Abstract
Sub-Saharan Africa is endemic for Kaposi’s sarcoma-associated herpesvirus (KSHV) and there is a high rate of early childhood infection; however, the transmission sources are not well characterized. We examined household members as potential KSHV transmission sources to young children in the KSHV-endemic country of Zambia. To this end, we enrolled and followed Zambian households with at least one KSHV-seropositive child and collected longitudinal buccal swab samples. KSHV burden was evaluated and K1 sequences from the children were determined and analyzed for differences to K1 sequences from household members. The K1 sequences were also analyzed for evolution over time. We generated K1 sequences from 31 individuals across 16 households. Nine households contained multiple KSHV-positive members, including at least one child. In 6 of 9 households, the child had 100% sequence identity to all household members. However, in two households the child and mother had distinct K1 sequences. In the remaining household, the children were the only KSHV-infected individuals. Furthermore, we report that 1 of 18 individuals had K1 sequence variation within the timespan analyzed. In the present study, we provide evidence that (1) early childhood KSHV transmission occurs from both within and outside the household, (2) intra-household transmission can occur via non-maternal sources, (3) viral shedding in the buccal cavity is highly variable, and (4) the dominant K1 sequence within an individual did not rapidly evolve over time. These results are important for developing KSHV intervention strategies.
Keywords: Kaposi’s sarcoma-associated herpesvirus, children, households, Zambia, K1, transmission
INTRODUCTION
Kaposi’s sarcoma-associated herpesvirus (KSHV), or human herpesvirus-8 (HHV-8), is the etiological agent of all forms of Kaposi’s sarcoma (KS), along with primary effusion lymphoma and multicentric Castleman’s disease.1–5 Global seroprevalence of KSHV is uneven; it is low in the United States and Western Europe, moderate in the Mediterranean, and as high as 80% in sub-Saharan Africa.6–-9 Zambia is one such country in sub-Saharan Africa stricken by high KSHV and KS prevalence, and with the onset of the HIV-1 epidemic, a drastic increase in KS cases was seen in both adults and children.10, 11 By 1992, KS accounted for approximately 25% of all childhood cancers diagnosed in Lusaka, the capitol of Zambia, making KS one of the most frequently diagnosed cancers.12
Since a KSHV vaccine is not likely to be available in the near future, elucidating the route and people involved as sources of transmission is imperative for the development of strategies to reduce KSHV spread and infection rates in endemic areas such as Zambia. Our previous study established that early childhood infection by KSHV is common in Zambia: approximately 40% of children were infected by four years of age.13 We and others have also shown that KSHV DNA is frequently detectable in saliva, indicating salivary contact as the major route of horizontal transmission to children in endemic regions.7, 14, 15 Nevertheless, the individuals involved as sources of KSHV transmission to children have yet to be identified. Previous studies of KSHV transmission have primarily focused on mother-to-child transmission, but salivary contact with children is not limited to the child’s mother. Moreover, we have shown that KSHV incidence is similar in children born to KSHV-positive and -negative mothers, suggesting that KSHV transmission can come from sources other than the mother.13 Therefore, to develop adequate KSHV intervention strategies in endemic areas, it is essential to evaluate the role of all personal contacts, including household members, as potential viral transmission sources during early childhood.
Despite a high level of genomic conservation, KSHV contains a gene on the extreme left-hand end of the genome, K1, that is highly variable.16 K1 is an 870-nucleotide gene that encodes a signal transducing, cell surface glycoprotein important in transformation of KSHV-infected and surrounding cells as well as induction of inflammatory proteins and downregulation of the B-cell receptor.17 The majority of K1 sequence variation is concentrated within two regions of the gene’s extracellular domain, variable region (VR) 1 and 2, which are 40- and 38-nucleotides respectively.16 Due to its high variability, K1 is routinely used to classify KSHV into at least five different genotypes (A, B, C, D, and E), and various sub-genotypes.16, 18
Using molecular analysis of K1, we examined a longitudinal cohort of KSHV-seropositive children and all members of their households in the KSHV endemic country of Zambia—making this study the first of its kind. Here, we provide evidence that transmission of KSHV to children can occur from both within and outside the household and intra-household transmission may occur via non-maternal sources. Furthermore, we report that superinfection was not detected in any individuals, and the dominant K1 sequence in the buccal cavity of an individual did not rapidly change over time. Additionally, we analyzed KSHV burden in the buccal cavity longitudinally and report that viral shedding is highly variable within an individual over time. Our findings have important implications for the development of strategies to prevent KSHV transmission to young children in endemic regions.
METHODS
Study Population
In the present study, participants were recruited from various compounds within Lusaka District, Zambia. We enrolled and followed all willing and eligible complete households (n = 134, 455 individuals) during September 2004 to November 2009. Eligible complete households were defined as family units in which (1) all related individuals who resided in the same dwelling agreed to participate in the study and (2) there was at least one KSHV-seropositive child under the age of four years (index child). Family units described in this study will herein be denoted as “households.” Venous blood and buccal swab samples were collected from each index child and mother every four months and from all other household members once a year at the University Teaching Hospital (UTH) in Lusaka for up to four years. KSHV serostatus was determined for each household member by monoclonal-enhanced immunofluorescence assay, as previously described.19 Additionally, presence of HIV-1 antibodies was determined for each individual as described previously.20 Trained nurses from the UTH provided information about the study and obtained written informed consent from participants or their guardians. This study was approved by the Institutional Review Board at the University of Nebraska-Lincoln and the Ethics Committee of the University of Zambia.
