Abstract
Background
A variety of species and experimental designs have been used to study genetic influences on alcohol dependence, ethanol response, and related traits. Integration of these heterogeneous data can be used to produce a ranked target gene list for additional investigation.
Results
In this study, we performed a unique multi-species evidence-based data integration using three microarray experiments in mice or humans that generated an initial alcohol dependence (AD) related genes list, human linkage and association results, and gene sets implicated in C. elegans and Drosophila. We then used permutation and false discovery rate (FDR) analyses on the genome-wide association studies (GWAS) dataset from the Collaborative Study on the Genetics of Alcoholism (COGA) to evaluate the ranking results and weighting matrices. We found one weighting score matrix could increase FDR based q-values for a list of 47 genes with a score greater than 2. Our follow up functional enrichment tests revealed these genes were primarily involved in brain responses to ethanol and neural adaptations occurring with alcoholism.
Conclusions
These results, along with our experimental validation of specific genes in mice, C. elegans and Drosophila, suggest that a cross-species evidence-based approach is useful to identify candidate genes contributing to alcoholism.
Background
Research on the genetics and neurobiology of alcoholism uses a variety of study designs and model organisms. A wealth of data are available, including linkage studies in human alcoholics, microarray studies of inbred mouse strains' brains and rat brains exposed to ethanol, and studies of loss or gain of function of genes in organisms such as C. elegans and Drosophila [1,2]. Although results or information across experiments are often compared by individual researchers in order to generate hypotheses, interpret results, or prioritize targets for follow up investigations [3], these analyses are not always done comprehensively and rarely include a cross-species approach [4-7]. While data integration itself can be challenging, how best to utilize combined results is also unclear. Although pooled results may yield valuable insights, there are potential benefits of using more systematic approaches to generate quantitative rankings that can then, in turn, guide additional studies. In particular, these rankings could be applied to choosing molecular targets for knockdown studies in model organisms or genetic association studies in humans. For this and other approaches, evaluation is often needed in order to determine whether the rankings are effective at the end of the data integration process.
Challenges exist for each stage of an integration process, including the creation of an empirical gene list across species and platforms, scoring the information, and then evaluating the scoring system itself. For example, once various data are collected, identifying the best way to integrate them poses a problem since the criteria for selecting gene lists often differ substantially across studies. Specifically in microarray studies, the expression of gene-specific transcripts is selected via statistical threshold(s), but individual genes can have multiple transcripts that may differ in their abundance [8]. Therefore, a given gene can yield multiple expression values through microarray or next generation RNA sequencing (RNA-Seq) analyses. Likewise, human genetic association studies test multiple genetic markers, usually single nucleotide polymorphisms (SNPs), across a gene. In contrast, the results of genetic linkage or quantitative trait locus (QTL) studies in humans or mice can span tens of megabases and contain potentially hundreds of genes. Furthermore, low replication rates and identification of non-functional markers in most studies makes the search for true genetic signals difficult [9-11]. While there are issues with data reduction or summarization, integration at the level of the gene can be used as a link across a number of commonly used approaches.
If genetic information is summarized at the gene level, then each gene in the genome can be assigned a score for each experiment or data set available. This measurement can be quantitative or qualitative. For example, p-values may be assigned to a gene within a quantitative trait locus (QTL) or a linkage region. However, differences in gene-specific p-values within an interval of interest may be misleading since linkage peaks can shift, and variants responsible for the linkage may not be at the peak itself. In contrast, large numbers of genes may show altered expression in microarray studies and represent real changes due to signal cascades affecting entire gene networks. These correlated expression networks, in which a large number of changes are expected, contrast with linkage regions, in which most if not all genes do not actually contain variant(s) linked to the disease. A combined p-value method can be used for quantitative analyses, but this approach may present its own challenges. The individual data sources may not be weighted equally since the relative magnitudes of the p-values can be vastly different across platforms (e.g., mouse and human QTL studies). To avoid such issues, qualitative scores that measure the presence or absence of evidence above a threshold may be used, but thresholds have their own problems. Regardless of scoring choice, and despite some problems associated with each, a combined gene rank score can be generated from data integration. These gene rank scores can be used to perform weighted analysis or to define gene subsets for further investigation.
