Abstract
Choline and trimethylamine (TMA) are small molecules that play central roles in biological processes throughout all kingdoms of life. These ubiquitous metabolites are linked through a single biochemical transformation, the conversion of choline to TMA by anaerobic microorganisms. This metabolic activity, which contributes to methanogenesis and human disease, has been known for over a century but has eluded genetic and biochemical characterization. We have identified a gene cluster responsible for anaerobic choline degradation within the genome of a sulfate-reducing bacterium and verified its function using both a genetic knockout strategy and heterologous expression in Escherichia coli. Bioinformatics and electron paramagnetic resonance (EPR) spectroscopy revealed the involvement of a C–N bond cleaving glycyl radical enzyme in TMA production, which is unprecedented chemistry for this enzyme family. Our discovery provides the predictive capabilities needed to identify choline utilization clusters in numerous bacterial genomes, underscoring the importance and prevalence of this metabolic activity within the human microbiota and the environment.
Keywords: fragmentation, choline trimethylamine-lyase, gastrointestinal tract, metabolism, trimethylaminuria
Choline and trimethylamine (TMA) are important nitrogen-containing metabolites that perform fundamental roles in biological pathways throughout nature. Choline is an essential nutrient for higher organisms, including humans, contributing to cell membrane function, methyl transfer events, and neurotransmission (1). The volatile odorant TMA is used as a carbon source by bacteria, is a precursor to the marine osmolyte trimethylamine-N-oxide (TMAO), and is converted to the powerful greenhouse gas methane by methanogenic archaea (2–4). The sole biochemical reaction directly connecting these two small molecules is the metabolism of choline to TMA by anaerobic microorganisms (Fig. 1A), a process that has not been genetically or biochemically characterized. In this report, we describe the discovery of a gene cluster involved in anaerobic choline utilization and the identification of the enzymes responsible for TMA formation. Unexpectedly, we have found that a glycyl radical enzyme promotes this C–N bond cleavage, which is completely unprecedented reactivity for this enzyme family.
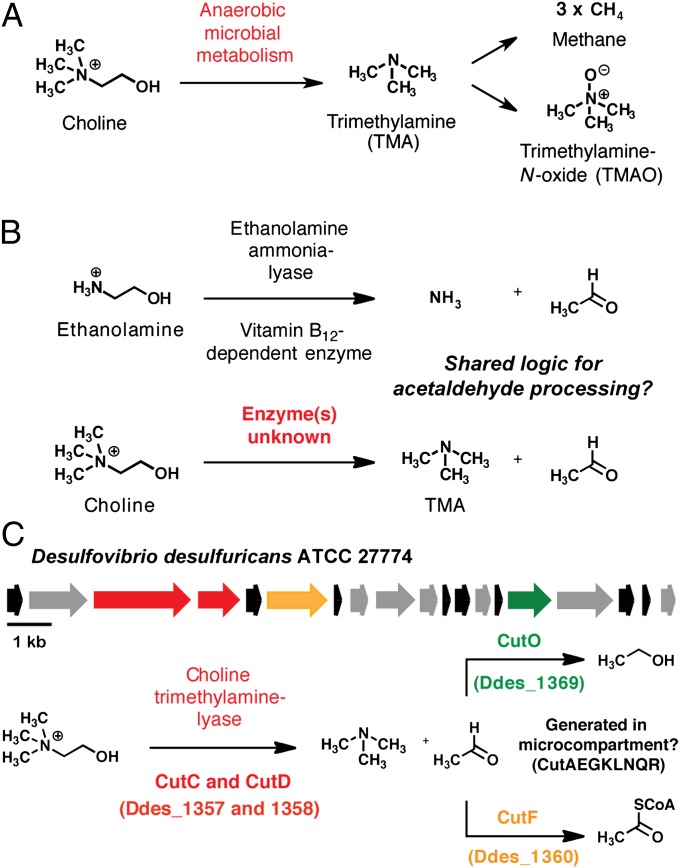
Fig. 1.
Discovery of a bacterial gene cluster for anaerobic choline utilization. (A) Generation of trimethylamine (TMA) from choline and its subsequent processing by other organisms. (B) Potential parallel logic between anaerobic choline utilization and bacterial ethanolamine utilization pathways. (C) Putative choline utilization (cut) gene cluster and proposed biochemical pathway for microbial choline metabolism.
Bacterial conversion of choline to TMA was first reported in 1910 (5). Experiments with whole cells and cell-free extracts of choline-fermenting anaerobes later revealed that this pathway involves an initial C–N bond cleavage of choline to produce TMA and acetaldehyde (6–8). This chemical transformation plays a significant role in many biological systems and impacts both the environment and human health. It has long been recognized that symbiotic gut microbes in humans (9–11) and other vertebrates (12–14) generate TMA from choline and that this metabolic activity is exclusively microbial. In humans, TMA is further processed to TMAO by the liver enzyme flavin-dependent monooxygenase 3 (FMO3) (15). This microbe–host metabolic interaction has been linked to multiple diseases, including the inherited metabolic disorder trimethylaminuria (fish-malodor syndrome) (16), nonalcoholic fatty liver disease (17), and most recently atherosclerosis and cardiovascular disease (18). Choline-derived TMA is also an important substrate for methanogenesis in both the gastrointestinal tracts of ruminants (12) and marine sediments (19, 20). Despite long-standing interest in this microbial metabolic pathway and its broad relevance to humans, nothing is currently known about its underlying genetics or biochemical mechanism, as the enzymes responsible for anaerobic choline utilization have not been identified.
Results
Discovery of a Choline Utilization Gene Cluster in Desulfovibrio desulfuricans and Identification of a Candidate Choline TMA-Lyase.
