The immune system acts as a shield against harmful microbes and requires exquisite control of the activation and resolution of multiple signaling pathways. Tight regulation of these processes is required not only to produce an effective response, but also to maintain immune homeostasis and prevent autoinflammatory disease. In the intestine, this process is further complicated by the residence of a beneficial community of microbes. One major question is how the immune system determines “friend from foe,” restricting antimicrobial responses to the intestinal microbiota but retaining protective antimicrobial responses against pathogens. Answering this question is critical to uncovering mechanisms that go awry in autoinflammatory disease. In PNAS, Lipinsky et al. (1) provide mechanistic insights into how polarized intestinal epithelial cells restrict antibacterial responses to the basolateral compartment through directing the subcellular localization the bacterial sensor NOD2 (nucleotide-binding oligomerization domain-2).
NOD2 is a member of the phylogenetically conserved NOD-like receptor family, which recognizes microbe-associated molecular patterns (i.e., bacterial peptidoglycan, flagellin, and so forth) and danger-associated molecular patterns (i.e., reactive oxygen species, ATP, and others.) in the cell cytosol (2). The NOD2 protein is primarily expressed in granulocytes, antigen-presenting cells, Paneth cells, and intestinal epithelial cells. NOD2 is activated by the muramyl dipeptide (MDP) component of the peptidoglycan cell wall present in most bacteria (3, 4). Like other NOD-like receptors, NOD2 forms a dynamic macromolecular platform with multiple proteins upon activation, which dictates cellular responses to bacteria. These responses include the production of cytokines, chemokines, and antimicrobial peptides, activation of autophagy, and stimulation of antigen presentation. The critical role of NOD2 in immune homeostasis is demonstrated by the association of both gain-of-function and loss-of-function NOD2 genetic variants with chronic inflammatory diseases, such as Crohn disease and Blau syndrome.
Because of its pivotal role in maintaining gut homeostasis, it is critical to understand the mechanisms involved in appropriate activation of NOD2-induced signaling. To this end, several global screening methodologies (i.e., yeast two-hybrid, biochemical purification, and RNAi) have been applied to identify an expanding pool of potential NOD2 modulators. Although these studies have identified some key players involved in NOD2 signaling, they generally fail to provide a good picture of how these regulators affect NOD2 function in a physiologic context. Commonly, the analyses are performed in easy to manipulate but not functionally relevant cell types using overexpressed proteins. The intracellular location and integration of regulators with other known NOD2 pathway components is also not usually examined. Although these more informative studies are technically challenging because of the low expression of NOD2 and the poor quality of NOD2-specific reagents (i.e., antibodies), these types of analyses are essential to provide significant, physiologic insights and are what make the study by Lipinski et al. (1) outstanding. The authors initially perform multiple, systematic layers of screening and confirmation to identify novel components of NOD2 signaling, followed by functional studies in relevant cell types using a combination of cell biology, molecular, and biochemical techniques. The authors then relate these findings to alterations in NOD2 signaling observed in the inflammatory bowel disease, Crohn disease.
In the report by Lipiniski et al. (1), the authors searched for proteins that modify NOD2 signaling using an RNAi library-screening approach. The authors performed an initial RNAi screen targeting a limited set of “druggable” genes in HEK293 cells expressing exogenous NOD2, and analyzed the effect of the RNAi knockdown on the activation of an NFκB reporter gene in response to the NOD2 ligand, MDP (screen 1). The initial pool of candidate genes was reconfirmed using the same system to rule out any nonreproducible results (screen 2). This process was followed by a third screen using a different RNAi library containing siRNAs designed by a different algorithm targeting the 69 reproducible candidates, resulting in 20 candidate-positive regulators of NFκB signaling (screen 3). Additionally, all of the data from these screens were subjected to network analysis to determine the relevance of the candidate genes to known NOD2 pathway components, which resulted in a stepwise increase in hits closer to the NOD2 network, as well as a decreased false-discovery rate.