DNA extraction and polymerase chain reaction for KSHV detection
DNA was extracted from buccal swab samples using the Puregene Genomic DNA Purification Kit (Qiagen) according to manufacturer’s protocol. Extracted DNA was subjected to polymerase chain reaction (PCR) using primers for human β-actin (Actin1 [5’-TTCTACAATGAGCTGCGTGT-3’] and Actin2 [5’-GCCAGACAGCACTGTGTTGG-3’]) or GAPDH (GAPDH1 [5’-CCATGGAGAAGGCTGGGG-3’] and GAPDH2 [5’-CAAAGTTGTCATGGATGACC-3’]). Subsequently, PCR-positive samples were analyzed by nested PCR for presence of KSHV DNA using previously described primers for the ORF26 gene.21 Each PCR reaction was performed in a total volume of 25 µl using 0.4 µM primers and TaKaRa Ex Taq DNA polymerase kit (TaKaRa Biotechnology) according to manufacturer’s protocol, with the exception of 2.5 units enzyme. For β-actin and GAPDH reactions, 1 µl genomic DNA was used. For first- and second-round ORF26 PCR, 2 µl genomic DNA and 2 µl PCR product were used, respectively. All reactions were performed using the following conditions: 95°C for 5 min, 35 cycles of 95°C for 30 sec, 58°C for 30 sec, 72°C for 30 sec, and one cycle of 72°C for 7 min.
K1 sequencing
Nested primers were used to PCR amplify the K1 gene from KSHV DNA-positive buccal swab samples: outer primers K1-R2 (5’-AGTACCAATCCACTGGTTGCG-3’) and K1-1Wh (5’-TGTCTTTCAGACCTTGTTGG-3’); and inner primers K1-F1 (5’-ATGTTCCTGTATGTTGTCTGC-3’) and K1-4Wh (5’-TGGTTGCGTATAGTCTTCCG-3’). K1 PCR reaction conditions were similar to those described above, except the elongation temperature was 68°C. K1 PCR products were gel purified using the QIAQuick Gel Extraction Kit (Qiagen) according to manufacturer’s instructions. Purified K1 PCR products were sequenced with primers K1-F1 and OLK1R2 (5’-GCACTGTTTTGTTTGAGTCAC-3’) using the BigDye Terminator v3.1 Cycle Sequencing Kit and ABI Prism 3100-Avant DNA Sequencer (Applied Biosystems) according to manufacturer’s protocols.
Sequence analysis
A 624-nucleotide sequence (positions 100–723) that encompasses VR1 and VR2 of the K1 gene was examined. All sequences were analyzed using BLAST (National Center for Biotechnology Information) to ensure the amplicon was K1. Subsequently, K1 sequences from KSHV-infected individuals were aligned to sequences from members of the same household and inspected for differences using Vector NTI software (v11.0, Invitrogen). A maximum likelihood tree was constructed for all households using MEGA (v5.04). Additionally, longitudinal K1 sequences from each individual were aligned and inspected for differences using Vector NTI software. KSHV genotype of each individual was determined by K1 sequence alignment to reference strains from GenBank and construction of a maximum likelihood tree using MEGA (v5.04). K1 reference sequences used for analysis were AF133038 (A1), AF130305 (A2), U86667 (A3), AF133039 (A4), AF178823 (A5), AF133040 (B), AF133041 (C1), AF133042 (C3), AF133043 (D1), AF133044 (D2), and AF220292 (E). The K1 sequences generated in this study are available from GenBank under accession numbers JQ422520 to JQ422550.
Quantitative real-time PCR
Longitudinal samples with K1 sequence data were also analyzed by a duplex quantitative real-time PCR (qPCR) assay developed to simultaneously quantitate the number of KSHV genomic copies and cellular equivalents in each sample. All qPCR template samples were subjected to phenol-chloroform purification, according to standard methods, to ensure maximum purity for accurate qPCR analysis. Briefly, at least three rounds of phenol-chloroform and one chloroform purification, followed by ethanol precipitation, were performed on each DNA sample. qPCR reactions were performed using Taqman chemistry (Applied Biosystems). Each 25 µl reaction mixture contained the following: 100 ng genomic DNA template, 300 nM and 200 nM ORF26 forward and reverse nested primers respectively, 200 nM ORF26 dual-labeled probe RT26p (5’-FAM-CCATGGTCGTGCCGCACGCA-BHQ1-3’), 30 nM each β-globin primer described elsewhere,22 200 nM β-globin dual-labeled probe BGX1 (5’-HEX-CTCCTGAGGAGAAGTCTGCCGTTACTGCC-BHQ1-3’), 1× Taqman buffer, 5 mM MgCl2, 0.4 mM each dNTP, 0.0125 units Amplitaq Gold, and 0.01 units uracil-DNA glycosylase (UNG). All reactions were performed with the BioRad iCyclerIQ using the following conditions: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 sec, 55°C for 1 min. The standard for KSHV was generated using a sequence of the ORF26 gene cloned into the pCR2.1 vector (pCR2.1.ORF26) according to the manufacturer’s protocol (Invitrogen). pCR2.1.ORF26 copy number was calculated based on molecular mass. The β-globin standard was generated using genomic DNA extracted from the 8E5 cell line and copy number was calculated by qPCR to a known laboratory standard. The duplex standard was generated by mixing calculated copy numbers of pCR2.1.ORF26 and 8E5 genomic DNA in Tris-EDTA, followed by five serial 10-fold dilutions. The cycle threshold values from PCR amplification of ORF26 and β-globin in the duplex standard were used to generate a curve for each amplicon. The duplex standard curve was run in parallel with buccal swab samples for each reaction. KSHV copy number was calculated for each well and normalized to the β-globin equivalent. All samples were run in triplicate and the mean KSHV copy number per 10,000 cellular equivalents was calculated.