Our search for candidate genes involved in microbial choline degradation was guided by the hypothesis that some of the enzymatic transformations in this pathway might resemble those used in catabolism of the structurally related metabolite ethanolamine (Fig. 1B). In bacteria, the enzymes required for ethanolamine utilization are encoded by the eut gene cluster (21). An initial C–N bond cleavage is performed by ethanolamine ammonia-lyase (EutBC), a vitamin B12-dependent enzyme that generates ammonia and acetaldehyde (22). The acetaldehyde produced in this initial step is further processed by alcohol dehydrogenase EutG and aldehyde oxidoreductase EutE to give ethanol and acetyl coenzyme A (acetyl CoA) (21). In addition to genes that code for catabolic enzymes, eut clusters also typically contain genes encoding bacterial microcompartment proteins, which may facilitate sequestration of the volatile acetaldehyde intermediate (23).
Recognizing that acetaldehyde is formed in the initial step of both ethanolamine and choline degradation pathways (8), we hypothesized that its conversion to downstream products might be similar. We used position-specific iterative (PSI)-BLAST to search for genes encoding homologs of EutG, EutE, and microcompartment protein EutM from Salmonella enterica in the genome of Desulfovibrio desulfuricans American Type Culture Collection (ATCC) 27774. This sulfate-reducing bacterium had been previously reported to metabolize choline to TMA (24), a finding that we independently reconfirmed (Fig. S1). Our search revealed homologs of all three eut genes clustered within a 16.6-kb genomic region that displays similar organization to the eut cluster (Fig. 1C, Table S1). We named this portion of the D. desulfuricans genome the choline utilization (cut) gene cluster. Remarkably, homologs of eutB and eutC, which encode ethanolamine ammonia-lyase, were absent from both this 19-gene cluster and the rest of the D. desulfuricans genome. Instead, the cut cluster contained a distinct set of genes predicted to encode a glycyl radical enzyme CutC (Ddes_1357) and a glycyl radical activating protein CutD (Ddes_1358).
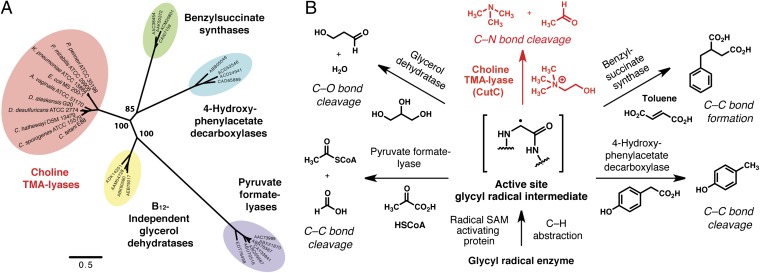
Bioinformatic analyses provided strong support for annotation of CutC as a member of the glycyl radical enzyme family. Both alignment of the CutC amino acid sequence with those of functionally characterized glycyl radical enzymes (Fig. S2A) and construction of a homology model (Fig. S2B) revealed conservation of key active site glycine and cysteine residues involved in radical catalysis (25). We hypothesized that CutC might catalyze the initial TMA-forming step in choline metabolism, although C–N bond cleavage by glycyl radical enzymes has not previously been reported. Maximum likelihood phylogenetic analysis (26) of CutC, its close homologs, and biochemically characterized glycyl radical enzymes revealed that these sequences cluster according to function and that CutC and its relatives form a separate clade (Fig. 2A). This finding supported our hypothesis that these proteins possess a biochemical function distinct from other glycyl radical enzymes, that of a choline TMA-lyase.
Fig. 2.
Bioinformatics identify choline trimethylamine-lyase CutC as a distinct type of glycyl radical enzyme. (A) Maximum likelihood phylogenetic tree showing the relationship between the amino acid sequences of choline TMA-lyases and other biochemically characterized members of the glycyl radical enzyme family. Bootstrap confidence values >50 are indicated on the nodes. (B) Biochemical transformations catalyzed by glycyl radical enzymes.
Disruption of Choline TMA-Lyase CutC Impairs Growth on Choline and Abolishes Production of TMA.
