuses at E14.5 were stained with Alcian blue, as described previously (48), except that the fetuses were skinned and eviscerated after fixation. Two or three complete skeletons of adult mice were stained for each genotype, and thereafter only the rib cage was stained. The skeletons of pups and fetuses were stained intact. Stained skeletons were photographed with a Coolpix 995 digital camera (Nikon). For scoring the number of rib fusions, fusion of one pair of ribs was counted as one fusion, and fusion of three consecutive ribs was counted as two fusions. Statistical analyses and graphing were performed using GraphPad Prism and QuickCalcs software (GraphPad Software).
In Situ Hybridization.
The template for the Sox9 riboprobe has been described previously (49). Templates for sense and antisense myogenin (Myog) riboprobes were produced from a cDNA clone (IMAGE: 6508229, GenBank: BC068019) by PCR with the T7 promoter sequence incorporated in either the forward or reverse primer. Primers to generate template for the antisense riboprobe were: 5′-GGCCAGTGGCAGGAACAAGC (Myog, forward) and 5′- CCAAGCTTCTAATACGACTCACTATAGGGAATTCGAGGCATATTATG (T7-Myog, reverse). Primers to generate template for the sense control probe were: 5′-CCAAGCTTCTAATACGACTCACTATAGGGCCAGTGGCAGGAACAAGC (T7-Myog, forward) and 5′-GGAATTCGAGGCATATTATG (Myog, reverse). Digoxigenin-labeled riboprobes were synthesized from these templates using the DIG RNA Labeling Mix (Roche Applied Science).
Embryos for whole-mount in situ hybridization were fixed overnight at 4 °C in 4% (wt/vol) paraformaldehyde dissolved in PBS, pH 7.4 (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4). After fixation, the embryos were dehydrated and stored in 100% methanol at −20 °C. In situ hybridization was performed as previously described (50) with the following modifications: PBS with Tween-20 contained 1% Tween-20 rather than 0.1%. Hybridization buffer contained 100 μg/mL sheared salmon sperm DNA. The embryos were not treated with RNase A after probe hybridization. After incubation with antidigoxygenin antibody (Roche Applied Science), embryos were washed overnight in Tris buffered saline (140 mM NaCl, 2.7 mM KCl, 25 mM Tris-HCl, pH7.5) with 1% Tween-20 at 4 °C. Hybridized probe was visualized with BM Purple Reagent (Roche Applied Science).
Acknowledgments
The authors thank Tom Sliwa for animal husbandry, Mitzie Walker for assistance with genotyping, and Jennifer Moran for SNP analysis. The template for the Sox9 in situ probe was generously provided by Masahiro Iwamoto (Thomas Jefferson University), and the Tg(Myog-cre)1Eno mice were generously provided by Eric Olson (University of Texas Southwestern Medical Center). This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (Project Z01-ES-101643).
Footnotes
The authors declare no conflict of interest.
References
- 1.Downes GB, Gautam N. The G protein subunit gene families. Genomics. 1999;62(3):544–552. doi: 10.1006/geno.1999.5992. [DOI] [PubMed] [Google Scholar]
- 2.Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiol Rev. 2005;85(4):1159–1204. doi: 10.1152/physrev.00003.2005. [DOI] [PubMed] [Google Scholar]
- 3.Wilkie TM, et al. Evolution of the mammalian G protein alpha subunit multigene family. Nat Genet. 1992;1(2):85–91. doi: 10.1038/ng0592-85. [DOI] [PubMed] [Google Scholar]
- 4.Muradov H, Kerov V, Boyd KK, Artemyev NO. Unique transducins expressed in long and short photoreceptors of lamprey Petromyzon marinus. Vision Res. 2008;48(21):2302–2308. doi: 10.1016/j.visres.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pineda VV, et al. Removal of G(ialpha1) constraints on adenylyl cyclase in the hippocampus enhances LTP and impairs memory formation. Neuron. 2004;41(1):153–163. doi: 10.1016/s0896-6273(03)00813-4. [DOI] [PubMed] [Google Scholar]
- 6.Rudolph U, et al. Ulcerative colitis and adenocarcinoma of the colon in G α i2-deficient mice. Nat Genet. 1995;10(2):143–150. doi: 10.1038/ng0695-143. [DOI] [PubMed] [Google Scholar]
- 7.Zuberi Z, Birnbaumer L, Tinker A. The role of inhibitory heterotrimeric G proteins in the control of in vivo heart rate dynamics. Am J Physiol Regul Integr Comp Physiol. 2008;295(6):R1822–R1830. doi: 10.1152/ajpregu.90625.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain M, et al. Targeted inactivation of Galpha(i) does not alter cardiac function or beta-adrenergic sensitivity. Am J Physiol Heart Circ Physiol. 2001;280(2):H569–H575. doi: 10.1152/ajpheart.2001.280.2.H569. [DOI] [PubMed] [Google Scholar]
