Abstract
The present work was conducted to understand the basis of adaptation in Caragana jubata in its niche environment at high altitude cold desert of Himalaya. Molecular data showed predominance of genes encoding chaperones and those involved in growth and development at low temperature (LT), a major cue operative at high altitude. Importantly, these genes expressed in C. jubata in its natural habitat. Their homologues in Arabidopsis thaliana, Oryza sativa, and Glycine max did not exhibit similar trend of gene expression at LT. Constitutive expression and a quick up-regulation of the above genes suggested the ability of C. jubata to adjust its cellular machinery to maintain growth and development in its niche. This was reflected in LT50 (the temperature at which 50% injury occurred) and LT mediated photosynthetic acclimatory response. Such molecular and physiological plasticity enables C. jubata to thrive in the high altitude cold desert of Himalayas.
High altitude ecosystem has traditionally been characterized as ‘harsh’1. Apart from the declining surface area2, both, species richness3 and diversity4 are known to decline at high altitude. This highlights the adaptive capability of only fewer species to perform against the increasing magnitude of environmental extremes, wherein low temperature (LT) is considered to be one of the critical cues. LT induces many anatomical and physiological alterations including reduced rates of biochemical reactions, shift in temperature optima of several physiological processes5, altered membrane fluidity and protein conformation6. LT is well known to suppress photosynthesis during winter in most woody plants, but many herbaceous plants are able to acclimate to avoid down-regulation of photosynthesis7. Several plants are able to modulate their metabolites levels which are believed to have varied roles8. Accumulation of cryoprotectant molecules such as soluble sugars, sugar alcohols, and low molecular weight nitrogenous compounds has been reported in response to LT9. A large number of molecular and metabolic modulations, including production of secondary messengers, reactive oxygen species (ROS), calcium-dependent protein kinases (CDPKs), and mitogen activated protein kinase (MAPK) cascade have been reported after LT exposure10. LT also modulates a large number of genes encoding specific proteins, which can be divided into two broad groups. The first group includes genes that encode proteins, such as late embryogenesis abundance (LEA) proteins, osmotin, antifreeze proteins, mRNA binding proteins, enzymes of osmolyte biosynthesis, water channel proteins, and detoxification enzymes, which have a role in protecting cells from LT damage. The second group consists of genes encoding transcription factors that regulate transcriptional activity under conditions of stress11. Although, these studies provide insight into the plant response to LT, their roles in imparting functional advantage under natural environment remains less understood.
Preponderance of a plant species in an ecological niche is a reflection of its adaptive suitability at varied level of plant organization, primarily controlled by gene expression. Hence, gene expression analysis is likely to provide important clues of adaptive advantage of a species over others. For example, down-regulation of genes involved in jasmonate cascade modulated herbivore attack, thereby modulating plant fitness in a given environment12. Understanding genomic response of an organism to environment can provide a valuable insight to extrapolate the population and community response as well, provided the experiments are conducted within the ecological framework13.
The present work was carried out in a high altitude cold desert plant Caragana jubata (Pall.) Poir, to understand LT responsive molecular mechanism and its adaptive capacity and fitness in its niche environment on the pretext of cross-talk of commonality of gene response to varied environmental cues14. C. jubata is a perennial shrub of family Fabaceae distributed widely in temperate/alpine environment of Eurasia15 as well as Himalaya16. The habitat of C. jubata in Himalaya is snow covered during winter (October to March/April), and the species has a period of only 5–6 months for active growth and development. Even during this later period, C. jubata experiences extreme diurnal variations in temperature ranging from about 5 to 25°C during the daytime and about 2 to (−) 10°C during night.
Results
Cloning, sequencing and annotation of suppression subtractive hybridization cDNA library (SSHL)
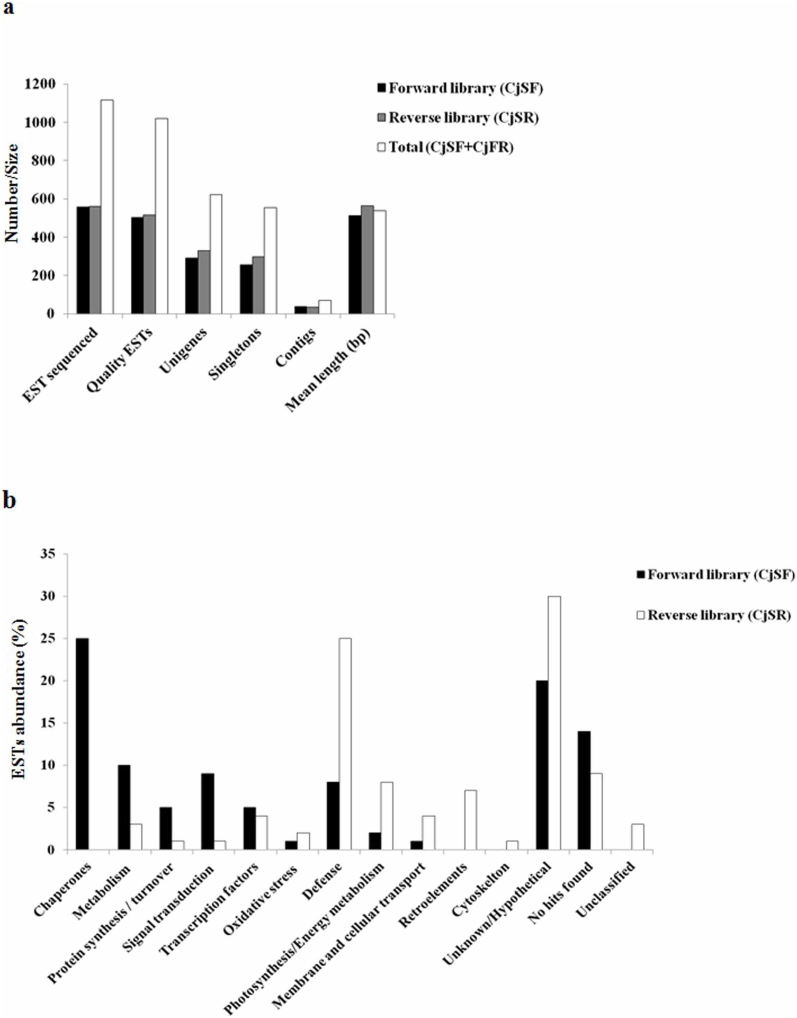
To understand the plant response to LT, SSHL was prepared using the RNA isolated from the leaf tissue of C. jubata exposed to 25°C (control, CO) and 4°C (LT). A total of 1116 expressed sequence tags (ESTs) were generated from forward (CjSF, 556 ESTs) and reverse (CjSR, 560 ESTs) SSHL. ESTs in CjSF represent the genes up-regulated in response to LT, whereas those in CjSR show the genes down-regulated in response to LT. Depending upon the quality of sequences in CjSF, 502 ESTs were analyzed for annotation and contigs assembly (Fig. 1a and Supplementary Table S1). Gene function analysis grouped ESTs into 256 singletons and 36 contigs (Fig. 1a and Supplementary Table S1), constituting a total of 292 unigenes. Out of these, 98 were hypothetical/unknown proteins (i.e. cDNA sequences without an analog in protein database), whereas 70 represented novel proteins i.e. with no significant homology in database (Supplementary Fig. S1a and Supplementary Table S2). ESTs were classified into 11 broad categories consisting of transcription factors, defense, signal transduction, oxidative stress, photosynthesis/energy metabolism, metabolism, protein synthesis/turnover, chaperones, membrane and cellular transport, hypothetical/unknown, and novel genes (Fig. 1b and Supplementary Table S2). ESTs for chaperones (e.g. LEA proteins) were the most abundant followed by those encoding early light inducible protein (ELIP), abscisic acid (ABA) inducible protein, seed maturation protein PM32 (PM32), ribulose-1,5-bisphosphate carboxylase (Rubisco), hypothetical chloroplast release factor 1, LEA-2, LEA-1 and RNA polymerase sigma subunit E (Fig. 1b and Supplementary Table S2).