The authors then went the extra mile to perform two other layers of analysis to identify specific candidate modulators of NOD2 signaling out of the final set of 20 regulators. First, a “counter screen” in HEK293 cells using TNF-α as the stimulus was performed to distinguish general NFκB regulators from specific NOD2 regulators (screen 4). Second, and perhaps more importantly, the 20 candidates were analyzed for effects on multiple NOD2-dependent functions in Caco-2 cells (screen 5). Caco-2 cells are a colonic epithelial cell line that can be polarized and express NOD2. Thus, through an extensive series of well-ordered screens, the authors identified six genes (four novel), which positively affect NOD2-dependent activation of NFκB, secretion of IL-8, and clearance of Listeria monocytogenes in a relevant cell type.
Membrane targeting of NOD2 is required for the recognition of MDP and subsequent activation of antibacterial responses in polarized intestinal epithelial cells (5–8). However, the precise mechanisms required for this localization are unclear. Previous studies demonstrated a requirement for cytoskeletal proteins in the membrane localization of NOD2 (8, 9) and suggest that a specific region in the carboxyl-terminal leucine-rich repeats of NOD2 is also involved (7). Other studies identified the basolateral protein ERBB2IP (ERBIN) as a negative regulator of NOD2 signaling, but interaction with ERBB2IP is not required to maintain basolateral localization of NOD2 (6, 10). It has been proposed that the basolateral localization of NOD2 is important not only to position NOD2 to sites of bacterial entry, but also to maintain intestinal epithelial cell tolerance to commensal bacteria present at the apical side of these cells.
One of the candidates identified by Lipinski et al. (1) is FRMPD2 (FERM and PDZ domain protein-containing 2), which is selectively localized at the basolateral membrane in polarized epithelial cells, where it is thought to be involved in maintenance of cell polarization and directing proteins to basolateral membrane.
FRMPD2 not only localizes NOD2 to basolateral membranes, but this interaction provides spatial specificity to NOD2 signaling.
This finding prompted the authors perform an elegant set of experiments to determine that FRMPD2 not only localizes NOD2 to basolateral membranes, but this interaction provides spatial specificity to NOD2 signaling (Fig. 1). In brief, a functional network analysis predicted a connection between FRMPD2, ERBB2IP, and NOD2. This connection was confirmed biochemically and a tripartite complex of these proteins was visualized in polarized epithelial cells. Additional biochemical analyses indicated that FRMPD2 interacts with the leucine-rich repeats of NOD2, suggesting that FRMPD2 may act as a membrane anchor for NOD2. When the authors silenced the expression of FRMPD2 in polarized epithelial cells, it resulted in the mislocalization of NOD2 to the cytosol and made these cells insensitive to MDP stimulation from the basal side. This study provides insight into the molecules required for functional NOD2 subcellular localization and the interplay of both positive and negative regulators in the NOD2 signalosome.
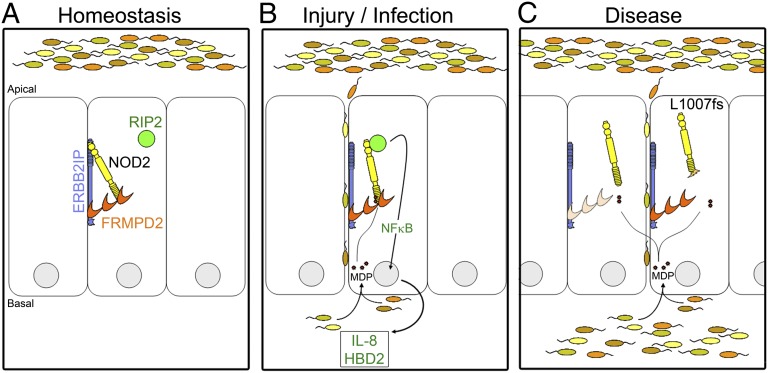
Fig. 1.