- 9.Yang J, et al. Signaling through Gi family members in platelets. Redundancy and specificity in the regulation of adenylyl cyclase and other effectors. J Biol Chem. 2002;277(48):46035–46042. doi: 10.1074/jbc.M208519200. [DOI] [PubMed] [Google Scholar]
- 10.Gohla A, et al. An obligatory requirement for the heterotrimeric G protein Gi3 in the antiautophagic action of insulin in the liver. Proc Natl Acad Sci USA. 2007;104(8):3003–3008. doi: 10.1073/pnas.0611434104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan H, et al. Lipopolysaccharide- and Gram-positive bacteria-induced cellular inflammatory responses: Role of heterotrimeric Galpha(i) proteins. Am J Physiol Cell Physiol. 2005;289(2):C293–C301. doi: 10.1152/ajpcell.00394.2004. [DOI] [PubMed] [Google Scholar]
- 12.Jiang M, et al. Mouse gene knockout and knockin strategies in application to alpha subunits of Gi/Go family of G proteins. Methods Enzymol. 2002;344:277–298. doi: 10.1016/s0076-6879(02)44721-0. [DOI] [PubMed] [Google Scholar]
- 13.Christ B, Huang R, Wilting J. The development of the avian vertebral column. Anat Embryol (Berl) 2000;202(3):179–194. doi: 10.1007/s004290000114. [DOI] [PubMed] [Google Scholar]
- 14.Huang R, et al. Ventral axial organs regulate expression of myotomal Fgf-8 that influences rib development. Dev Biol. 2003;255(1):30–47. doi: 10.1016/s0012-1606(02)00051-9. [DOI] [PubMed] [Google Scholar]
- 15.Tallquist MD, Weismann KE, Hellström M, Soriano P. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 2000;127(23):5059–5070. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- 16.Vinagre T, et al. Evidence for a myotomal Hox/Myf cascade governing nonautonomous control of rib specification within global vertebral domains. Dev Cell. 2010;18(4):655–661. doi: 10.1016/j.devcel.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Laclef C, et al. Altered myogenesis in Six1-deficient mice. Development. 2003;130(10):2239–2252. doi: 10.1242/dev.00440. [DOI] [PubMed] [Google Scholar]
- 18.Vivian JL, Olson EN, Klein WH. Thoracic skeletal defects in myogenin- and MRF4-deficient mice correlate with early defects in myotome and intercostal musculature. Dev Biol. 2000;224(1):29–41. doi: 10.1006/dbio.2000.9788. [DOI] [PubMed] [Google Scholar]
- 19.Li S, et al. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc Natl Acad Sci USA. 2005;102(4):1082–1087. doi: 10.1073/pnas.0409103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akiyama H, et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci USA. 2005;102(41):14665–14670. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burke AC, Nowicki JL. A new view of patterning domains in the vertebrate mesoderm. Dev Cell. 2003;4(2):159–165. doi: 10.1016/s1534-5807(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 22.Nowicki JL, Takimoto R, Burke AC. The lateral somitic frontier: Dorso-ventral aspects of anterio-posterior regionalization in avian embryos. Mech Dev. 2003;120(2):227–240. doi: 10.1016/s0925-4773(02)00415-x. [DOI] [PubMed] [Google Scholar]
- 23.Spörle R. Epaxial-adaxial-hypaxial regionalisation of the vertebrate somite: Evidence for a somitic organiser and a mirror-image duplication. Dev Genes Evol. 2001;211(4):198–217. doi: 10.1007/s004270100139. [DOI] [PubMed] [Google Scholar]
- 24.Sudo H, et al. Inductive signals from the somatopleure mediated by bone morphogenetic proteins are essential for the formation of the sternal component of avian ribs. Dev Biol. 2001;232(2):284–300. doi: 10.1006/dbio.2001.0198. [DOI] [PubMed] [Google Scholar]
- 25.Chen JM. Studies on the morphogenesis of the mouse sternum. II. Experiments on the origin of the sternum and its capacity for self-differentiation in vitro. J Anat. 1952;86(4):387–401. [PMC free article] [PubMed] [Google Scholar]
- 26.Durland JL, Sferlazzo M, Logan M, Burke AC. Visualizing the lateral somitic frontier in the Prx1Cre transgenic mouse. J Anat. 2008;212(5):590–602. doi: 10.1111/j.1469-7580.2008.00879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo G, et al. BMP-7 is an inducer of nephrogenesis, and is also required for eye development and skeletal patterning. Genes Dev. 1995;9(22):2808–2820. doi: 10.1101/gad.9.22.2808. [DOI] [PubMed] [Google Scholar]
- 28.Katagiri T, Boorla S, Frendo JL, Hogan BLM, Karsenty G. Skeletal abnormalities in doubly heterozygous Bmp4 and Bmp7 mice. Dev Genet. 1998;22(4):340–348. doi: 10.1002/(SICI)1520-6408(1998)22:4<340::AID-DVG4>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 29.Mine N, Anderson RM, Klingensmith J. BMP antagonism is required in both the node and lateral plate mesoderm for mammalian left-right axis establishment. Development. 2008;135(14):2425–2434. doi: 10.1242/dev.018986. [DOI] [PubMed] [Google Scholar]