Figure 1. Functional analyses of expressed sequence tags (ESTs) obtained from suppression subtractive hybridization library (SSHL) prepared from leaf tissue of Caragana jubata exposed to 25°C (control, CO) and 4°C (low temperature, LT).
Panel ‘a’ shows EST details of CO and LT SSHL. Major abundant transcripts in CO and LT SSHL are shown in panel ‘b’. Transcripts were functionally classified according to Munich Information Center for Protein Sequences (MIPS, http://www.helmholtz-muenchen.de/en/ibis). Details on abundance of all the ESTs are shown in Supplementary Table S2 and Supplementary Table S3.
In CjSR, 515 ESTs were selected for annotation and contigs assembly (Fig. 1a and Supplementary Table S1). These ESTs were clustered into 297 singletons and 34 contigs, constituting a total of 331 unigenes. All the ESTs were assigned putative function and categorized into functional categories consisting of transcription factors, signal transduction, oxidative stress, photosynthesis/energy metabolism, metabolism, protein synthesis/turnover, cytoskeleton, membrane and cellular transport, defense, chaperones, retroelements, unclassified (i.e. ESTs with homology in the database but could not be assigned any function), hypothetical/unknown, and novel genes (Supplementary Fig. S1b). ESTs for putative pathogenesis related protein 1 precursor were most abundant in CjSR (Fig. 1b and Supplementary Table S3). This was followed by ESTs encoding glycine-rich RNA-binding protein 2, retrotransposon protein, chlorophyll a/b binding protein, RNA polymerase beta' subunit, chlorophyll a/b binding protein of LHCII type III, chloride channel calcium activated 4, retroelements pol polyprotein, unclassified and putative ammonium transporters, respectively.
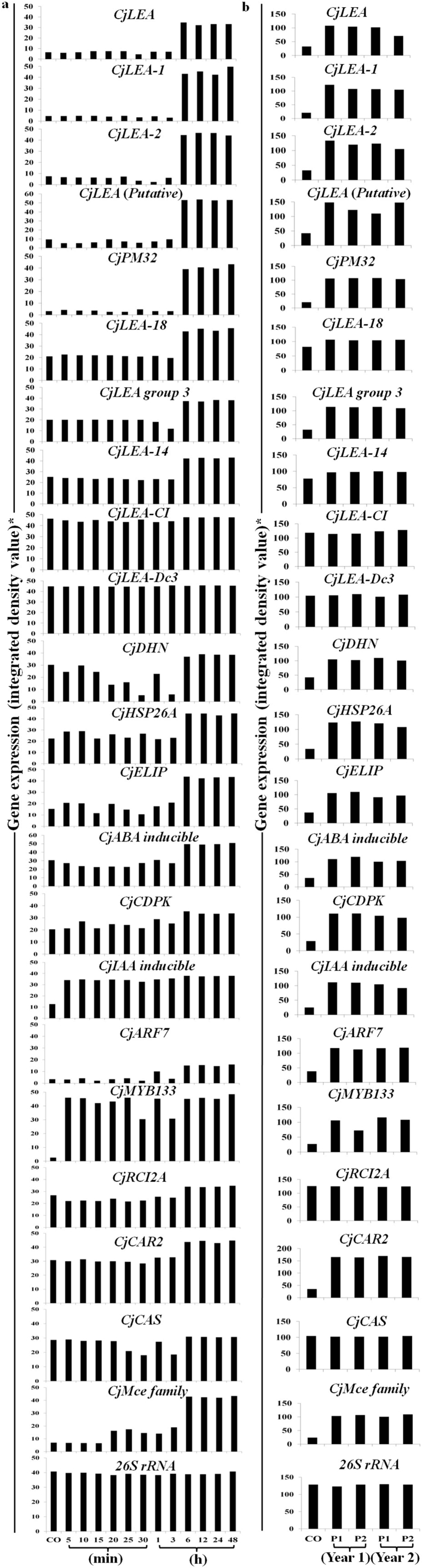
One of the interesting features of the SSHL analysis was the abundance of genes encoding chaperones [CjLEA-1 (group 1), CjLEA-2 (group 2), CjLEA-14 (group 2), CjDHN (group 2), CjLEA-Dc3 (group 3), CjLEA group 3 (group 3), LEA-cold inducible (CjLEA-CI) (group 4), CjLEA (group 4), CjLEA (Putative) (group 5), CjPM32 (group 6), CjLEA-18 (group 6) and CjHSP26A; LEA proteins were classified into various groups depending upon the presence of specific sequence motifs or biased amino acid composition17,18] and those associated with growth and development [early light inducible protein (CjELIP), CjABA inducible, CjCDPK, indole acetic acid inducible protein (CjIAA inducible), auxin responsive factor 7 (CjARF7), MYB transcription factor 133 (CjMYB133), rare cold inducible 2A (CjRCI2A), cold acclimation responsive2 (CjCAR2), cold acclimation specific (CjCAS), mammalian cell entry family protein (CjMce family)19,20] in CjSF (Supplementary Table S2). Since these genes could impart adaptive advantage, a detailed kinetic study was carried out at 13 time points spanning between 0–48 h, to identify the early and late responsive genes in response to LT (Fig. 2a and Supplementary Fig. S2a). Further, to understand the adaptive significance of these genes, their expression was also followed in two populations of C. jubata collected from the niche environment for two separate years (Fig. 2b, Supplementary Fig. S2b and Supplementary Fig. S3). A comparative expression analysis of gene homologues in Arabidopsis21 (Arabidopsis thaliana; a cold-tolerant plant species), rice22 (Oryza sativa; a cold-sensitive plant species) and soybean23 (Glycine max; family Fabaceae), using publicly available microarray data, was also performed in silico (Supplementary Fig. S4, Supplementary Fig. S5 and Supplementary Table S6).
Figure 2. Kinetics of gene expression in response to low temperature as a function of time, as well as at low and high altitude in the leaf tissue of Caragana jubata.
For panel ‘a’, plants were shifted from 25±3°C (control, CO) to 4±3°C (low temperature, LT) and the samples were harvested at different time intervals as shown in the figure. Gene expression was studied by reverse transcriptase-polymerase chain reaction (RT-PCR) that was standardized to get amplification in the exponential phase. RT-PCR was also performed for the plants housed at 25°C at the same time intervals as performed for those kept at LT (data not shown since there was no change in gene expression over a span of 48 h). Name of the gene is shown in each panel and the corresponding accession number is shown in Supplementary Table S2. Details of primers and PCR conditions are mentioned in Supplementary Table S7. For panel ‘b’, leaves of two populations (P1 and P2) of C. jubata growing in niche environment of high altitude in Kibber-Gete area (Lahaul and Spiti district, Himachal Pradesh, India) were collected during September/October in two separate years (year 1 and year 2). Samples were also collected for the plants growing at 25±3°C (CO) at low altitude in the institute (altitude, 1300 m; Palampur, Himachal Pradesh, India). Leaf tissues were harvested, stored immediately in liquid nitrogen on site and stored at (−) 80°C till further use. 26S rRNA served as internal control for both panel ‘a’ and ‘b’. Gel figures for RT-PCR are shown in Supplementary Fig. S2a,b. *To calculate integrated density value (IDV), PCR products were separated on 1.2% agarose gel, stained with ethidium bromide and quantified using Alpha DigiDoc 1000 software supplied along with gel documentation system (Alpha DigiDocTM, Alpha Innotech, USA). IDV thus obtained was divided by 1,000 for ease of writing on the y axis; thus, the IDV needs to be multiplied by 1,000 to get the original value of gene expression.