RESULTS
KSHV screening and genotype analysis
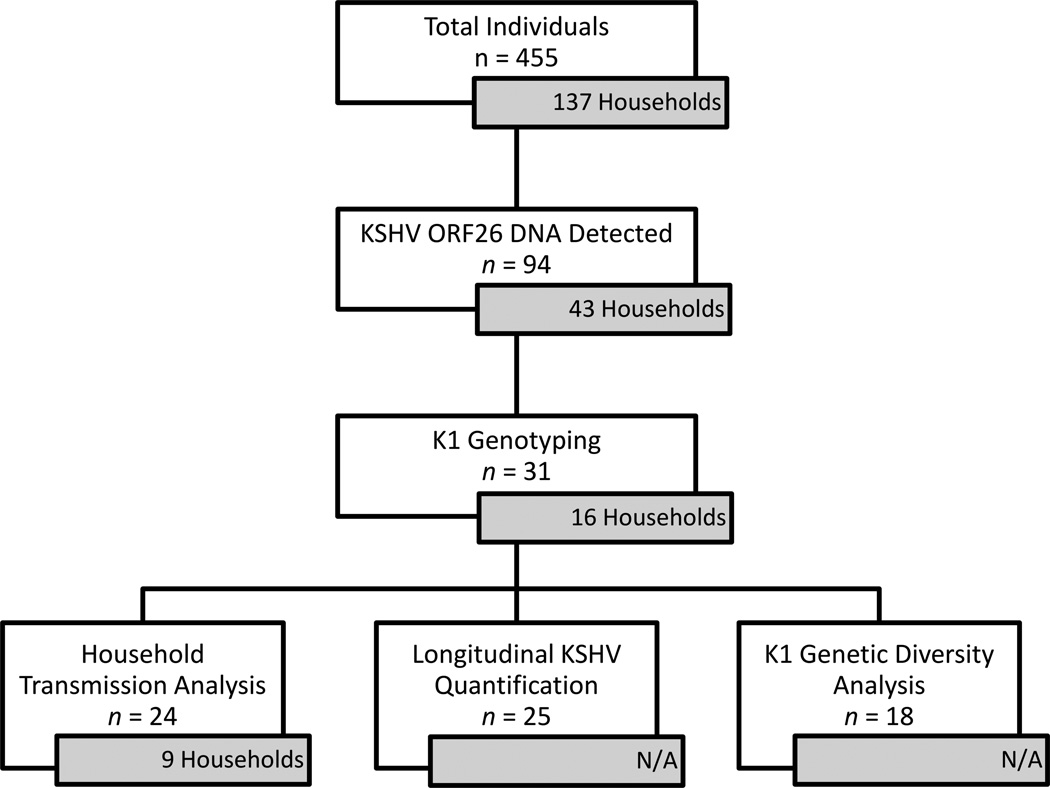
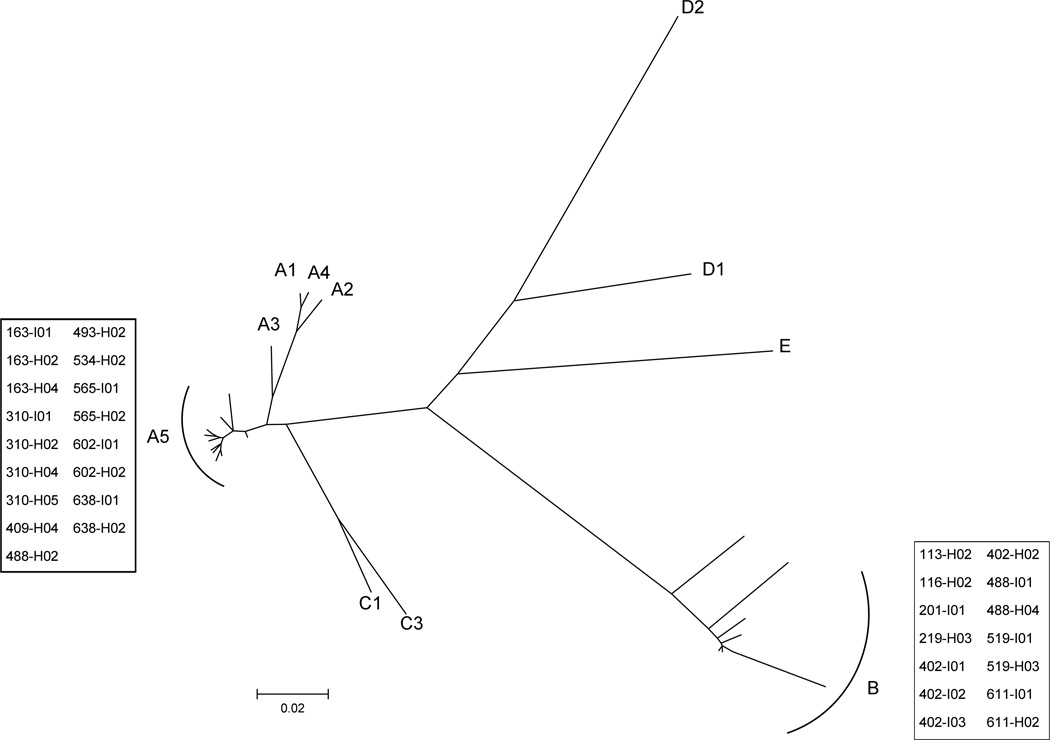
In the present study, we examined buccal swab samples from individuals (n = 455) of 137 complete households. Among these, 94 individuals from 43 households had KSHV DNA detectable by PCR in the buccal cavity and we attempted to amplify K1 for sequence analysis from these samples. Of the 94 KSHV DNA-positive individuals, the K1 gene was successfully amplified and sequenced in 31 individuals (12 index children and 19 household members) across 16 households (Fig. 1). A maximum likelihood tree with 11 K1 reference sequences was generated for KSHV genotype analysis of the 31 K1 sequences (Fig. 2). As expected, sequences from all 31 individuals clustered with genotypes A5 (n = 17) or B (n = 14), which are the most prevalent KSHV genotypes in Africa.23
Figure 1.
Flowchart summarizing sample analysis from a longitudinal cohort of Zambian households with at least one KSHV-positive child. Inset boxes indicate number of households comprised of analyzed individuals. N/A, not applicable.
Figure 2.
Phylogenetic analysis of K1 amino acid sequence to determine KSHV genotype of each individual. A maximum likelihood tree was generated with K1 sequences generated in this study and 11 prototypic reference sequences obtained from GenBank. Sequences generated in this study are boxed and reference sequences are labeled A1, A2, A3, A4, A5, B, C1, C3, D1, D2, and E.