FRMPD2 localizes NOD2 to the basolateral membrane in polarized intestinal epithelial cells and dictates spatial specificity to NOD2 signaling. (A) Under normal conditions in the gut, NOD2 localizes to the basolateral membrane through interactions with FRMPD2 and is nonresponsive to commensal bacteria present on apical surfaces through an interaction with ERBB2IP. (B) When injury or pathogenic infection occurs, cells are exposed to bacteria (MDP) basolaterally. Membrane-associated NOD2 senses MDP and is released from ERBB2IP repression, which results in recruitment of RIP2 to the membrane, initiation of signaling through NFκB, and stimulation of a proinflammatory and antibacterial response. (C) In a disease context [i.e., FRMPD2 expression is decreased by inflammatory signals (Left) or a Crohn disease-associated NOD2 variant (L1007fs) is expressed], NOD2 is mislocalized and is unable to respond effectively and clear the bacterial infection.
Finally, the authors link their observations to NOD2 functional defects observed in Crohn disease. NOD2 was the first Crohn disease susceptibility gene identified and contributes the largest proportion of genetic risk to developing this disease (11–13). Three main NOD2 risk variants (R702W, G908R, and L1007fs) result in reduction or loss of NOD2 function. It is unclear how these genetic changes reduce NOD2 function, but previous studies demonstrate that the L1007fs variant fails to localize to the cell membrane (5, 7). Interestingly, this NOD2 variant does not interact with FRMPD2 and these findings suggest a potential mechanism for the functional defect of this risk variant. Interestingly, the authors also examine the expression levels of FRMPD2 in inflammatory conditions and found them reduced in the intestinal mucosa of Crohn disease patients and experimental colitis models (1). It will be interesting to see if these alterations in FRMPD2 expression result in NOD2 mislocalization in the context of disease.
The study by Lipinski et al. (1) reinforces the need to understand not only what proteins are involved in regulating antibacterial responses, but where these activities occur within relevant cells to shape appropriate tissue responses. Their findings also highlight the importance of examining not only the contribution of a single regulator to a pathway, but also the complex interplay between molecules within a signaling pathway. Understanding the complexity is what ultimately provides valuable information as to how cellular responses are altered in health and disease.
Footnotes
The authors declare no conflict of interest.
See companion article on page 21426.
References
- 1.Lipinski S, et al. RNAi screening identifies mediators of NOD2 signaling: Implications for spatial specificity of MDP recognition. Proc Natl Acad Sci USA. 2012;109:21426–21431. doi: 10.1073/pnas.1209673109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elinav E, Strowig T, Henao-Mejia J, Flavell RA. Regulation of the antimicrobial response by NLR proteins. Immunity. 2011;34(5):665–679. doi: 10.1016/j.immuni.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Girardin SE, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278(11):8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 4.Inohara N, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J Biol Chem. 2003;278(8):5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 5.Lécine P, et al. The NOD2-RICK complex signals from the plasma membrane. J Biol Chem. 2007;282(20):15197–15207. doi: 10.1074/jbc.M606242200. [DOI] [PubMed] [Google Scholar]
- 6.McDonald C, et al. A role for Erbin in the regulation of Nod2-dependent NF-kappaB signaling. J Biol Chem. 2005;280(48):40301–40309. doi: 10.1074/jbc.M508538200. [DOI] [PubMed] [Google Scholar]
- 7.Barnich N, Aguirre JE, Reinecker HC, Xavier R, Podolsky DK. Membrane recruitment of NOD2 in intestinal epithelial cells is essential for nuclear factor-kappaB activation in muramyl dipeptide recognition. J Cell Biol. 2005;170(1):21–26. doi: 10.1083/jcb.200502153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevens C, et al. The intermediate filament protein, vimentin, is a regulator of NOD2 activity. Gut. 2012 doi: 10.1136/gutjnl-2011-301775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eitel J, et al. Beta-PIX and Rac1 GTPase mediate trafficking and negative regulation of NOD2. J Immunol. 2008;181(4):2664–2671. doi: 10.4049/jimmunol.181.4.2664. [DOI] [PubMed] [Google Scholar]
- 10.Kufer TA, Kremmer E, Banks DJ, Philpott DJ. Role for erbin in bacterial activation of Nod2. Infect Immun. 2006;74(6):3115–3124. doi: 10.1128/IAI.00035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411(6837):599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 12.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411(6837):603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 13.Jostins L, et al. International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]