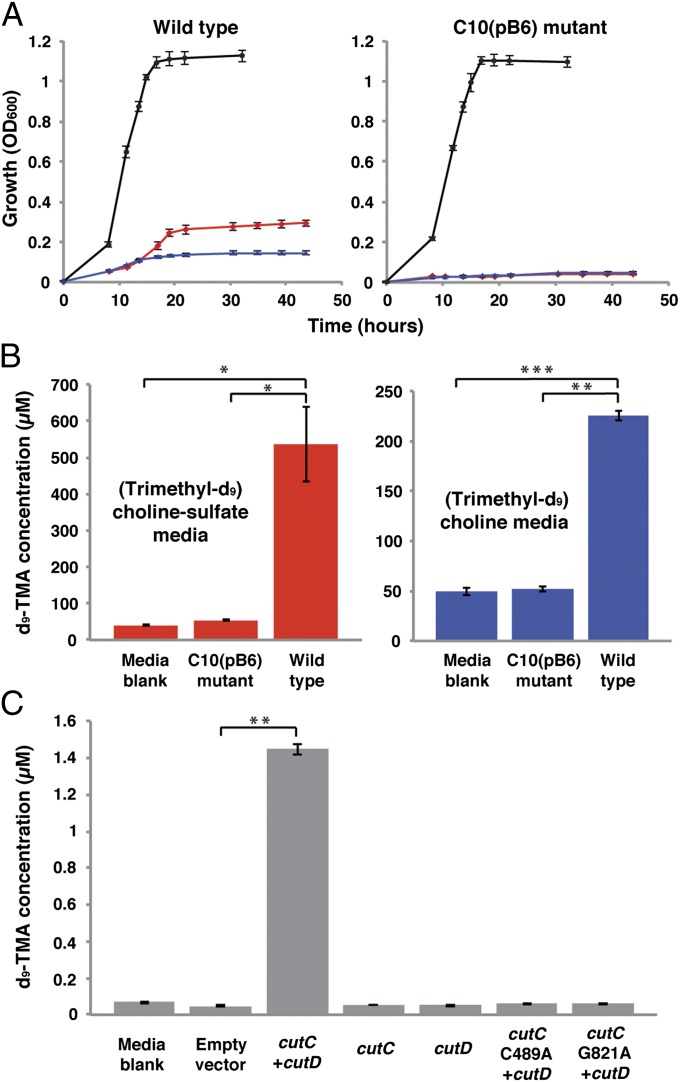
We devised two complementary approaches to evaluate whether the cut gene cluster was involved in choline utilization: genetic knockout in a choline-degrading organism and heterologous expression of putative choline TMA-lyase CutC and activating protein CutD in a noncholine-using host. Among sequenced bacteria containing homologs of the putative choline degradation gene cluster was Desulfovibrio alaskensis G20, a sulfate-reducing bacterium capable of anaerobic choline utilization (27). A G20 mutant strain [C10(pB6)] with a disruption in the gene encoding predicted choline TMA-lyase CutC (Dde_3282) had been generated previously using transposon mutagenesis, but a mutant phenotype had not been clearly identified (28). We obtained both wild-type D. alaskensis G20 and strain C10(pB6) and evaluated their ability to grow using lactate and choline as carbon sources (Fig. 3A). Consistent with earlier studies (28), we observed no difference in growth between wild type and C10(pB6) using media containing lactate. In contrast, the two strains exhibited a marked discrepancy in their ability to grow on choline. Whereas the wild-type strain could both ferment choline and use it for growth with sulfate as a terminal electron acceptor, strain C10(pB6) was unable to grow in either choline-containing media. These results clearly connect CutC to anaerobic choline metabolism.
Fig. 3.
The cut gene cluster is responsible for choline metabolism and TMA production. (A) Growth of D. alaskensis G20 wild-type and C10(pB6) strains at 37 °C on lactate sulfate (black), choline sulfate (red), and choline fermentation (blue) media. The data shown are the average OD600 values of four cultures. Error bars represent SEM. (B) LC-MS quantification of d9-TMA produced by D. alaskensis G20 wild type and C10(pB6) during incubations in (trimethyl-d9)-choline sulfate and (trimethyl-d9)-choline fermentation media containing (trimethyl-d9)-choline chloride (60 mM). Bar graphs represent the mean ± SEM of four cultures. *P < 0.02; **P < 10−5; ***P < 10−6. (C) LC-MS quantification of d9-TMA produced by E. coli BL21(DE3) during incubations in Luria–Bertani (LB) medium containing (trimethyl-d9)-choline chloride (100 µM). Bar graphs represent the mean ± SEM of four cultures. **P < 10−5.
We performed further experiments to validate the involvement of the cut gene cluster in TMA production. To verify that choline utilization in D. alaskensis proceeds via formation of TMA, we used liquid chromatography-mass spectrometry (LC-MS) to measure the d9-TMA present after inoculation and incubation of wild-type and C10(pB6) strains in media containing (trimethyl-d9)-choline. Use of isotopically labeled substrate ensured quantification of only TMA derived directly from choline. Growth of the wild-type organism was accompanied by the formation of d9-TMA, whereas levels for the C10(pB6) mutant did not exceed that of an uninoculated media control (Fig. 3B). Detection of TMA during growth on choline and the lack of TMA production from the mutant confirm that the identified gene cluster encodes a pathway that processes choline via C–N bond cleavage.
Heterologous Expression of Choline TMA-Lyase.
In parallel with these analyses we evaluated the in vivo activity of the putative TMA-forming enzymes CutC and CutD using a heterologous expression strategy. A 3.4-kb fragment of the D. alaskensis G20 gene cluster encoding both choline TMA-lyase CutC (Dde_3282) and its activating protein CutD (Dde_3281) was cloned into Escherichia coli BL21(D3). Using LC-MS, we measured formation of d9-TMA from (trimethyl-d9)-choline in vivo by strains harboring either the choline-degrading enzymes or an empty expression vector. Only E. coli coexpressing CutC and CutD produced d9-TMA at levels above an uninoculated media control (Fig. 3C), verifying that these enzymes are responsible for the initial C–N bond cleavage in anaerobic choline degradation. Expression of either enzyme on its own completely abolished TMA production, which is consistent with their annotations as a glycyl radical enzyme and activating protein; generation of the key active site glycyl radical involved in catalysis by glycyl radical enzymes requires the action of a separate radical S-adenosylmethionine (SAM) enzyme (Fig. 2B) (29). Site-directed mutagenesis experiments with CutC provided further support for a radical-based mechanism of C–N cleavage, as mutation of either Gly821 or Cys489 to Ala prevented TMA formation. These universally conserved active site residues are the locations of the two protein-bound radicals involved in catalysis and are essential for glycyl radical enzyme function (25).
Growth of D. desulfuricans on Choline Involves a Glycyl Radical.