KSHV transmission in complete households
Of the 16 households containing individuals in which K1 was sequenced, nine were examined for intra-household transmission of KSHV to children because K1 sequence data from the index child and at least one other household member was obtained. Table 1 summarizes demographic information collected from all members of these nine households (12 index children and 30 household members). Of note, all 12 children had detectable levels of KSHV antibodies in sera and 11 had detectable KSHV DNA shedding in the buccal cavity. Twenty-five of the 30 household members were KSHV-seropositive, of which 15 had detectable levels of KSHV DNA in the buccal cavity for at least one time point during the study. We were then able to obtain K1 sequence from all 11 KSHV DNA-positive index children and 13 of the 15 household members for at least one time point. Household members from whom K1 sequences were obtained were mothers and older siblings as KSHV DNA was not frequently detected in the buccal cavities of other household members.
Table 1.
Demographic Characteristics of Complete Households with K1 Sequence Data from Index Child and At Least One Other Household Member
|
Study ID |
Relation to Index Child |
Sex |
Age at Baseline |
HIV-1 Status |
KSHV Serostatus |
KSHV Buccal Shedding |
KSHV K1 Sequence |
KSHV Genotype |
|---|---|---|---|---|---|---|---|---|
| 163-I01 | Index | Male | 17 mo | − | + | + | + | A5 |
| 163-I02 | Index | Female | 7 mo | + | + | − | − | |
| 163-H02 | Mothera | Female | 30 yr | + | + | + | + | A5 |
| 163-H03 | Sister | Female | 12 yr | − | + | − | − | |
| 163-H04 | Brother | Male | 9 yr | − | + | + | + | A5 |
| 163-H05b | Father | Male | 45 yr | + | + | N/A | − | |
| 310-I01 | Index | Male | 12 mo | − | + | + | + | A5 |
| 310-H02 | Mothera | Female | 28 yr | − | + | + | + | A5 |
| 310-H03 | Father | Male | 30 yr | − | − | − | − | |
| 310-H04 | Brother | Male | 4 yr | − | + | + | + | A5 |
| 310-H05 | Brother | Male | 7 yr | − | + | + | + | A5 |
| 402-I01 | Index | Male | 21 mo | − | + | + | + | B |
| 402-I02 | Index | Female | 21 mo | − | + | + | + | B |
| 402-I03 | Index | Female | 9 mo | Indeterminate | + | + | + | B |
| 402-H02 | Mothera | Female | 23 yr | + | + | + | + | B |
| 402-H03 | Father | Male | 32 yr | − | + | + | − | |
| 488-I01 | Index | Male | 8 mo | − | + | + | + | B |
| 488-H02 | Mothera | Female | 30 yr | + | + | + | + | A5 |
| 488-H03 | Brother | Male | 8 yr | − | + | + | − | |
| 488-H04 | Sister | Female | 5 yr | − | + | + | + | B |
| 519-I01 | Index | Male | 21 mo | − | + | + | + | B |
| 519-H02 | Mothera | Female | 24 yr | + | − | − | − | |
| 519-H03 | Brother | Male | 3 yr | − | + | + | + | B |
| 565-I01 | Index | Male | 24 mo | + | + | + | + | A5 |
| 565-H02 | Mothera | Female | 29 yr | + | + | + | + | A5 |
| 565-H03 | Brother | Male | 7 yr | − | + | − | − | |
| 565-H04 | Sister | Female | 4 yr | + | + | − | − | |
| 602-I01 | Index | Female | 17 mo | + | + | + | + | A5 |
| 602-H02 | Mothera | Female | 33 yr | + | + | + | + | A5 |
| 602-H03 | Father | Male | 47 yr | + | + | − | − | |
| 602-H04 | Brother | Male | 10 yr | − | + | − | − | |
| 602-H05 | Brother | Male | 7 yr | − | − | − | − | |
| 602-H06 | Brother | Male | 4 yr | − | + | − | − | |
| 611-I01 | Index | Male | 18 mo | − | + | + | + | B |
| 611-H02 | Mothera | Female | 29 yr | − | + | + | + | B |
| 638-I01 | Index | Male | 18 mo | − | + | + | + | A5 |
| 638-H02 | Mothera | Female | 18 yr | + | + | + | + | A5 |
| 638-H03 | Grandmother | Female | 45 yr | + | − | − | − | |
| 638-H04 | Uncle | Male | 4 yr | − | − | − | − | |
| 638-H05 | Aunt | Female | 23 yr | − | + | − | − | |
| 638-H06 | Grandfather | Male | 44 yr | + | + | − | − | |
| 638-H07 | Cousin | Male | 2 yr | − | + | − | − | |
Abbreviations: HIV-1, human immunodeficiency virus type 1; KSHV, Kaposi's sarcoma-associated herpesvirus; mo, month; yr, year; N/A, not applicable.
Primary caregiver to index child.
Buccal swabs were not collected from this individual.