Genes encoding six groups of LEA proteins showed up-regulation in response to LT and in the niche environment
CjSF yielded 11 LEAs belonging to six groups. Differential expression of genes encoding different LEA proteins was confirmed by detailed kinetics of expression analysis at different time points (Fig. 2a and Supplementary Fig. S2a). Data showed that CjLEA-Dc3 was constitutively expressed (Fig. 2a) whereas, CjLEA, CjLEA-1, CjLEA-2, CjLEA (putative) and CjPM32, exhibited a strong up-regulation at 6 h of exposure to LT (Fig. 2a). CjLEA-18, CjLEA group 3, CjLEA-14, CjLEA-CI and CjDHN exhibited constitutive expression in CO and were slightly down-regulated during early period of LT (upto 3 h) but up-regulation was recorded at 6 h of exposure to LT and maintained thereafter during the entire period of experiment (Fig. 2a). All the CjLEAs exhibited expression in the two populations of C. jubata at high altitude during both the years of analyses (Fig. 2b, Supplementary Fig. S2b and Supplementary Fig. S3).
It was important to note that in Arabidopsis, only three LEAs (homologues to CjLEA, CjLEA-14, CjDHN) exhibited up-regulation in response to LT (Supplementary Fig. S4 and Supplementary Table S4). In rice, expression of only one LEA (Os03g20680, a CjLEA homologue) was up-regulated under LT, while up-regulation of rest of the LEAs was not observed (Supplementary Fig. S5 and Supplementary Table S5). In soybean, only three LEAs (homologues to CjLEA-18, CjLEA-14 and CjLEA-CI) exhibited up-regulation in response to LT (Supplementary Table S6).
CjHSP26A showed up- and down-regulation of expression during early period (upto 3 h) of exposure to LT. An evident up-regulation was observed at 6 h exposure to LT and maintained thereafter during the whole period of experiment (Fig. 2a and Supplementary Fig. S2a). Also, the gene was found to be up-regulated in populations of C. jubata growing in the high altitude (Fig. 2b, Supplementary Fig. S2b and Supplementary Fig. S3). In Arabidopsis, its homologue was induced within 30 min following the exposure to LT (Supplementary Fig. S4 and Supplementary Table S4). Expression analysis in soybean revealed up-regulation of CjHSP26A homologue under LT (Supplementary Table S6).
Genes involved in growth and development exhibited up-regulation in response to LT and in the niche environment
Gene encoding ELIP was present in abundance (5.2%) in CjSF during LT (Supplementary Fig. S1a). CjELIP exhibited both up- and down-regulation of expression pattern during early period (upto 3 h) followed by up-regulation at 6 h of exposure to LT and maintained thereafter during the entire period of experiment (Fig. 2a and Supplementary Fig. S2a). Up-regulation of CjELIP was also observed in populations growing at high altitude (Fig. 2b Supplementary Fig. S2b and Supplementary Fig. S3). ELIP homologue in Arabidopsis exhibited up-regulation at 3 h of cold treatment (Supplementary Fig. S4 and Supplementary Table S4), while there was no change in the expression of rice ELIP homologue (Supplementary Fig. S5 and Supplementary Table S5). In soybean, ELIP homologue exhibited down-regulation during LT (Supplementary Table S6).
CjCDPK also exhibited a pattern of up- and down-regulation of expression during early period (upto 3 h) of exposure to LT. Up-regulation was observed at 6 h and onwards of exposure to LT (Fig. 2a and Supplementary Fig. S2a). Up-regulation of CjCDPK was also observed in populations growing in high altitude (Fig. 2b, Supplementary Fig. S2b and Supplementary Fig. S3). Expression of CDPK homologue was unaltered in rice (Supplementary Fig. S5 and Supplementary Table S5) while in soybean, its homologue exhibited up-regulation at LT (Supplementary Table S6).
Gene encoding abscisic acid (ABA) inducible proteins was present to an extent of 4.9% in CjSF (Supplementary Fig. S1a). Expression analysis showed that CjABA inducible gene exhibited constitutive expression during early period of exposure to LT (upto 3 h) and was up-regulated at 6 h and onwards of exposure (Fig. 2a and Supplementary Fig. S2a). The gene also expressed under natural environment at high altitude (Fig. 2b, Supplementary Fig. S2b and Supplementary Fig. S3). Importantly, its homologue in Arabidopsis was down-regulated in response to LT (Supplementary Fig. S4 and Supplementary Table S4).
Expression of CjIAA inducible gene was not visible in CO, but at 5 min of exposure to LT, gene expression was noticeable. An evident up-regulation was observed at 6 h of exposure to LT and maintained thereafter (Fig. 2a and Supplementary Fig. S2a). Up-regulation of CjIAA inducible gene was also observed in populations growing in niche environment of high altitude (Fig. 2b, Supplementary Fig. S2b and Supplementary Fig. S3). Expression of its homologues in Arabidopsis and rice was unaltered (Supplementary Fig. S4 and S5 and Supplementary Table S4 and S5). In soybean, its homologue exhibited up-regulation during LT (Supplementary Table S6).
Auxin responsive factor 7 (CjARF7) was up-regulated at 6 h of exposure to LT in C. jubata (Fig. 2a and Supplementary Fig. S2a) and was expressing under natural conditions at high altitude (Fig. 2b, Supplementary Fig. S2b and Supplementary Fig. S3). Its homologue in Arabidopsis exhibited down-regulation at 1 h and up-regulation at 12 h of LT treatment (Supplementary Fig. S4 and Supplementary Table S4). Also in soybean, its homologue exhibited down-regulation in response to LT (Supplementary Table S6).
Transcript of gene encoding MYB transcription factor (CjMYB133) was not detected under CO condition, but the expression was evident at 5 min of exposure to LT (Fig. 2a and Supplementary Fig. S2a). Also, the gene expression was evident in plants in its niche environment at high altitude (Fig. 2b, Supplementary Fig. S2b and Supplementary Fig. S3). In Arabidopsis, its homologue was up-regulated at 6 h of LT treatment (Supplementary Fig. S4 and Supplementary Table S4), while the expression of its homologue in rice was unaltered (Supplementary Fig. S5 and Supplementary Table S5).
Expression of COR gene encoding cold acclimation responsive (CjCAR2) protein was unaltered till 3 h of LT treatment, but up-regulated at 6 h and the up-regulation was maintained thereafter. Expression of genes encoding rare cold inducible (CjRCI2A) and cold acclimation sensitive (CjCAS) protein was down-regulated up to 3 h, but the expression regained to the constitutive level at 6 h of LT treatment (Fig. 2a and Supplementary Fig. S2a). Up-regulation of these COR genes was also observed in populations growing in the niche environment of high altitude (Fig. 2b, Supplementary Fig. S2b and Supplementary Fig. S3). In Arabidopsis, homologue of CjCAR2 was up-regulated only at 12 h of LT treatment (Supplementary Fig. S4 and Supplementary Table S4), while homologue of CjCAS showed down-regulation at 30 min following LT treatment, and up-regulation was observed at 24 h of LT exposure (Supplementary Fig. S4 and Supplementary Table S4). In rice, homologues of both, CjCAR2 and CjCAS did not show any change in expression at LT (Supplementary Fig. S5 and Supplementary Table S5) while, in soybean, homologues of both CjCAR2 and CjCAS exhibited up-regulation at LT (Supplementary Table S6).