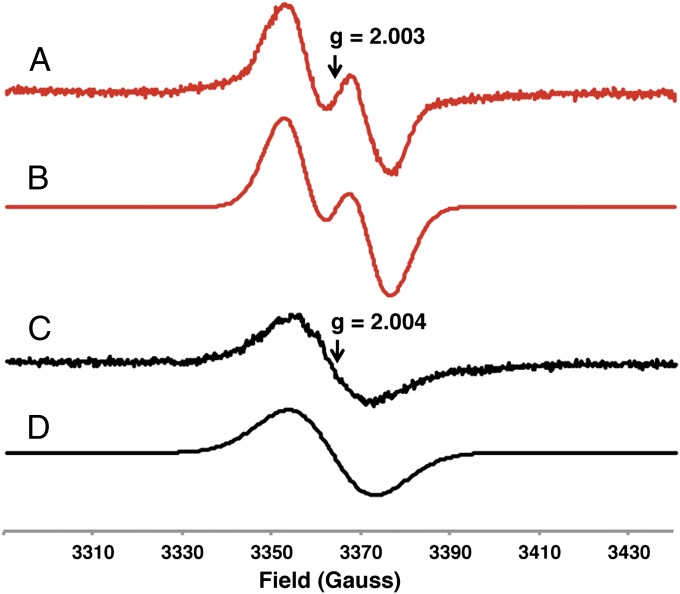
We obtained additional evidence connecting a glycyl radical enzyme to choline metabolism using electron paramagnetic resonance (EPR) spectroscopy. We prepared cell suspensions of D. desulfuricans ATCC 27774 grown on either choline or pyruvate fermentation media and recorded their EPR spectra (Fig. 4). The characteristic signal of a glycine-centered radical was observed only for cells grown on choline. The g value (2.003) and strong hyperfine coupling to a single proton (1.3 mT) indicate the presence of an organic radical located at the α-carbon of an amino acid and are consistent with previously reported EPR spectra of cells expressing glycyl radical enzymes (30, 31). The weaker nonhyperfine isotropic signal observed for cells from pyruvate cultures likely originates from flavoproteins, which are produced by a related Desulfovibrio strain during growth on pyruvate (32). Overall, these EPR experiments link choline utilization to a glycine-centered radical and indicate that choline TMA-lyase CutC is a member of the glycyl radical enzyme family.
Fig. 4.
Growth of D. desulfuricans on choline involves a glycyl radical. EPR spectra of D. desulfuricans ATCC 27774 cell suspensions grown on choline (red) and pyruvate (black) fermentation media. (A) Experimental spectrum of cells grown on choline. (B) Simulation assuming an isotropic signal with g = 2.003, line width (W) = 1.14 mT, and isotropic hyperfine interaction with one proton (A) = 1.3 mT. (C) Experimental spectrum of cells grown on pyruvate. (D) Simulation assuming an isotropic signal with g = 2.004 and line width (W) = 1.94 mT.
Discussion
The search for genes involved in bacterial choline utilization has led to the discovery of an additional member of the glycyl radical enzyme family, choline TMA-lyase. At the outset of our studies, it was not obvious what type of enzyme would be involved in producing TMA from choline. From the standpoint of biochemical logic, the initial C–N bond cleavage event in choline utilization is most similar to catabolic pathways involving vitamin B12-dependent radical rearrangements (ethanolamine and propanediol utilization) (33). However, no gene cluster encoding a vitamin B12-dependent enzyme has been linked to choline degradation and no enzyme from this family has ever been shown to catalyze this transformation. A critical distinction between choline and metabolites processed by B12-dependent pathways, such as ethanolamine, is the presence of a trimethylammonium substituent. Choline lacks N–H bonds and therefore cannot make all of the hydrogen bonding contacts within an enzyme-active site that are considered important for catalysis of B12-dependent eliminations (22, 33). Although this crucial structural difference called into question whether analogies in pathway logic between choline and ethanolamine utilization would extend to the C–N bond cleaving event, it was evident that powerful enzymatic chemistry would be needed to perform this transformation. The annotation of CutC as a glycyl radical enzyme provided a potential solution to supplying the requisite catalytic activity for C–N bond scission.
Glycyl radical enzymes are widespread in anaerobic microbes and include the well-characterized enzymes pyruvate formate-lyase and class III ribonucleotide reductase (34). Members of this enzyme family use highly reactive protein-based radical intermediates to promote a diverse set of chemical transformations, including C–C bond formation (benzylsuccinate synthase), C–C bond cleavage (pyruvate formate-lyase, 4-hydroxyphenylacetate decarboxylase), and dehydration (glycerol dehydratase) (Fig. 2B). However, genome sequencing and bioinformatics have revealed numerous uncharacterized glycyl radical enzymes within microbial genomes, suggesting that the functional diversity of these enzymes is underappreciated (25). Our discovery that choline metabolism involves a glycyl radical enzyme confirms this prediction, as C–N bond cleavage represents an entirely unique activity for this enzyme family.
In addition to describing a glycyl radical enzyme of unique function, our findings also reveal further parallel logic between the chemistry of glycyl radical enzymes and vitamin B12-dependent enzymes. Shared reactivity between these two enzyme families was previously limited to diol dehydration and ribonucleotide reduction. C–N bond cleavage is therefore an additional example of convergent evolution of function for these enzyme classes, which use different types of radical intermediates for catalysis (protein vs. cofactor based). Notably, recent computational studies suggest that B12-dependent and -independent glycerol dehydratases catalyze eliminations using different mechanisms, a finding that could also apply to ethanolamine ammonia-lyase and choline TMA-lyase (35). Further biochemical characterization and mechanistic studies of choline TMA-lyase will enable a better understanding of both pathway logic and the chemical mechanism involved in this intriguing enzymatic transformation.