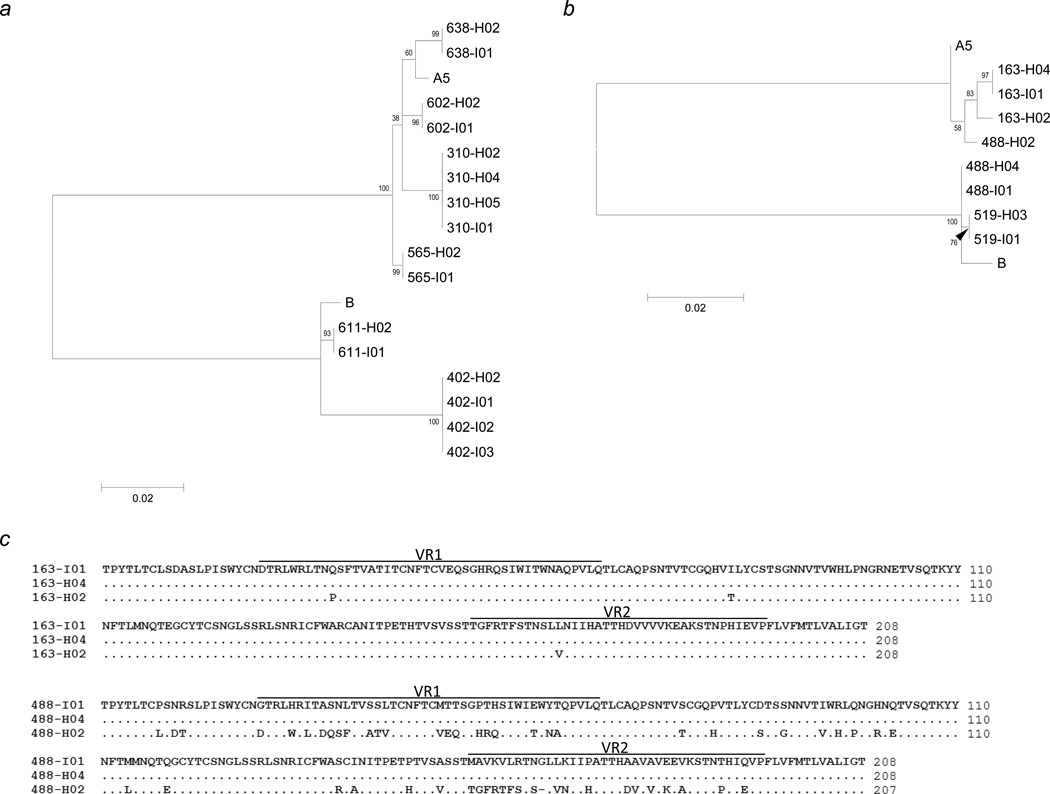
K1 sequences from the 11 index children and 13 household members of these nine households were aligned with A5 and B reference sequences and differences represented phylogenetically (Figs. 3a and 3b). In six households (310, 402, 565, 602, 611, and 638), the index child had 100% K1 sequence identity to all analyzed household members (Fig. 3a). Additionally, members from each household clustered to a distinct phylogenetic group with no overlap between households, providing evidence that cross-contamination between samples did not occur.
Figure 3.
Alignment of K1 sequences from members of nine households where sequence data was obtained from the index child and at least one other household member. Maximum likelihood phylogenetic tree of K1 nucleotide sequence from individuals indicating intra-household (a) and extra-household (b) transmission of KSHV to children. (c) K1 amino acid alignment for individuals of households 163 and 488. H02 designates the mother in each household.
Interestingly, in three other households (163, 488, and 519) the index child had 100% K1 sequence identity to a sibling but not the mother. For household 163, the K1 sequence of the mother was distinct from her two children, with three amino acid substitutions. For household 488, the mother’s K1 sequence differed from her children’s by 56 amino acids, consequently belonging to a different KSHV genotype: A5 versus B (Figs. 3b and 3c). For the remaining household (519), the two children had identical KSHV K1 sequence, despite the fact that the only other household member was KSHV-negative as determined by both antibody and DNA detection (Table 1 and Fig. 3b).
Longitudinal quantification of KSHV in the buccal cavity
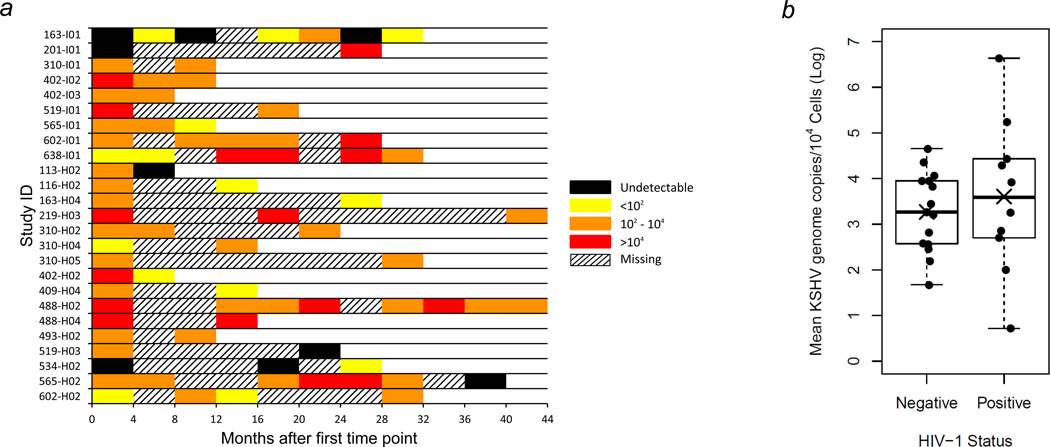
To further explore factors that may affect KSHV transmission within the household, we used qPCR to quantify KSHV burden in 25 of the 31 KSHV genotyped individuals, each of whom had KSHV-positive buccal swab samples at more than one time point (Fig. 1). Lack of sufficient quantities of sample or absence of KSHV DNA in previous detection assays prevented us from quantifying KSHV copy numbers for all time points. Figure 4a illustrates the KSHV shedding levels in the buccal cavity for each of the 25 individuals analyzed. Of note, 76% of individuals analyzed had levels of KSHV shedding that varied substantially. Furthermore, six individuals had undetectable KSHV shedding for at least one time point, which was preceded or succeeded by detectable levels of KSHV.
Figure 4.
Longitudinal quantification of KSHV burden by quantitative real-time PCR analysis of buccal swab samples. Buccal swab samples were collected from each index child and mother every four months and from all other household members once a year at UTH in Lusaka for up to four years. (a) Twenty-five individuals who had KSHV-positive buccal swab samples by PCR at two or more time points were analyzed. Index children are denoted with the letter “I”, and household members are denoted with the letter “H”. KSHV burden is reported as the number of KSHV genomes per 104 cellular genomes with a detection limit of 10 KSHV genomes. (b) Box and scatter plots of mean KSHV burden in buccal swab samples from 25 HIV-1 negative and positive individuals. Mean (X) and median (―) values are shown. p = 0.57, two-sided Student’s t-test.
To examine whether HIV-1 infection increases KSHV shedding in the buccal cavity, the mean number of KSHV copies/104 cells for all time points of each individual was compared between HIV-1 infected (n = 10) and uninfected (n = 15) individuals (Fig. 4b). Individuals who were HIV-1 positive had a marginally higher mean and median level of KSHV buccal shedding compared to uninfected individuals. However, there was no significant difference (p = 0.57) due to high variation and a small number of samples.