Putative Mce family protein (CjMce family) showed an early up-regulation at 5 min of LT treatment followed by a strong up-regulation at 6 h (Fig. 2a and Supplementary Fig. S2a). Also, the gene expressed in the samples of C. jubata collected from its niche environment (Fig. 2b, Supplementary Fig. S2b and Supplementary Fig. S3). Its homologue was not found in rice and soybean.
C. jubata acclimated quickly to LT
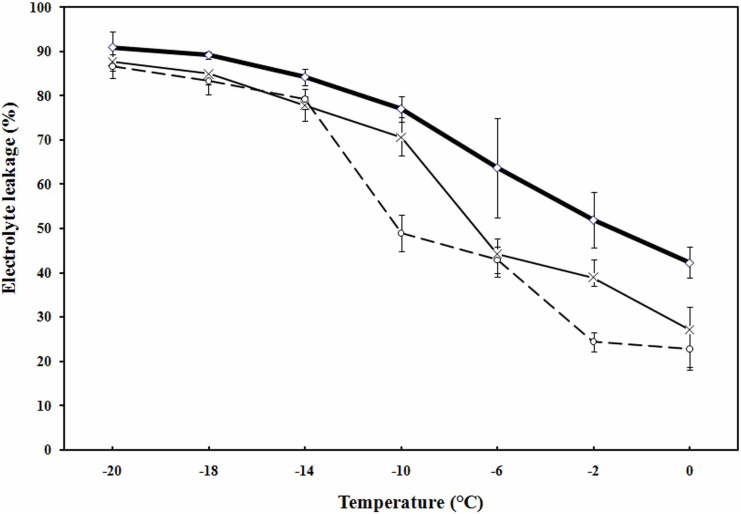
Freezing tolerance (LT50, the temperature at which 50% injury occurred) was evaluated through measurement of electrolyte leakage in C. jubata. Data showed that freezing tolerance of non-acclimated (NA) plant was (−) 1.8°C, which decreased to (−) 7°C and (−) 10°C at 3 h and 12 h, respectively upon exposure to 4°C (Fig. 3).
Figure 3. Freezing tolerance of non-acclimated and acclimated leaf tissue of Caragana jubata as measured by electrolyte leakage.
Control plants (◊, non-acclimated) were maintained in a plant growth chamber at 25°C, light intensity of 200 μmol m−2 s−1 and 60–80% relative humidity. For cold acclimation, plants were exposed to 4°C for 3 h (×) and 12 h (o). Each value is a mean of three separate biological replicates with standard error represented as error bar.
Photosynthetic acclimation to LT
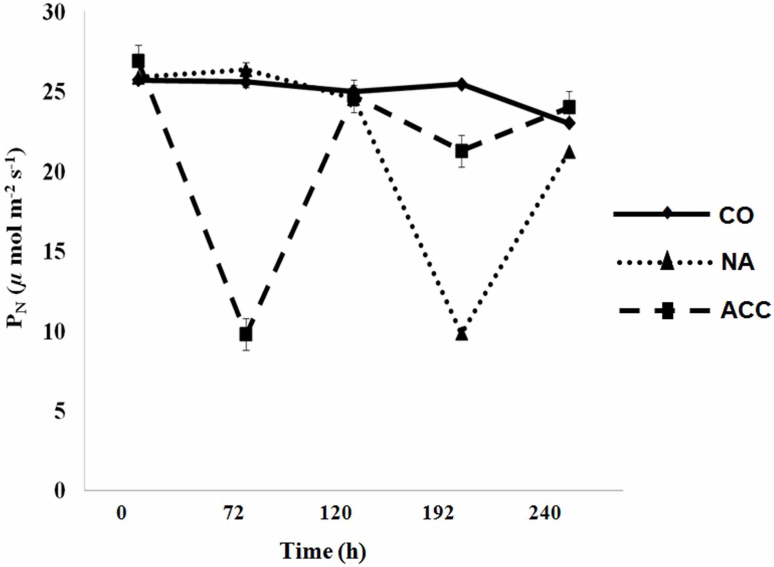
To study species acclimation response to LT, net photosynthetic rates (PN) were studied in three sets of plants of C. jubata grown in growth chamber. In this experiment (lasting for 240 h), twelve weeks old plants were grouped into three sets, control (CO; 25°C), non-acclimated (NA) and acclimated (ACC) (Supplementary Fig. S6). NA and ACC plants were exposed to LT as shown in Supplementary Fig. S6. Each of the LT treatment (lasting for 72 h) was terminated by subjecting plants to 25°C for 48 h. PN showed no significant change throughout the period of experiment in CO plants, but declined significantly during LT treatment (at 72 h in ACC plants and 192 h in NA plants; Fig. 4 and Supplementary Fig. S6). Exposing ACC plants to 25°C for 48 h restored PN. At 120 h of experimentation, ACC plants were re-exposed to LT (for 72 h). PN recorded at 192 h showed decline, but the percent decline was less than half to that recorded after the first LT treatment suggesting acclimation in PN (Fig. 4 and Supplementary Fig. S6).
Figure 4. Low temperature mediated phosynthetic acclimation in Caragana jubata.
Net photosynthetic rates (PN) were measured in control (CO,  ), non-acclimated (NA,
), non-acclimated (NA,  ), and acclimated (ACC,
), and acclimated (ACC,  ) plants. CO plants were exposed to 25°C throughout the experiment. NA plants were initially exposed to 25°C for 120 h followed by exposure to low temperature (4°C) upto 192 h for the next duration of 72 h; thereafter, plants were exposed to 25°C again (Supplementary Fig. S6). ACC plants were exposed to a cycle of 4°C, 25°C, 4°C, 25°C, at 0 h, 72 h, 120 h and 192 h, respectively (Supplementary Fig. S6). Each value is a mean of 45 observations on 5 different biological replicates with standard error shown as vertical bar.
) plants. CO plants were exposed to 25°C throughout the experiment. NA plants were initially exposed to 25°C for 120 h followed by exposure to low temperature (4°C) upto 192 h for the next duration of 72 h; thereafter, plants were exposed to 25°C again (Supplementary Fig. S6). ACC plants were exposed to a cycle of 4°C, 25°C, 4°C, 25°C, at 0 h, 72 h, 120 h and 192 h, respectively (Supplementary Fig. S6). Each value is a mean of 45 observations on 5 different biological replicates with standard error shown as vertical bar.
Discussion
Simultaneous up-regulation of all the six groups of LEA protein genes in response to LT in C. jubata merits exceptional attention. Till date there is no report that describes LT related up-regulation of all the six groups of LEA protein genes simultaneously in any plant system. LEAs are typically highly hydrophilic proteins that provide stability to other proteins under dehydration, freezing, high salinity or drought and impart stress tolerance24. Homologues of all the six groups of LEAs identified in the present work are reported to be associated with stress tolerance in plants17. For example, CAP85 and CAP160, the members of the LEA/dehydrin superfamily, enhanced freezing tolerance in tobacco25. Wheat dehydrin gene WCOR410 enhanced tolerance of strawberry leaves to freezing26. Dehydrins contributed to freezing tolerance, partly due to their protective effect on membranes27. Group 3 LEA protein gene WCS19 improved freezing tolerance of the transgenic Arabidopsis, however only in the ACC plants suggesting a need of other endogenous components that only accumulate in response to low temperature28. Thus, a role of LEAs can be attributed to acclimation in C. jubata leading to freezing tolerance as indicated by LT50 (Fig. 3).