Our discovery of a choline utilization gene cluster has implications beyond elucidating the biochemical basis for TMA formation; it also greatly enhances our understanding of the phylogenetic and environmental distribution of this metabolic activity by ascribing function to previously uncharacterized gene sequences. Searches of the nonredundant protein database in the National Center for Biotechnology Information (NCBI) for CutC homologs revealed cut gene clusters in a total of 89 bacterial genomes (Table S2), including organisms closely related to known choline degraders (Desulfovibrio, Clostridia, Streptococcus, Klebsiella, and Proteus) as well as members of many genera not previously reported to use choline. Multiple sequence alignments of the predicted choline TMA-lyases from these clusters showed conservation of predicted active site residues, supporting the hypothesis of shared function (Fig. S2C). We identified choline degradation pathways in a range of obligate and facultative anaerobes, including human commensals, human pathogens, and environmental isolates (Fig. S3A). The environmental strains are largely from marine sediments, habitats that are rich in TMA-based methanogenesis, confirming that choline metabolism contributes to TMA production in these ecosystems. The majority of the human commensal strains with cut clusters are gastrointestinal tract isolates (20 of 34). This finding strengthens earlier hypotheses that anaerobic choline degradation is a major source of TMA formation within this environment (9) and provides initial insights regarding which symbiotic microbes may harbor this metabolic activity. Notably, choline utilization does not appear to be evenly distributed between common phyla found in the human gut (Fig. S3B). Whereas cut gene clusters are found in Firmicutes, Proteobacteria, and Actinobacteria, they are completely absent from Bacteroidetes, a major component of the gut microbiota. In addition to providing insights from genomes of cultured microbes, the ability to identify the choline utilization pathway in sequencing data will also permit characterization of its role in natural microbial communities through use of culture-independent methods such as metagenomics and metatranscriptomics (36).
Large-scale microbial genome sequencing has afforded an unprecedented opportunity to uncover the genetic basis for important activities associated with complex microbial communities. Our discovery of the genes involved in anaerobic choline utilization illustrates the power of combining biochemical knowledge with bioinformatics to correlate sequence data with function. These findings have immediately increased our understanding of this metabolic pathway’s distribution in biology and expanded the scope of known radical-mediated enzymatic chemistry. Ultimately, our work will enable a better understanding of anaerobic choline metabolism, both in the environment, where it impacts the global carbon cycle and greenhouse gas production, and within the human body, where it contributes to disease-associated interspecies metabolism.
Materials and Methods
General Procedures.
All general procedures for microbial cultivation and LC-MS analyses are detailed in SI Materials and Methods.
Identification of the cut (Choline Utilization) Gene Cluster in D. desulfuricans ATCC 27774.
A putative choline utilization gene cluster was identified within the sequenced genome of D. desulfuricans ATCC 27774 using the BLAST search tool at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Specifically, the following protein sequences from the ethanolamine utilization operon of Salmonella enterica subsp. Enterica serovar Typhimurium were used as input for individual PSI-BLAST searches of the D. desulfuricans ATCC 2774 genome: alcohol dehydrogenase EutG (AAA80211), aldehyde oxidoreductase EutE (AAA80209), and microcompartment protein cchA/EutM (AAA80207). The genes encoding the closest homologs of each of these three protein sequences were colocalized within the D. desulfuricans genome. Annotation of the cut gene cluster is found in Table S1.
Identification of cut Gene Clusters in Other Sequenced Organisms.
Additional choline utilization gene clusters were located using BLASTp and tBLASTn searches to identify homologs of choline TMA-lyase CutC (Ddes1357) (ACL49259) within the nonredundant protein and nucleotide databases in NCBI. The genomic contexts of all potential hits were inspected individually, and homologs situated within putative choline utilization clusters were tabulated (Table S2). A cluster was considered a potential cut cluster if, in addition to the choline TMA-lyase, it contained genes predicted to encode a glycyl radical activating enzyme, an aldehyde dehydrogenase, an alcohol dehydrogenase, and bacterial microcompartment proteins. The ordering of genes within clusters showed some variation between different strains. Organisms containing cut gene clusters were categorized according to the source of the isolate as identified in the Genomes OnLine Database (GOLD) (http://www.genomesonline.org/cgi-bin/GOLD/index.cgi) or the Human Microbiome Project (HMP) Data Analysis and Coordination Center (http://www.hmpdacc.org/) (Fig. S3A). Human-associated commensals isolated from the gastrointestinal tract were further classified by phylum (Fig. S3B). The cut gene cluster in D. alaskensis G20 was identified as part of the search described in the previous section. The arrangement of the genes in this cluster is identical to that of the D. desulfuricans ATCC 27774 cluster. The protein sequences are also highly similar, with amino acid identities ranging from 41% to 95%. A detailed annotation of this cluster is found in Table S3.
Bioinformatic Analyses of Choline TMA-Lyase CutC.
The amino acid sequences of CutC from D. desulfuricans ATCC 27774 (Ddes_1357) and D. alaskensis G20 (Dde_3282) were aligned with the amino acid sequences of biochemically characterized glycyl radical enzymes using ClustalW (37). The resulting alignment indicated that the essential glycine and cysteine residues involved in radical catalysis are conserved in both CutC sequences (Fig. S2A) (25). A CutC (Ddes_1357) homology model was generated with the HHPred interactive server (http://toolkit.tuebingen.mpg.de/hhpred) (38) using the crystal structure of B12-independent glycerol dehydratase (39) as a template [Protein DataBase (PDB) ID 1R9D]. Overlay of the CutC homology model with the glycerol dehydratase crystal structure revealed conservation of active site architecture and the essential catalytic glycine and cysteine residues (Fig. S2B). Multiple sequence alignment of Ddes_1357 with CutC homologs from other organisms containing cut gene clusters showed conservation of active site residues predicted from the CutC homology model (Fig. S2C). The maximum likelihood phylogenetic tree of representative glycyl radical enzymes was reconstructed using PHYML (26) with the general time reversible (GTR) nucleotide substitution model, the proportion of invariable sites and the γ parameter of across-site rate variation (using four rate categories) was estimated from the dataset. Bootstrap support values were calculated with the same parameters (100 replicates). The sequence dataset used for the analysis was obtained starting from choline TMA-lyase Ddes_1357, other functionally characterized representatives of the glycyl radical enzyme family, and close BLAST hits from the GenBank nucleotide database (Table S4). These sequences were aligned using ClustalW and trimmed manually using MacClade.