Lack of K1 genetic diversity over time within KSHV-infected individuals
We also sought to determine whether the dominant KSHV genotype in an individual changes over time, whether these changes correlate with transmission of variant genotypes within the same household and whether superinfection by other KSHV variants can occur. For each individual that we obtained K1 sequences from at least two samples over a minimum of 12 months (n = 18), we inspected sequences for evolution over time. Table 2 summarizes the number of samples analyzed, timespan of samples, and whether K1 sequence varies over time within each individual. Surprisingly, 17 of the 18 individuals had no variation in the dominant K1 sequence over the period analyzed, even up to 40 months. Only one individual (163-I01) had detectable K1 sequence variation: a single adenosine/guanosine mixed nucleotide that resulted in a cysteine/tyrosine mixture at the 100th amino acid position. However, the sequence variation was only observed at the sixth of seven follow up time points over the course of a 28-month period. At the subsequent time point, the variant reverted to the original sequence resulting in no net change in the dominant K1 sequence.
Table 2.
K1 Evolution Over Time
|
Study ID |
KSHV Genotype |
Samples |
Timespan (Months) |
K1 Sequence Variation |
|---|---|---|---|---|
| 116-H02 | B | 2 | 12 | − |
| 409-H04 | A5 | 2 | 12 | − |
| 519-I01 | B | 2 | 16 | − |
| 488-H04 | B | 2 | 16 | − |
| 310-H02 | A5 | 3 | 20 | − |
| 519-H03 | B | 3 | 20 | − |
| 201-I01 | B | 2 | 24 | − |
| 488-I01 | B | 2 | 24 | − |
| 163-H04 | A5 | 2 | 24 | − |
| 534-H02 | A5 | 3 | 24 | − |
| 602-I01 | A5 | 5 | 24 | − |
| 310-H05 | A5 | 2 | 28 | − |
| 602-H02 | A5 | 4 | 28 | − |
| 163-I01 | A5 | 7 | 28 | +a |
| 638-I01 | A5 | 7 | 28 | − |
| 565-H02 | A5 | 7 | 32 | − |
| 219-H03 | B | 3 | 40 | − |
| 488-H02 | A5 | 8 | 40 | − |
Variation was not sustained over time.
DISCUSSION
The principal objective of the present study was to elucidate the role of household members as transmission sources for early childhood infection of KSHV in an endemic setting. To this end, we used molecular analysis of viral sequences to link donor and recipient pairs. Our analysis of household K1 sequences revealed that in 6 of 9 households the index child had 100% K1 sequence identity to all other household members examined. This suggests that intra-household transmission is frequent and household practices common to all members within the household are the means by which KSHV contaminated saliva is transmitted. Indeed, Butler et al.24 showed that sharing food and/or sauce plates with other household members is associated with a child being KSHV seropositive. Therefore, if an infected household member is shedding KSHV in saliva, sharing household food/sauce plates may provide a route for transmission to all other household members.
Additionally, three households provided evidence of extra-household KSHV transmission. For two households (488 and 519), we are confident that the index child and sibling did not acquire KSHV infection from the mother as there was high K1 sequence divergence within household 488 and no maternal KSHV infection for household 519. For household 163, the K1 sequences obtained from the children and mother varied by <1.5%. Differences in K1 sequences could be a result of viral evolution in the children as suggested by Mbulaiteye et al.25 or a minor KSHV variant, undetectable by direct sequencing, may have been transmitted. Nevertheless, the observation that both children in each household had identical K1 sequences distinct from their mothers’ suggests that these children did not acquire KSHV infection from their mothers. Due to dense living conditions within compounds of Lusaka District, it is likely that these children acquired distinct KSHV variants from personal contact with community members outside of the household, which were not analyzed in the present study. Thus, our findings provide molecular evidence to support what previous reports have suggested: household members other than the mother, as well as personal contacts from outside the household may play an important role in KSHV transmission to children.24, 26, 27
We also report that viral burden in the buccal cavity is highly variable over time (e.g. from undetectable to high levels, and vice versa) for the majority of household members, highlighting the importance of longitudinal observation in KSHV transmission studies. This is consistent with reports in longitudinal cohorts of men who have sex with men in low infection prevalence settings.28, 29 Within our cohort, adults had periodic and robust lytic KSHV infection in the buccal cavity, which could increase the opportunity for transmission to children. Additionally, the high variability of viral shedding may partially explain a lack and/or inconsistency of correlation between KSHV infection and specific behaviors in which children are exposed to saliva, making it more difficult to identify the common practices that may increase the spread of KSHV in endemic areas.
Given the sequence diversity observed within K1, it is unclear whether viral quasispecies can develop in individuals during the course of KSHV infection. In the present study, we found that the dominant sequence of K1 in the buccal cavity of an individual did not change rapidly within the time analyzed. We observed sequence variation in only 1 of 18 individuals, suggesting that K1 evolution can occur within an individual; however, this variation was not sustained over time. Forward and reverse sequences were of excellent quality, indicating that the variation detected was genuine. However, it is possible that the mutation detected was a PCR artifact. In support of our finding that K1 evolution is possible within an individual, two recent studies have demonstrated that multiple KSHV variants and even closely related genotypes can exist within the same individual at a given time point.25, 30 In contrast to our study, these groups sequenced individual K1 clones. This method is advantageous for detecting minor sequence variants; nevertheless, a small number of clones can misrepresent the proportion of variants present in the individual. Hence, high quality direct sequencing of PCR product is ideally suited to determine the dominant sequences present within a sample. Our findings suggest that although ongoing evolution of the K1 gene can occur in an individual, the level of variants is minor in the scope of the individual’s entire viral population and that K1 evolution does not induce a shift in the dominant viral genotype within an individual over the analyzed time.