- 30.Solloway MJ, Robertson EJ. Early embryonic lethality in Bmp5;Bmp7 double mutant mice suggests functional redundancy within the 60A subgroup. Development. 1999;126(8):1753–1768. doi: 10.1242/dev.126.8.1753. [DOI] [PubMed] [Google Scholar]
- 31.Murtaugh LC, Chyung JH, Lassar AB. Sonic hedgehog promotes somitic chondrogenesis by altering the cellular response to BMP signaling. Genes Dev. 1999;13(2):225–237. doi: 10.1101/gad.13.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogden SK, et al. G protein Galphai functions immediately downstream of Smoothened in Hedgehog signalling. Nature. 2008;456(7224):967–970. doi: 10.1038/nature07459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Philipp M, Caron MG. Hedgehog signaling: Is Smo a G protein-coupled receptor? Curr Biol. 2009;19(3):R125–R127. doi: 10.1016/j.cub.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 34.Barzi M, Kostrz D, Menendez A, Pons S. Sonic Hedgehog-induced proliferation requires specific Gα inhibitory proteins. J Biol Chem. 2011;286(10):8067–8074. doi: 10.1074/jbc.M110.178772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long F, Zhang XM, Karp S, Yang Y, McMahon AP. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 2001;128(24):5099–5108. doi: 10.1242/dev.128.24.5099. [DOI] [PubMed] [Google Scholar]
- 36.Mak KK, Chen M-H, Day TF, Chuang P-T, Yang Y. Wnt/β-catenin signaling interacts differentially with Ihh signaling in controlling endochondral bone and synovial joint formation. Development. 2006;133(18):3695–3707. doi: 10.1242/dev.02546. [DOI] [PubMed] [Google Scholar]
- 37.Tavella S, et al. Targeted expression of SHH affects chondrocyte differentiation, growth plate organization, and Sox9 expression. J Bone Miner Res. 2004;19(10):1678–1688. doi: 10.1359/JBMR.040706. [DOI] [PubMed] [Google Scholar]
- 38.Schulte G, Bryja V. The Frizzled family of unconventional G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28(10):518–525. doi: 10.1016/j.tips.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 39.Reed SC. The inheritance and expression of Fused, a new mutation in the house mouse. Genetics. 1937;22(1):1–13. doi: 10.1093/genetics/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pickett EA, Olsen GS, Tallquist MD. Disruption of PDGFRalpha-initiated PI3K activation and migration of somite derivatives leads to spina bifida. Development. 2008;135(3):589–598. doi: 10.1242/dev.013763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghosh P, Garcia-Marcos M, Bornheimer SJ, Farquhar MG. Activation of Galphai3 triggers cell migration via regulation of GIV. J Cell Biol. 2008;182(2):381–393. doi: 10.1083/jcb.200712066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kreuzer J, et al. Platelet-derived growth factor activates production of reactive oxygen species by NAD(P)H oxidase in smooth muscle cells through Gi1,2. FASEB J. 2003;17(1):38–40. doi: 10.1096/fj.01-1036fje. [DOI] [PubMed] [Google Scholar]
- 43.Marty C, Ye RD. Heterotrimeric G protein signaling outside the realm of seven transmembrane domain receptors. Mol Pharmacol. 2010;78(1):12–18. doi: 10.1124/mol.110.063453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pyne NJ, et al. Experimental systems for studying the role of G-protein-coupled receptors in receptor tyrosine kinase signal transduction. Methods Enzymol. 2004;390:451–475. doi: 10.1016/S0076-6879(04)90028-6. [DOI] [PubMed] [Google Scholar]
- 45.Rudolph U, Bradley A, Birnbaumer L. Targeted inactivation of the Gi2 α gene with replacement and insertion vectors: Analysis in a 96-well plate format. Methods Enzymol. 1994;237:366–386. doi: 10.1016/s0076-6879(94)37076-1. [DOI] [PubMed] [Google Scholar]
- 46.Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev. 2002;119(Suppl 1):S97–S101. doi: 10.1016/s0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 47.Peters PWJ. Double staining of fetal skeletons for cartilage and bone. In: Neubert D, Merker H-J, Kwasigroch TE, editors. Methods in Prenatal Toxicology. Stuttgart, Germany: Georg Thieme; 1977. pp. 153–154. [Google Scholar]
- 48.Jegalian BG, De Robertis EM. Homeotic transformations in the mouse induced by overexpression of a human Hox3.3 transgene. Cell. 1992;71(6):901–910. doi: 10.1016/0092-8674(92)90387-r. [DOI] [PubMed] [Google Scholar]
- 49.Tamamura Y, et al. Developmental regulation of Wnt/β-catenin signals is required for growth plate assembly, cartilage integrity, and endochondral ossification. J Biol Chem. 2005;280(19):19185–19195. doi: 10.1074/jbc.M414275200. [DOI] [PubMed] [Google Scholar]
- 50.Wilkinson DG. In Situ Hybridization: A Practical Approach. Oxford: IRL; 1992. [Google Scholar]