Growth of D. alaskensis G20 Wild-Type and C10(pB6) Strains on Choline and Quantitation of TMA Production from Choline.
Desulfovibrio alaskensis G20 wild-type and C10(pB6) mutant strains were obtained from Lee Krumholz (University of Oklahoma, Norman, OK). The G20 strain (40) is a spontaneously nalidixic acid-resistant derivative of strain G100A, a known choline degrader (27). The C10(pB6) mutant strain was generated previously using signature-tagged mutagenesis; this mutant contains a transposon insertion that disrupts the gene encoding predicted choline TMA-lyase CutC (Dde_3282) and was initially identified in a screen for D. alaskensis genes conferring sediment fitness (28). Both strains were grown anaerobically at 37 °C on lactate-sulfate (LS), choline-sulfate (CS), and choline fermentation (C) media as described in SI Materials and Methods. Kanamycin (1,050 μg/mL) was added to all cultures of the C10(pB6) mutant. For each media type, four 18 × 150 mm Hungate tubes containing 10 mL of medium were inoculated with 0.05 mL (LS media) or 0.1 mL (CS and C media) of an overnight LS starter culture (OD600 = 1.0–1.1) and incubated at 37 °C until reaching stationary phase (∼48 h).
For quantitation of d9-TMA formation, the strains were grown on CS and C media containing (trimethyl-d9)-choline (60 mM). This substrate was added as an anaerobic, filter-sterilized aqueous solution shortly before inoculation. Kanamycin (1,050 μg/mL) was added to all cultures of the C10(pB6) mutant. Four 10-mL cultures of each media type (LS, CS, and C) were grown side by side at 37 °C until reaching stationary phase (∼48 h). The concentration of d9-TMA in culture media and blanks was determined using LC-MS as described in SI Materials and Methods, except that the derivatization reaction contained 100 μL of 22 μM aqueous TMA solution for the C10(pB6) mutant samples and 100 μL of 2.2 mM aqueous TMA solution for the wild-type samples. The quenched derivatization reactions for the C10(pB6) mutant were analyzed without dilution, whereas for the wild-type samples, a 10-μL aliquot was diluted 100-fold before analysis. The transitions used for quantitation were m/z 155.1→ m/z 127.1 for d9-TMA and m/z 146.1→ m/z 118.1 for TMA. Media blanks consisted of 10 mL of uninoculated CS and C media containing (trimethyl-d9)-choline (60 mM). Four tubes per type of media were incubated at 37 °C alongside the G20 wild-type and C10(pB6) mutant cultures until they reached stationary phase.
Cloning of CutC (Dde_3282) and CutD (Dde_3281) from D. alaskensis G20 and construction of CutC C489A and G821A Mutants.
All cloning and molecular biology procedures including primers used (Table S5) are detailed in SI Materials and Methods.
Heterologous Expression of CutC and CutD and Quantitation of TMA Production from Choline.
Individual cultures of LB medium (5 mL) containing 50 µg/mL kanamycin were inoculated with the pET-29b-CutC/CutD, pET-29b-CutD, pET-28a-CutC, pET-29b-CutC/CutD-C489A, and pET-29b-CutC/CutD-G821A expression strains, as well as a control strain of E. coli BL21(DE3) containing an empty pET-29b(+) vector, and incubated at 37 °C overnight. Aliquots (500 μL) of each overnight culture (OD600 = 1.6–1.8), were used to inoculate four 18 × 150 mm modified Hungate tubes containing 10 mL of LB medium (sparged with argon for 25 min) supplemented with NaCl (0.4 M), kanamycin (50 μg/mL), (trimethyl-d9)-choline chloride (105 µM), and iron (III) ammonium citrate (0.069 mg/mL). High salt conditions were used to ensure expression of high-affinity choline transporter, betT, which is required for transport of choline into E. coli (41). The cultures were incubated at 37 °C with shaking (190 rpm), and protein expression was induced with 500 μM Isopropyl β-D-1-thiogalactopyranoside (IPTG) at an OD600 of ∼0.3. Incubation continued for an additional 16 h at 37 °C. Four media blanks consisting of 18 × 150 mm modified Hungate tubes containing the growth media described above were incubated at 37 °C alongside the E. coli cultures.
The concentrations of d9-TMA in the media blanks and growth media of E. coli cultures were determined using LC-MS as described in SI Materials and Methods. The transition used for quantitation was m/z 155.1→ 66.2. The derivatization reaction mixture contained the same components, except that the standard solution of TMA was not included. Quantitation was performed using a standard curve. Calibration standards were prepared by adding different concentrations of d9-TMA hydrochloride solution to uninoculated LB media supplemented with 0.4 M NaCl (0.4 M), kanamycin (50 μg/mL), choline chloride (100 µM), and iron (III) ammonium citrate (0.066 mg/mL), such that the concentration of the derivatized d9-TMA in the solution obtained after quenching would be in the 5 to 50 nM range. To ensure that the C498A and G821A CutC mutants were produced to the same extent as wild type, cell lysates from the heterologous expression experiments detailed above were analyzed using SDS-PAGE (Fig. S4).
Electron Paramagnetic Resonance Spectroscopy.