Constitutive expression of LEAs (e.g. CjLEA-Dc3, CjLEA-CI, CjLEA-14) are suggestive of their role in taking care of initial shock (e.g. might be involved in imparting tolerance upon sudden exposure to LT, particularly for the period in which other chaperones were not induced in response to LT), and with increasing the protective demand at LT, inducible LEAs might function synergistically with other members. Since C. jubata in its niche environment is expected to be exposed to different environmental cues including LT, constitutively expressing LEAs could render a protective role. Similar protective role could be assigned to gene encoding low molecular weight heat shock protein CjHSP26A, which is up-regulated in response to LT as well as at high altitude (Fig. 2a,b and Supplementary Fig. S3). The protein has a protective effect through membrane protection, refolding of denatured proteins, and preventing protein aggregation at LT29.
ELIPs are chloroplast proteins, the genes of which are transiently transcribed during the greening process of etiolated plants. Genes encoding ELIPs have been reported to be up-regulated in Arabidopsis, pea (Pisum sativum), wheat (Triticum aestivum), tea (Camellia sinensis) and barley (Hordeum vulgare) in response to LT30,31,32. By using an Arabidopsis mutant (chaos), a photo-protective role of ELIP was shown at LT in the presence of high light intensity33. In C. jubata, up-regulation of ELIP in response to LT as well as at high altitude (Fig. 2a,b and Supplementary Fig. S3) is likely to provide protection against photo-oxidative damage and support plant growth.
CDPKs play a key role in Ca2+-mediated signal transduction pathway. Genes encoding CDPKs are reported to be induced in other plant species including rice and maize (Zea mays) in response to LT and other abiotic stresses34,35,36. Over-expression of CDPK enhanced freezing tolerance in transgenic rice37. An early up-regulation of CjCDPK in C. jubata (Fig. 2a,b and Supplementary Fig. S3) suggested its role in freezing tolerance, through signaling pathway.
ABA inducible proteins have been reported from various plant species including rice, wheat, and barley during different environmental stresses, with possible role in optimizing plant performance under stress conditions38. Up-regulation of CjABA inducible (Fig. 2a,b and Supplementary Fig. S3) suggested its role in growth and development of C. jubata at high altitude.
Indole acetic acid (IAA) modulates plant growth and development through IAA inducible proteins that are short lived nuclear proteins and function as transcriptional regulators. These proteins do not interact with DNA but exert their regulatory activity through another group of proteins called auxin responsive factors. IAA inducible proteins have been reported in Arabidopsis (AXR2/IAA17, AXR3/IAA17) and pea (IAA4, IAA6), which play a role in auxin mediated signaling39. Further, ARF gene family encodes transcriptional regulators that are involved in auxin signaling. These transcription factors bind to auxin-responsive cis-acting elements present in the promoter region of auxin-responsive genes through its DNA binding domain40. ARF7 and ARF19 play essential roles in auxin-mediated plant development by regulating auxin responsive genes in Arabidopsis. Auxin response factor 7/NONPHOTOTROPHIC HYPOCOTYL4 (ARF7/NPH4) was up-regulated in Arabidopsis during LT41. Data (Fig. 2a,b and Supplementary Fig. S3) suggested a role of CjIAA inducible and the CjARF genes in maintaining plant growth and development under LT as well as at high altitude through the mechanisms detailed above.
MYB proteins are a superfamily of transcription factors that play regulatory roles in developmental processes and defense responses. Plant MYB proteins are classified into three major groups: R2R3-MYB, R1R2R3-MYB and MYB related proteins42. Two R2R3-MYB transcription factors, HOS10 in Arabidopsis and Osmyb4 in rice imparted cold tolerance through CBF-independent pathway43. Over expression of R1R2R3-MYB transcription factor, OsMYB3R-2 from rice increased freezing tolerance in transgenic Arabidopsis44. Arabidopsis MYB 15 (R2R3-MYB) is also involved in enhancing freeze tolerance45. With this background on the importance of MYBs in imparting stress tolerance and maintenance of developmental processes, it is likely that up-regulated CjMYB133 in C. jubata (Fig. 2a,b and Supplementary Fig. S3) has similar roles at LT and other stresses at high altitude.
The COR proteins are highly hydrophilic and have role in cold acclimation, freezing tolerance46 and in maintenance of growth particularly at LT20. Constitutive over-expression of COR15a enhanced freezing tolerance in non-acclimated transgenic plants of Arabidopsis47. In Medicago truncatula and M. falcata, low expression of COR gene CAS15 was reported in plants maintained at 25°C, but an evident up-regulation was observed at 6 h of exposure to low temperature48. An early induction of these genes (CjRCI2A, CjCAR2 and CjCAS; upto 3 h) at LT coupled with expression in the niche environment (Fig. 2a,b and Supplementary Fig. S3) is suggestive of their roles in cold acclimation, freezing tolerance and in maintaining growth in C. jubata.
Mce operon encodes a subfamily of ABC transporters that has a role in remodeling the cell envelop in Mycobacterium tuberculosis49. This family of proteins is not well characterized in plant system. An evident up-regulation of CjMce family at LT and in the niche environment (Fig. 2a,b and Supplementary Fig. S3) requires detailed investigation to decipher its possible role in plants in response to environmental cue.
C. jubata in its natural habitat is exposed to a wide temperature range, from about 25°C during day to sub-zero temperature during night. Gene expression data showed the presence of the genes encoding chaperones and those associated with growth and development within 5 min to 3 h of exposure to LT (Fig. 2a). Importantly, all these genes also expressed in C. jubata in its natural habitat (Fig. 2b and Supplementary Fig. S3). Given the biological function of chaperones in cold acclimation and freezing tolerance, and the kinetics of their gene expression, data supported quick acclimation of the species to LT (Fig. 3). In Arabidopsis, LT50 of (−) 7°C was achieved at day 7 of cold acclimation50. In M. truncatula, leaf LT50 was (−) 11°C when cold acclimated at 2°C for 4 weeks48. C. jubata, thus quickly acclimated to LT as compared to Arabidopsis or M. truncatula. Similarly, expression of genes associated with growth and development would support C. jubata to thrive in its natural habitat.
Photosynthesis is generally suppressed at LT, as was also observed in our study (Fig. 4). This suppression could be due to reduction in activities of enzymes of carbon metabolism, thylakoid electron transport rate, photochemical efficiency of the photosystem (PS) II, and stomatal conductance51. LT mediated reduction in PN in C. jubata can be explained based upon down regulation of genes associated with photosynthesis. These include those encoding for chlorophyll a/b-binding protein, chlorophyll a–b binding protein of LHCII type III, PS I light-harvesting proteins, Rubisco subunits, NADH dehydrogenase subunits (Supplementary Table S3 and Supplementary Fig. S9).
Photosynthetic acclimation is an important attribute that enhances plant fitness in changing environments52. LT-mediated photosynthetic acclimation in C. jubata (Fig. 4) can be related to up-regulation of chaperones such as CjLEA-18, a group 6 of LEA protein gene (Fig. 2a,b and Supplementary Fig. S3). Over-expression of CaLEA6, a group 6 of LEA protein gene of Capsicum annuum, in transgenic Arabidopsis conferred protection in maintaining photosynthetic capacity under stress53. The functional advantage of photosynthetic acclimation to LT could be seen in Saxifraga paniculata, where plant achieved values around 4.0 μmol CO2 m−2 s−1 even after night temperature dropped to (−)22°C7.