One limitation of our study is the small final sample size. In our cohort, we detected KSHV DNA via PCR in 94 out of 455 individuals. This is not unexpected as not all household members were KSHV-seropositive. Moreover, a number of previous reports demonstrated that KSHV DNA is not always detectable in the buccal cavity of KSHV-seropositive individuals.14, 15, 25 Further limiting the number of households available for analysis, the K1 gene has high sequence variability resulting in PCR detection and sequencing that is not as sensitive as for the conserved ORF26 gene. Another limitation of our study is the use of households where children were already KSHV-seropositive, preventing us from directly correlating levels of household member KSHV shedding in the buccal cavity to the time of childhood infection.
Despite these limitations, our study is significant as we were able to analyze samples from complete households in a longitudinal cohort within an endemic area. The cohort analyzed was representative of urban families in Zambia, and to our knowledge child care practices do not differ significantly in the rural setting. However, to further substantiate our findings in other endemic areas, it will be important to correlate KSHV infection in household/community members with viral transmission to young children in those areas. The findings of our study are important for developing strategies—such as behavior modification—to prevent KSHV transmission during early childhood and therefore reduce KS incidence in endemic areas.
ACKNOWLEGMENTS
We thank all study members from Lusaka for their participation in the study. We thank Veenu Minhas, Kay Crabtree, and all community health workers and laboratory staff at UTH for their contributions to recruitment, data management, and sample processing. We also thank Sandra Gonzalez for technical assistance and Jesse Thompson for helpful discussion. This work was supported by the National Institutes of Health (NIH; RO1 CA75903, T32 AIO60547, and P30 GM103509 to C.W.) and the Fogarty International Center (D43 TW01492 to C.W.). L.N.O. is supported by the NIH under a Ruth L. Kirschstein National Research Service Award from the National Institute of Allergy and Infectious Diseases, and C.G. is a Fogarty Fellow.
Footnotes
The present study is the first to describe a molecular analysis of Kaposi’s sarcoma-associated herpesvirus (KSHV) transmission to children in a longitudinal cohort of KSHV-positive children and their entire households in Zambia—a country endemic for KSHV and Kaposi’s sarcoma. Our results have important implications for the development of strategies to prevent KSHV transmission to young children and reduce Kaposi’s sarcoma incidence in endemic areas.
Contributor Information
Landon N. Olp, Nebraska Center for Virology and School of Biological Sciences, University of Nebraska-Lincoln, Lincoln, NE 68583 USA
Danielle M. Shea, Nebraska Center for Virology and School of Biological Sciences, University of Nebraska-Lincoln, Lincoln, NE 68583 USA
Maxine K. White, Nebraska Center for Virology and School of Biological Sciences, University of Nebraska-Lincoln, Lincoln, NE 68583 USA
Clement Gondwe, Department of Paediatrics and Child Health, University Teaching Hospital, 10101 Lusaka, Zambia.
Chipepo Kankasa, Department of Paediatrics and Child Health, University Teaching Hospital, 10101 Lusaka, Zambia.
Charles Wood, Nebraska Center for Virology and School of Biological Sciences, University of Nebraska-Lincoln, Lincoln, NE 68583 USA.
REFERENCES
- 1.Huang YQ, Li JJ, Kaplan MH, Poiesz B, Katabira E, Zhang WC, Feiner D, Friedman-Kien AE. Human herpesvirus-like nucleic acid in various forms of Kaposi's sarcoma. Lancet. 1995;345:759–761. doi: 10.1016/s0140-6736(95)90641-x. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 3.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 4.Schulz TF. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) J Gen Virol. 1998;79(Pt 7):1573–1591. doi: 10.1099/0022-1317-79-7-1573. [DOI] [PubMed] [Google Scholar]
- 5.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay MF, Clauvel JP, Raphael M, Degos L, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 6.Dedicoat M, Newton R. Review of the distribution of Kaposi's sarcoma-associated herpesvirus (KSHV) in Africa in relation to the incidence of Kaposi's sarcoma. Br J Cancer. 2003;88:1–3. doi: 10.1038/sj.bjc.6600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mbulaiteye SM, Pfeiffer RM, Engels EA, Marshall V, Bakaki PM, Owor AM, Ndugwa CM, Katongole-Mbidde E, Goedert JJ, Biggar RJ, Whitby D. Detection of kaposi sarcoma-associated herpesvirus DNA in saliva and buffy-coat samples from children with sickle cell disease in Uganda. J Infect Dis. 2004;190:1382–1386. doi: 10.1086/424489. [DOI] [PubMed] [Google Scholar]
- 8.Gao SJ, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo CR, Saah A, Phair J, Detels R, Chang Y, et al. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 9.Simpson GR, Schulz TF, Whitby D, Cook PM, Boshoff C, Rainbow L, Howard MR, Gao SJ, Bohenzky RA, Simmonds P, Lee C, de Ruiter A, et al. Prevalence of Kaposi's sarcoma associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet. 1996;348:1133–1138. doi: 10.1016/S0140-6736(96)07560-5. [DOI] [PubMed] [Google Scholar]
- 10.Patil P, Elem B, Zumla A. Pattern of adult malignancies in Zambia (1980–1989) in light of the human immunodeficiency virus type 1 epidemic. J Trop Med Hyg. 1995;98:281–284. [PubMed] [Google Scholar]
- 11.Patil PS, Elem B, Gwavava NJ, Urban MI. The pattern of paediatric malignancy in Zambia (1980–1989): a hospital-based histopathological study. J Trop Med Hyg. 1992;95:124–127. [PubMed] [Google Scholar]