D. desulfuricans ATCC 27774 was grown in serum bottles (OD × H 51.7 mm × 94.5 mm, Supelco; 33110-U) containing 100 mL of choline fermentation (C) and pyruvate fermentation (P) media using a 1-mL inoculum from an overnight LS culture (OD = 0.84–0.85). A total culture volume of 500 mL was used to prepare each cell suspension. The cultures were incubated at 37 °C in an anaerobic chamber (Coy Laboratory Products) under an atmosphere of 98% nitrogen and 2% hydrogen until reaching mid-to-late exponential phase. The cultures were combined in 2 × 250 mL polypropylene centrifuge tubes with plug seal caps (Corning; 430776) and centrifuged at 6,766 × g for 20 min at 4 °C. Cell pellets were resuspended in 0.1 M 3-(N-morpholino)propanesulfonic acid (MOPS) buffer (pH 7.2) containing 5 mM DTT to a final volume of 220–300 μL in the anaerobic chamber. The final volume of cell suspension was scaled according to the final average OD of the bacterial cultures. The cell suspensions were transferred to 4-mm thin-wall Suprasil screw-cap EPR sample tubes (7” length, Wilmad-LabGlass; 734-TR-7). The contents of the tubes were frozen in an acetone/dry-ice bath and placed in the cavity of the EPR instrument at 120 K. EPR spectra were recorded and modeled as described in SI Materials and Methods, General Procedures.
Supplementary Material
Acknowledgments
We thank J. N. Carita (Universidade Nova de Lisboa, Lisbon) for providing D. desulfuricans ATCC 27774 and L. R. Krumholz and P. D. Bradstock (University of Oklahoma, Norman, OK) for providing D. alaskensis G20 wild-type and C10(pB6) strains. We received assistance with LC-MS experiments from A. Saghatelian and T. Zhang, assistance with anaerobic culturing from P. Turnbaugh and H. Haiser, and advice about EPR experiments from T. Betley, G. Sazama, A. Fout, E. Nolan, and J. Hayden. Financial support was provided by Harvard University, the Smith Family Award for Excellence in Biomedical Research, the Milton Fund, and the Corning Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 21184.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215689109/-/DCSupplemental.
References
- 1.Zeisel SH, da Costa KA. Choline: An essential nutrient for public health. Nutr Rev. 2009;67(11):615–623. doi: 10.1111/j.1753-4887.2009.00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y, Patel NA, Crombie A, Scrivens JH, Murrell JC. Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc Natl Acad Sci USA. 2011;108(43):17791–17796. doi: 10.1073/pnas.1112928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seibel BA, Walsh PJ. Trimethylamine oxide accumulation in marine animals: Relationship to acylglycerol storage. J Exp Biol. 2002;205(Pt 3):297–306. doi: 10.1242/jeb.205.3.297. [DOI] [PubMed] [Google Scholar]
- 4.Hippe H, Caspari D, Fiebig K, Gottschalk G. Utilization of trimethylamine and other N-methyl compounds for growth and methane formation by Methanosarcina barkeri. Proc Natl Acad Sci USA. 1979;76(1):494–498. doi: 10.1073/pnas.76.1.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ackermann D, Schutze H. The formation of trimethylamine by Bacterium prodigiosum. Zentralb Physiol. 1910;24:210–211. [Google Scholar]
- 6.Hayward HR, Stadtman TC. Anaerobic degradation of choline. I. Fermentation of choline by an anaerobic, cytochrome-producing bacterium, Vibrio cholinicus n. sp. J Bacteriol. 1959;78:557–561. doi: 10.1128/jb.78.4.557-561.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayward HR, Stadtman TC. Anaerobic degradation of choline. II. Preparation and properties of cell-free extracts of Vibrio cholinicus. J Biol Chem. 1960;235:538–543. [PubMed] [Google Scholar]
- 8.Hayward HR, Stadtman TC. Anaerobic degradation of choline. III. Acetaldehyde as an intermediate in the fermentation of choline by extracts of Vibrio cholinicus. J Biol Chem. 1960;235:3292–3596. [PubMed] [Google Scholar]
- 9.Bain MA, Fornasini G, Evans AM. Trimethylamine: Metabolic, pharmacokinetic and safety aspects. Curr Drug Metab. 2005;6(3):227–240. doi: 10.2174/1389200054021807. [DOI] [PubMed] [Google Scholar]
- 10.de la Huerga J, Popper H. Urinary excretion of choline metabolites following choline administration in normals and patients with hepatobiliary diseases. J Clin Invest. 1951;30(5):463–470. doi: 10.1172/JCI102463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeisel SH, Wishnok JS, Blusztajn JK. Formation of methylamines from ingested choline and lecithin. J Pharmacol Exp Ther. 1983;225(2):320–324. [PubMed] [Google Scholar]
- 12.Neill AR, Grime DW, Dawson RMC. Conversion of choline methyl groups through trimethylamine into methane in the rumen. Biochem J. 1978;170(3):529–535. doi: 10.1042/bj1700529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.al-Waiz M, Mikov M, Mitchell SC, Smith RL. The exogenous origin of trimethylamine in the mouse. Metabolism. 1992;41(2):135–136. doi: 10.1016/0026-0495(92)90140-6. [DOI] [PubMed] [Google Scholar]
- 14.Niizeki N, et al. Mechanism of biosynthesis of trimethylamine oxide from choline in the teleost tilapia, Oreochromis niloticus, under freshwater conditions. Comp Biochem Physiol B Biochem Mol Biol. 2002;131(3):371–386. doi: 10.1016/s1096-4959(01)00508-5. [DOI] [PubMed] [Google Scholar]