Comparative expression analyses amongst C. jubata, Arabidopsis, rice and soybean showed that most of the genes exhibited either constitutive expression or early induction following LT treatment in C. jubata, only 12 out of 22 genes showed induction in Arabidopsis (cold-tolerant), one in rice (cold-sensitive) and 8 in soybean (a species of the same family as that of C. jubata). Though, Arabidopsis is considered a cold tolerant species, it is not distributed naturally in extremes of the environment such as cold desert. The data suggests adaptive advantage of C. jubata to LT over other plants such as Arabidopsis, rice and soybean. The LT responsive genes encoding chaperones and CORs in the present work are also known to confer tolerance to plants under other abiotic stresses18,54 and are the likely candidates to protect the plants under the environment of high altitude.
To conclude, the study suggested that: (1) C. jubata has an exceptional capability to express genes encoding chaperones and those associated with growth and development. Some of these were constitutively expressed, whereas others were strongly up-regulated within 5 min to 3 h of exposure to LT. Also these genes were expressed in the species in its niche environment, suggesting the capability of C. jubata to thrive in the environment of high altitude; (2) The temperate/alpine environment, where C. jubata is distributed naturally, experiences frequent fluctuations in temperature16 and calls for quick acclimation. Indeed, the species exhibited a quick acclimation response to LT (in terms of LT50) with concomitant expression of the relevant genes as compared to the other species reported so far; (3) Photosynthetic acclimation of the species to LT was an important trait for functional advantage in the cold environment of high altitude.
Methods
Plant material and growth conditions
Seeds of C. jubata were collected from the plants growing in its niche in Kibber-Gete (32° 18′ 21″ N, 78° 01′ 33″ E; altitude 4,300 m; Lahaul and Spiti district, Himachal Pradesh), India and brought to the institute at Palampur (32° 06′ N, 76° 33′ E; altitude 1300 m; Kangra district, Himachal Pradesh), India for further studies. Seeds were sown in pots and placed in a greenhouse maintained at a temperature of 25±3°C and relative humidity of 60–80%. Light intensity ranged between 500–800 μmol m−2 s−1. At twelve weeks after sowing, plants were placed in two separate growth chambers (light intensity, 200 μmol m−2 s−1; RH, 70±10%) maintained at 25±3°C (control, CO) and 4±3°C (low temperature, LT). Leaves were harvested for the preparation of suppression subtractive hybridization cDNA libraries (SSHL) and also for gene expression analysis.
In a separate experiment, leaf tissue was harvested from the plants growing at Kibber-Gete where air temperature was 12±3°C and light intensity was 2500±100 μmol m−2 s−1 at the time of sampling. The air temperature was measured by a digital thermometer and light intensity was measured using quantum sensor housed on a portable photosynthesis system LI 6400 (Li-COR, Lincoln, USA). Two populations of C. jubata were identified which were present in different hillocks separated by a water stream of melting snow water. Plants growing under similar climate conditions in a patch of 100 m × 100 m were considered as one population. Sampling was done during the period of September/October for two separate years (Supplementary Fig. S7), frozen immediately in liquid nitrogen, and stored at (−) 80°C till further use. Leaf and apical buds of minimum 15 plants of C. jubata were pooled together.
Construction of SSHL
SSHL was constructed using PCR-select™ cDNA SSH Kit (Clontech, USA) essentially as previously described55 except that forward subtraction was performed using the cDNA synthesized from the mRNA of LT treated tissue (4°C for 48 h) as tester and driver cDNA was synthesized from the mRNA of CO (25°C). For reverse subtraction, tester cDNA was synthesized from mRNA of CO and driver cDNA was produced from the mRNA of LT treated tissue. For the purpose, total RNA was isolated from the leaves of CO and LT treated plants as previously described56. Poly (A)+ RNA (mRNA) was purified from total RNA using GenElute™ mRNA Miniprep Kit (Sigma, USA) as per the manufacturer's recommendations. Yield and quality of RNA was determined spectrophotometrically by recording absorbance at 260 and 280 nm. Also, RNA was run on a denaturing agarose/formaldehyde gel and visualized using a gel documentation system (Alpha DigiDocTM, Alpha Innotech, USA) to verify the integrity.
Sequencing and analysis of cloned expressed sequence tags (ESTs)
Plasmids isolated by GenElute™ Plasmid Miniprep Kit (Sigma, USA) were single pass sequenced using BigDye terminator (version 3.1) Cycle Sequencing Mix (Applied Biosystems, USA) on an automated DNA sequencer (3130xl, Genetic Analyzer, Applied Biosystems, USA) with M13 forward primer (5′-GTAAAACGACGGCCAGTG-3′).
The sequences were analysed and submitted at NCBI dbEST database [dbEST accession no. FE968693-FE968985 for forward library (CjSF) and dbEST accession no. GW327090-GW327420 for reverse library (CjSR)]. Details on the method of analyses are mentioned in the legend of Supplementary Table S2.
Differential screening of SSHL
Expressed sequence tags (ESTs) thus generated were screened by slot blot hybridization (reverse northern analysis) as described previously56, except that labeled [α-P32] probes were synthesized from the unsubtracted driver and tester cDNA of C. jubata (Supplementary Fig. S8 and Supplementary Fig. S9).
Gene expression analysis by reverse transcriptase polymerase chain reaction (RT-PCR)
RT-PCR was performed using Superscript III (Invitrogen, USA) essentially as described previously56. Details of RNA isolation, cDNA synthesis, PCR, and quantification of amplicons using integrated density value are given in Supplementary Fig. S2. Gene expression was also validated by quantitative real-time PCR (qRT-PCR; details mentioned in Supplementary Fig. S3) for the experiment on C. jubata growing at low and high altitudes.
In silico expression analysis of homologues genes of Arabidospsis (Arabidopsis thaliana), rice (Oryza sativa) and soybean (Glycine max) in response to LT
In silico expression analysis was carried out to study the response of C. jubata gene homologues in Arabidopsis, rice and soybean using the publicly available LT responsive expression data. Gene homologues were identified through BLASTX in TAIR 10 (www.arabidopsis.org) (Supplementary Table S4), RGAP 757 (Supplementary Table S5) and soybean genome (http://www.phytozome.net/soybean.php) (Supplementary Table S6). For soybean protein sequences, BLASTX with significant E-value of less than 0.01 was considered (Supplementary Table S6). Microarray data for genes of Arabidopsis at LT was retrieved from AtGenExpress (http://jsp.weigelworld.org/expviz/expviz.jsp). Datasets obtained were corresponding to LT (4°C) at different time points viz. 0.5, 1, 3, 6, 12, and 24 h for aerial tissues. Microarray data for Arabidopsis genes At4g04720 and At3g51290, homologues of CjCDPK and CjMce family, respectively were not available in database and hence could not be included in the analysis (Supplementary Fig. S4 and Supplementary Table S4).
Similarly, microarray data of rice genes at LT (experiment ID GSE6901) were retrieved from rice oligonucleotide array database (www.ricearray.org). In the experiment, seven-day-old light-grown rice seedlings were subjected to LT (4°C) for 3 h. The seedlings kept in water for 3 h at 28°C, served as control58. The values were log2 transformed and used to plot graphs for gene expression. Expression of each clone was tested in at least three independent LT treatment experiments. Microarray data for rice genes Os01g21250, Os05g38870, Os06g09660, Os05g04700, and Os11g05935, homologues of CjLEA-CI, CjABA inducible, CjARF7, CjRCI2A, and CjMce family, respectively were not available in the database and hence could not be included in the analysis (Supplementary Fig. S5 and Supplementary Table S5).