- 12.Chintu C, Athale UH, Patil PS. Childhood cancers in Zambia before and after the HIV epidemic. Arch Dis Child. 1995;73:100–104. doi: 10.1136/adc.73.2.100. discussion 04-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minhas V, Crabtree KL, Chao A, M'Soka TJ, Kankasa C, Bulterys M, Mitchell CD, Wood C. Early childhood infection by human herpesvirus 8 in Zambia and the role of human immunodeficiency virus type 1 coinfection in a highly endemic area. Am J Epidemiol. 2008;168:311–320. doi: 10.1093/aje/kwn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brayfield BP, Kankasa C, West JT, Muyanga J, Bhat G, Klaskala W, Mitchell CD, Wood C. Distribution of Kaposi sarcoma-associated herpesvirus/human herpesvirus 8 in maternal saliva and breast milk in Zambia: implications for transmission. J Infect Dis. 2004;189:2260–2270. doi: 10.1086/421119. [DOI] [PubMed] [Google Scholar]
- 15.de Souza VA, Sumita LM, Nascimento MC, Oliveira J, Mascheretti M, Quiroga M, Freire WS, Tateno A, Boulos M, Mayaud P, Pannuti CS. Human herpesvirus-8 infection and oral shedding in Amerindian and non-Amerindian populations in the Brazilian Amazon region. J Infect Dis. 2007;196:844–852. doi: 10.1086/520549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zong JC, Ciufo DM, Alcendor DJ, Wan X, Nicholas J, Browning PJ, Rady PL, Tyring SK, Orenstein JM, Rabkin CS, Su IJ, Powell KF, et al. High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi's sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J Virol. 1999;73:4156–4170. doi: 10.1128/jvi.73.5.4156-4170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brinkmann MM, Schulz TF. Regulation of intracellular signalling by the terminal membrane proteins of members of the Gammaherpesvirinae. J Gen Virol. 2006;87:1047–1074. doi: 10.1099/vir.0.81598-0. [DOI] [PubMed] [Google Scholar]
- 18.Cook PM, Whitby D, Calabro ML, Luppi M, Kakoola DN, Hjalgrim H, Ariyoshi K, Ensoli B, Davison AJ, Schulz TF. Variability and evolution of Kaposi's sarcoma-associated herpesvirus in Europe and Africa. International Collaborative Group. AIDS. 1999;13:1165–1176. doi: 10.1097/00002030-199907090-00004. [DOI] [PubMed] [Google Scholar]
- 19.Minhas V, Crosby LN, Crabtree KL, Phiri S, M'Soka TJ, Kankasa C, Harrington WJ, Mitchell CD, Wood C. Development of an immunofluorescence assay using recombinant proteins expressed in insect cells to screen and confirm presence of human herpesvirus 8-specific antibodies. Clin Vaccine Immunol. 2008;15:1259–1264. doi: 10.1128/CVI.00487-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minhas V, Crabtree KL, Chao A, Wojcicki JM, Sifuniso AM, Nkonde C, Kankasa C, Mitchell CD, Wood C. The Zambia Children's KS-HHV8 Study: rationale, study design, and study methods. Am J Epidemiol. 2011;173:1085–1092. doi: 10.1093/aje/kwq465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mantina H, Kankasa C, Klaskala W, Brayfield B, Campbell J, Du Q, Bhat G, Kasolo F, Mitchell C, Wood C. Vertical transmission of Kaposi's sarcoma-associated herpesvirus. Int J Cancer. 2001;94:749–752. doi: 10.1002/ijc.1529. [DOI] [PubMed] [Google Scholar]
- 22.Gorry PR, Bristol G, Zack JA, Ritola K, Swanstrom R, Birch CJ, Bell JE, Bannert N, Crawford K, Wang H, Schols D, De Clercq E, et al. Macrophage Tropism of Human Immunodeficiency Virus Type 1 Isolates from Brain and Lymphoid Tissues Predicts Neurotropism Independent of Coreceptor Specificity. Journal of Virology. 2001;75:10073–10089. doi: 10.1128/JVI.75.21.10073-10089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayward GS, Zong JC. Modern evolutionary history of the human KSHV genome. Curr Top Microbiol Immunol. 2007;312:1–42. doi: 10.1007/978-3-540-34344-8_1. [DOI] [PubMed] [Google Scholar]
- 24.Butler LM, Were WA, Balinandi S, Downing R, Dollard S, Neilands TB, Gupta S, Rutherford GW, Mermin J. Human herpesvirus 8 infection in children and adults in a population-based study in rural Uganda. J Infect Dis. 2011;203:625–634. doi: 10.1093/infdis/jiq092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mbulaiteye S, Marshall V, Bagni RK, Wang CD, Mbisa G, Bakaki PM, Owor AM, Ndugwa CM, Engels EA, Katongole-Mbidde E, Biggar RJ, Whitby D. Molecular evidence for mother-to-child transmission of Kaposi sarcoma-associated herpesvirus in Uganda and K1 gene evolution within the host. J Infect Dis. 2006;193:1250–1257. doi: 10.1086/503052. [DOI] [PubMed] [Google Scholar]
- 26.Butler LM, Neilands TB, Mosam A, Mzolo S, Martin JN. A population-based study of how children are exposed to saliva in KwaZulu-Natal Province, South Africa: implications for the spread of saliva-borne pathogens to children. Trop Med Int Health. 2010;15:442–453. doi: 10.1111/j.1365-3156.2010.02474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mbulaiteye SM, Pfeiffer RM, Whitby D, Brubaker GR, Shao J, Biggar RJ. Human herpesvirus 8 infection within families in rural Tanzania. J Infect Dis. 2003;187:1780–1785. doi: 10.1086/374973. [DOI] [PubMed] [Google Scholar]
- 28.Casper C, Krantz E, Selke S, Kuntz SR, Wang J, Huang ML, Pauk JS, Corey L, Wald A. Frequent and asymptomatic oropharyngeal shedding of human herpesvirus 8 among immunocompetent men. J Infect Dis. 2007;195:30–36. doi: 10.1086/509621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Widmer IC, Erb P, Grob H, Itin P, Baumann M, Stalder A, Weber R, Cathomas G. Human herpesvirus 8 oral shedding in HIV-infected men with and without Kaposi sarcoma. J Acquir Immune Defic Syndr. 2006;42:420–425. doi: 10.1097/01.qai.0000226790.31463.e6. [DOI] [PubMed] [Google Scholar]
- 30.Al-Otaibi LM, Al-Sulaiman MH, Teo CG, Porter SR. Extensive oral shedding of human herpesvirus 8 in a renal allograft recipient. Oral Microbiol Immunol. 2009;24:109–115. doi: 10.1111/j.1399-302X.2008.00481.x. [DOI] [PubMed] [Google Scholar]