- 15.Krueger SK, Williams DE. Mammalian flavin-containing monooxygenases: Structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther. 2005;106(3):357–387. doi: 10.1016/j.pharmthera.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mackay RJ, McEntyre CJ, Henderson C, Lever M, George PM. Trimethylaminuria: Causes and diagnosis of a socially distressing condition. Clin Biochem Rev. 2011;32(1):33–43. [PMC free article] [PubMed] [Google Scholar]
- 17.Dumas ME, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci USA. 2006;103(33):12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King GM. Metabolism of trimethylamine, choline, and glycine betaine by sulfate-reducing and methanogenic bacteria in marine sediments. Appl Environ Microbiol. 1984;48(4):719–725. doi: 10.1128/aem.48.4.719-725.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiebig K, Gottschalk G. Methanogenesis from choline by a coculture of Desulfovibrio sp. and Methanosarcina barkeri. Appl Environ Microbiol. 1983;45(1):161–168. doi: 10.1128/aem.45.1.161-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garsin DA. Ethanolamine utilization in bacterial pathogens: Roles and regulation. Nat Rev Microbiol. 2010;8(4):290–295. doi: 10.1038/nrmicro2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibata N, et al. Crystal structures of ethanolamine ammonia-lyase complexed with coenzyme B12 analogs and substrates. J Biol Chem. 2010;285(34):26484–26493. doi: 10.1074/jbc.M110.125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kofoid E, Rappleye C, Stojiljkovic I, Roth J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol. 1999;181(17):5317–5329. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao CK, Zeisel SH. Formation of trimethylamine from dietary choline by Streptococcus sanguis I, which colonizes the mouth. J Nutr Biochem. 1990;1(2):89–97. doi: 10.1016/0955-2863(90)90055-p. [DOI] [PubMed] [Google Scholar]
- 25.Lehtiö L, Goldman A. The pyruvate formate lyase family: Sequences, structures and activation. Protein Eng Des Sel. 2004;17(6):545–552. doi: 10.1093/protein/gzh059. [DOI] [PubMed] [Google Scholar]
- 26.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 27.Weimer PJ, Van Kavelaar MJ, Michel CB, Ng TK. Effect of phosphate on the corrosion of carbon steel and on the composition of corrosion products in two-stage continuous cultures of Desulfovibrio desulfuricans. Appl Environ Microbiol. 1988;54(2):386–396. doi: 10.1128/aem.54.2.386-396.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo Q, Groh JL, Ballard JD, Krumholz LR. Identification of genes that confer sediment fitness to Desulfovibrio desulfuricans G20. Appl Environ Microbiol. 2007;73(19):6305–6312. doi: 10.1128/AEM.00715-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vey JL, Drennan CL. Structural insights into radical generation by the radical SAM superfamily. Chem Rev. 2011;111(4):2487–2506. doi: 10.1021/cr9002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Unkrig V, Neugebauer FA, Knappe J. The free radical of pyruvate formate-lyase. Characterization by EPR spectroscopy and involvement in catalysis as studied with the substrate-analogue hypophosphite. Eur J Biochem. 1989;184(3):723–728. doi: 10.1111/j.1432-1033.1989.tb15072.x. [DOI] [PubMed] [Google Scholar]
- 31.Rabus R, et al. Anaerobic initial reaction of n-alkanes in a denitrifying bacterium: Evidence for (1-methylpentyl)succinate as initial product and for involvement of an organic radical in n-hexane metabolism. J Bacteriol. 2001;183(5):1707–1715. doi: 10.1128/JB.183.5.1707-1715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pereira PM, et al. Energy metabolism in Desulfovibrio vulgaris Hildenborough: Insights from transcriptome analysis. Antonie van Leeuwenhoek. 2008;93(4):347–362. doi: 10.1007/s10482-007-9212-0. [DOI] [PubMed] [Google Scholar]
- 33.Toraya T. Radical catalysis in coenzyme B12-dependent isomerization (eliminating) reactions. Chem Rev. 2003;103(6):2095–2127. doi: 10.1021/cr020428b. [DOI] [PubMed] [Google Scholar]
- 34.Selmer T, Pierik AJ, Heider J. New glycyl radical enzymes catalysing key metabolic steps in anaerobic bacteria. Biol Chem. 2005;386(10):981–988. doi: 10.1515/BC.2005.114. [DOI] [PubMed] [Google Scholar]
- 35.Feliks M, Ullmann GM. Glycerol dehydratation by the B12-independent enzyme may not involve the migration of a hydroxyl group: A computational study. J Phys Chem B. 2012;116(24):7076–7087. doi: 10.1021/jp301165b. [DOI] [PubMed] [Google Scholar]
- 36.Su C, Lei L, Duan Y, Zhang K-Q, Yang J. Culture-independent methods for studying environmental microorganisms: Methods, application, and perspective. Appl Microbiol Biotechnol. 2012;93(3):993–1003. doi: 10.1007/s00253-011-3800-7. [DOI] [PubMed] [Google Scholar]
- 37.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244-248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Brien JR, et al. Insight into the mechanism of the B12-independent glycerol dehydratase from Clostridium butyricum: Preliminary biochemical and structural characterization. Biochemistry. 2004;43(16):4635–4645. doi: 10.1021/bi035930k. [DOI] [PubMed] [Google Scholar]
- 40.Wall JD, Murnan T, Argyle J, English RS, Rapp-Giles BJ. Transposon mutagenesis in Desulfovibrio desulfuricans: Development of a random mutagenesis tool from Tn7. Appl Environ Microbiol. 1996;62(10):3762–3767. doi: 10.1128/aem.62.10.3762-3767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamark T, et al. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol Microbiol. 1991;5(5):1049–1064. doi: 10.1111/j.1365-2958.1991.tb01877.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.