Microarray data of soybean under cold stress was available at http://www.ebi.ac.uk/arrayexpress/experiments/E-MEXP-3164. In these experiments, soybean plants grown in soil-filled plastic pots were kept for 3 weeks under a 12 h light/12 h dark at 28°C. Thereafter these were transferred to 4°C and sampled at day 1 of the treatment23. (Supplementary Table S6 shows various gene homologues in soybean that could be retrieved from the database).
Evaluation of freezing tolerance of C. jubata through measurement of electrolyte leakage
Non-acclimated (NA) plants were maintained in a plant growth chamber at 25°C at light intensity of 200 μmol m−2 s−1 and 60–80% relative humidity. For cold acclimation (ACC), plants were exposed to 4°C for 3 h and 12 h. Electrolyte leakage was measured essentially as previously described50 with minor modifications. Leaf tissue (100 ± 5 mg) was placed in a 15 ml plastic tube (Tarsons, India), which was capped and submerged in a refrigerated circulator bath (CoolTech 320, Thermo Scientific, Germany) filled with 60% glycerol. The bath was set at 0°C for 1 h, the temperature was lowered to (−) 2°C and held for 30 min. Small pieces of ice (4 crystals; each prepared with 200 μl of sterile demineralized water) were added in each tube to initiate nucleation. The temperature was maintained at (−) 2°C for 1 h and then lowered at a rate of 2°C h−1. Samples were removed at different temperature points and allowed to thaw on ice for overnight59. After thawing, sterile demineralized water was added to each tube to a final volume of 12 ml and the tubes were shaken for 4 h. Leaves were removed and frozen at (−) 80°C overnight in a deep freezer (Thermo Electron Corporation, USA). In the meantime, conductivity (C1) of the supernatant was measured using a conductivity meter (CyberScan PC 510, Eutech Instruments, Singapore) and the supernatant was stored in cold room (4°C) for further use. Leaf tissues placed in deep freezer were removed and transferred to their respective tubes containing the supernatant and shaken for another 4 h to measure the conductivity (C2). Electrolyte leakage was measured using the equation: (C1/C2) × 100.
Assessment of photosynthetic acclimation in response to low temperature
Twelve weeks old plants were grouped into three sets: control (CO), non-acclimated (NA) and acclimated (ACC) (details for CO, NA and ACC are mentioned in the legend of Fig. 4 and Supplementary Fig. S6). Net photosynthetic rate (PN) was measured using a portable open gas exchange system LI 6400 (Li-COR, Lincoln, USA), as previously described60. The gas exchange system was equipped with a programmable light, temperature and CO2 control. For all measurements, irradiance of 500 μmol m−2 s−1 was maintained by a cool light source (6400-02 LED) fitted on top of the leaf chamber, and data was recorded after 3–4 minutes when a steady state was achieved. Temperature of the chamber was maintained at 25°C through a Peltier cooling and heating system. CO2 within the chamber was maintained at 400 μmol mol−1 using 6400-01 CO2 injector and a CO2 mixer supplied along with the instrument. Data were computed by one-way analysis of variance (ANOVA), using general linear model procedures in SPSS statistical package (SPSS, Chicago, USA). The differences were calculated at the significance level of p < 0.05. Significant results were compared using Gabriel post hoc comparison.
Author Contributions
PKB carried out the SSH cDNA library construction, expression analysis, data collection, data analysis, preliminary drafting of the manuscript. RK, DM performed cold acclimation experiment. GB performed expression analysis (RT-PCR and qRT-PCR) with samples obtained from the niche environment. AKS performed in silico analysis of gene expression in Arabidopsis and rice. VA performed in silico analysis of gene expression in soybean. SKV performed experiments on photosynthesis. RK, DM, AKS, VA, and SKV assisted in writing the manuscript. PSA supported the work. SK spotted the plant species and its niche, conceived the study, designed the experiments, analysed the data, wrote, integrated and finalised the manuscript including title. All authors read and approved the final manuscript.
Supplementary Material
Supplementary information
Acknowledgments
Authors thank (i) Council of Scientific and Industrial Research (CSIR) for funding through mission mode project on “exploratory studies on climate change and adaptation of species complex” NWP020, (ii) Department of Biotechnology (DBT), Government of India for funding the project entitled “Bioprospecting genes for adaptive and growth strategies in cold desert plant species of Spiti” and “Novel Products from Western Himalayas: Antioxidant Formulations, Cloning of Low Temperature Related cDNA, and Development of Microbial Formulations”, and (iii) Indo-German Science and Technology Centre (IGSTC), India. PKB acknowledges CSIR for awarding Senior Research Fellowship. RK and DM thank DBT for providing financial assistance. GB acknowledges IGSTC for financial support. Help rendered by Mr. Digvijay Singh Naruka for sequencing is duly acknowledged. The manuscript represents IHBT publication number 2089.
References
- Billings W. D. Arctic and alpine vegetations: Similarities, differences, and susceptibility to disturbance. BioScience 23, 697–704 (1973). [Google Scholar]
- Körner C. The use of ‘altitude’ in ecological research. Trends Ecol. Evol. 22, 569–574 (2007). [DOI] [PubMed] [Google Scholar]
- Bhattarai K. R. & Vetaas O. R. Can Rapoport's rule explain tree species richness along the Himalayan elevation gradient, Nepal? Divers. Distrib. 12, 373–378 (2006). [Google Scholar]
- Wang Z., Brown J. H., Tang Z. & Fang J. Temperature dependence, spatial scale, and tree species diversity in eastern Asia and North America. Proc. Natl. Acad. Sci. USA 106, 13388–13388 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz C. Cell physiology of plants growing in cold environments. Protoplasma 244, 53–73 (2010). [DOI] [PubMed] [Google Scholar]
- Margesin R., Fonteyne P. A., Schinner F. & Sampaio J. P. Rhodotorula psychrophila sp nov., Rhodotorula psychrophenolica sp nov., and Rhodotorula glacialis sp nov., novel psychrophilic basidiomycetous yeast species isolated from alpine environments. Int. J. Syst. Evol. Microbiol. 57, 2179–2184 (2007). [DOI] [PubMed] [Google Scholar]
- Hacker J. & Neuner G. Photosynthetic capacity and PS II efficiency of the evergreen alpine cushion plant Saxifraga paniculata during winter at different altitudes. Arct. Antarct. Alp. Res. 38, 198–205 (2006). [Google Scholar]
- Streb P., Aubert S., Gout E. & Bligny R. Reversibility of cold-and light-stress tolerance and accompanying changes of meta-bolite and antioxidant levels in the two high mountain plant species Soldanella alpina and Ranunculus glacialis. J. Exp. Bot. 54, 405–418 (2003). [DOI] [PubMed] [Google Scholar]
- Guy C. L. Cold-acclimation and freezing stress tolerance: role of protein metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41, 187–223 (1990). [Google Scholar]
- Janska A., Marsik P., Zelenkova S. & Ovesna J. . Cold stress and acclimation- what is important for metabolic adjustment? Plant Biol. 12, 395–405 (2010). [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K. & Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803 (2006). [DOI] [PubMed] [Google Scholar]
- Kessler A., Halitschke R. & Baldwin I. T. Silencing the jasmonate cascade: induced plant defenses and insect populations. Science 305, 665–668 (2004). [DOI] [PubMed] [Google Scholar]
- Ungerer M. C., Johnson L. C. & Herman M. A. Ecological genomics: understanding gene and genome function in the natural environment. Heredity 100, 178–183 (2008). [DOI] [PubMed] [Google Scholar]
- Knight H. & Knight M. R. Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci. 3, 32–36 (2001). [DOI] [PubMed] [Google Scholar]
- Xiwu N. & Hongwen G. The distribution, characteristics, use and economical function of Caragana in China. International conference on grassland science and industry Hailar City, China. (2001). [Google Scholar]
- Bhardwaj P. K., Ahuja P. S. & Kumar S. Characterization of gene expression of QM from Caragana jubata, a plant species that grows under extreme cold. Mol. Biol. Rep. 37, 1003–1010 (2010). [DOI] [PubMed] [Google Scholar]
- Battaglia M. et al. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148, 6–24 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark M. & Hincha D. K. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9, 118 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J. et al. Arabidopsis calcium-dependent protein kinase CPK10 Functions in abscisic acid- and Ca2+-mediated stomatal regulation in response to drought Stress. Plant Physiol. 154, 1232–1243 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M. F. Role of cold-responsive genes in plant freezing tolerance. Plant Physiol. 118, 1–7 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M. W. et al. Costs and benefits of cold tolerance in transgenic Arabidopsis thaliana. Mol. Ecol. 13, 3609–3615 (2004). [DOI] [PubMed] [Google Scholar]
- Kaw R. N. & Khush G. S. Combining ability for low-temperature tolerance in rice. Proceedings of the International Rice Genetics Symposium. IRRI, Los Baños, Philippines. 593–612 (1986). [Google Scholar]
- Maruyama K. et al. Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 19, 37–49 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram J. & Bartels D. The Molecular basis of dehydration tolerance in plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 47, 377–403 (1996). [DOI] [PubMed] [Google Scholar]
- Kaye C. et al. Characterization of a gene for spinach CAP160 and expression of two spinach cold-acclimation proteins in tobacco. Plant Physiol. 116, 1367–1377 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde M., Dallaire S., N'Dong D. & Sarhan F. Overexpression of the acidic dehydrin WCOR410 improves freezing tolerance in transgenic strawberry leaves. Plant Biotechnol. J. 2, 381–387 (2004). [DOI] [PubMed] [Google Scholar]
- Puhakainen T. et al. Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol. Biol. 54, 743–753 (2004). [DOI] [PubMed] [Google Scholar]
- NDong C. et al. Cold-regulated cereal chloroplast late embryogenesis abundant-like proteins. Molecular characterization and functional analysis. Plant Physiol. 129, 1368–1381 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timperio A. M., Egidi M. G. & Zolla L. Proteomics applied on plant abiotic stresses: role of heat shock proteins (HSP). J. Proteomics 71, 391–411 (2008). [DOI] [PubMed] [Google Scholar]
- Wang L. et al. Identification of genes induced in response to low-temperature treatment in tea leaves. Plant Mol. Biol. Rep. 27, 257–265 (2009). [Google Scholar]
- Shimosaka E., Sasanuma T. & Handa H. A wheat cold-regulated cDNA encoding an early light-inducible protein (ELIP): its structure, expression and chromosomal location. Plant Cell Physiol. 40, 319–325 (1999). [DOI] [PubMed] [Google Scholar]
- Montané M. H., Dreyer S., Triantaphylides C. & Kloppstech K. Early light-induced proteins during long-term acclimation of barley to photooxidative stress caused by light and cold: high level of accumulation by post transcriptional regulation. Planta 202, 293–302 (1997). [Google Scholar]
- Hutin C. et al. Early light-induced proteins protect Arabidopsis from photooxidative stress. Proc. Natl. Acad. Sci. USA 100, 4921–4926 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274, 1900–1902 (1996). [DOI] [PubMed] [Google Scholar]
- Llop-Tous I., Dominguez-Puigjaner E. & Vendrell M. Characterization of a strawberry cDNA clone homologous to calcium-dependent protein kinases that is expressed during fruit ripening and affected by low temperature. J. Exp. Bot. 53, 2283–2285 (2002). [DOI] [PubMed] [Google Scholar]
- Saijo Y. et al. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 23, 319–327 (2000). [DOI] [PubMed] [Google Scholar]
- Komatsu S. et al. Over-expression of calcium-dependent protein kinase 13 and calreticulin interacting protein 1 confers cold tolerance on rice plants. Mol. Genet. Genomics 277, 713–723 (2007). [DOI] [PubMed] [Google Scholar]
- Swamy P. M. & Smith B. Role of abscisic acid in plant stress tolerance. Curr. Sci. 76, 1220–1227 (1999). [Google Scholar]
- Dreher K. A., Brown J., Saw R. E. & Callis J. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. The Plant Cell 18, 699–714 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G. & Guilfoyle T. J. ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865 (1997). [DOI] [PubMed] [Google Scholar]
- Lee B., Henderson D. A. & Zhu J. K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. The Plant Cell 17, 3155–3175 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanhui C. et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60, 107–24 (2006). [DOI] [PubMed] [Google Scholar]
- Zhu J. et al. HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc. Natl. Acad. Sci. USA 102, 9966–9971 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dai X. et al. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 143, 1739–1751 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal M. et al. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 49, 37636–37645 (2006). [DOI] [PubMed] [Google Scholar]
- Hannah M. A., Heyer A. G. & Hincha D. K. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet. 1, e26 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus N. N. et al. Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc. Natl. Acad. Sci. USA 93, 13404–13409 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennycooke J. C., Cheng H. & Stockinger E. J. Comparative genomic sequence and expression analyses of Medicago truncatula and alfalfa subspecies falcata COLD-ACCLIMATION-SPECIFIC genes. Plant Physiol. 146, 1242–1254 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali N. & Riley L. W. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics 8, 60 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S. J., Hajela R. K. & Thomashow M. F. Cold acclimation in Arabidopsis thaliana. Plant Physiol. 87, 745–750 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huner N. P. A., Oquist G. & Sarhan F. Energy balance and cold acclimation to light and cold. Trends Plant Sci. 3, 224–230 (1998). [Google Scholar]
- Athanasiou K., Dyson B. C., Webster R. E. & Johnson G. N. Dynamic acclimation of photosynthesis increases plant fitness in changing environments. Plant Physiol. 152, 366–373 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. S. et al. Molecular and functional characterization of CaLEA6, the gene for a hydrophobic LEA protein from Capsicum annuum. Gene 344, 115–123 (2005). [DOI] [PubMed] [Google Scholar]
- Wang H., Zhang H., Gao F., Li J. & Li Z. Comparison of gene expression between upland and lowland rice cultivars under water stress using cDNA microarray. Theor. Appl. Genet. 115, 1109–1126 (2007). [DOI] [PubMed] [Google Scholar]
- Ghawana S. et al. An RNA isolation system for plant tissues rich in secondary metabolites. BMC Res. Notes 4, 85 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A. & Kumar S. Response to winter dormancy, temperature and plant hormones shares gene networks, Funct. Integr. Genomics 11, 659–664 (2011). [DOI] [PubMed] [Google Scholar]
- Ouyang S. et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 35, D883–D887 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R. et al. S MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics. 8, 242 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitch L. V. et al. The effect of overexpression of two Brassica CBF/DREB1-like transcription factors on photosynthetic capacity and freezing tolerance in Brassica napus. Plant Cell Physiol. 46, 1525–1539 (2005). [DOI] [PubMed] [Google Scholar]
- Kumar N. et al. Effect of altitude on the primary products of photosynthesis and the associated enzymes in barley and wheat. Photosynth. Res. 88, 63–71 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information