The effectiveness of such ranking can be verified by conducting further testing on genes ranked highly in the analyses. Alternatively, because the design of genome-wide association studies (GWAS) is hypothesis free, this approach offers opportunities to empirically test a ranking method and provide insight into further refinement, and all or most potential candidate genes can be tested in one experiment. If higher ranked genes contain more significant SNPs than a random set of genes, then the utility of a cross-species and platform integration and ranking approach would be demonstrated [12]. In this report, we attempt to implement and evaluate the utility of the approach outlined above by collecting data across species and approaches, summarizing at the gene level, ranking the genes, and testing the rankings in complex traits related to alcoholism and ethanol response. We included data generated from ethanol response experiments because this trait is one of the contributing factors for alcoholism [13].
Results
Ranked gene list
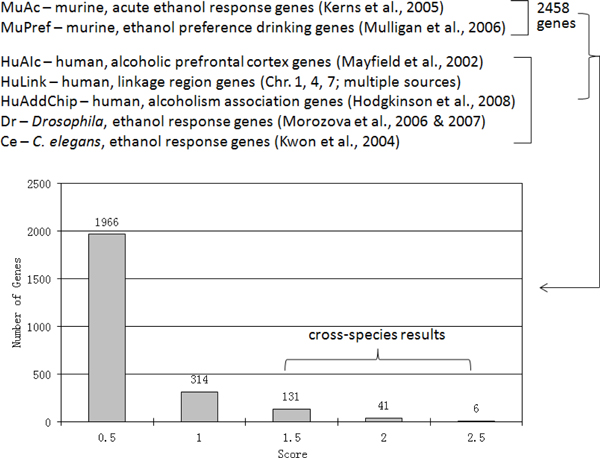
An initial list of 2458 genes that show altered expression in mouse brain in response to ethanol in two previous studies [3,14] was used as a starting point. These datasets were abbreviated as MuAc and MuPref (see Figure 1). Five additional data sources were used to construct a score for these genes, including 1) genes showing altered expression in the prefrontal cortex of human alcoholics (abbreviated as HuAlc) [15], 2) linkage intervals from published studies of the Collaborative Study on the Genetics of Alcoholism (COGA) and the Irish Study of Alcoholism samples (abbreviated as HuLink) [16-18], 3) genes contained on a human addiction/alcoholism array (abbreviated as HuAddChip) [19], 4) those from a smaller list of ethanol-related genes compiled from Drosophila (abbreviated as Dr) [20,21], and 5) a short list of ethanol-related genes compiled from C. elegans (abbreviated as Ce) [22]. Additionally, genes having cross-species hits acquired bonus scores (the "Cross" score in our algorithm, see Table 1), as cross-species evidence was regarded as an important factor in gene salience. Here, we used score to estimate the evidence of a gene, rather than using a quantitative measurement (e.g., significance level, see section Materials and methods). We proposed 10 weighting score matrices (Table 1). The corresponding ranking results are shown in Table 2.
Figure 1.
Data sources and ranking score results using weighting score matrix 3. The details of weighting score matrix 3 are provided in Table 1.
Table 1.
Ten weighting score matrices used in cross-species data integration and gene ranking.
| Weighting score matrix (WSM) | Description |
|---|---|
| WSM1: (0.5,0.5,1,1,0.5,0.5,0.5,1) * | HuAlc, HuAddChip, and Cross ** given 1, all others 0.5 |
| WSM2: (0.5,0.5,1,1,0.5,0.5,0.5,0.5) | HuAlc and HuAddChip given 1, all others 0.5 |
| WSM3: (0.5,0.5,0.5,1,0.5,0.5,0.5,0.5) | HuAddChip given 1, all others 0.5 |
| WSM4: (0.5,0.5,1,1,1,0.5,0.5,0.5) | All human data given 1, all others 0.5 |
| WSM5: (1,1,1,1,0.5,0.5,0.5,0.5) | All mouse data, human HuAlc and HuAddChip given 1, all others 0.5 |
| WSM6: (1,1,0.5,0.5,0.5,0.5,0.5,0.5) | All mouse data given 1, all others 0.5 |
| WSM7: (1,1,1,1,1,0.5,0.5,0.5) | All mouse and human data given 1, all others 0.5 |
| WSM8: (0.5,0.5,0.5,1,0.5,0.5,0.5,1) | HuAddChip and Cross given 1, all others 0.5 |
| WSM9: (1,1,1,1,1,1,1,1) | All given 1 |
| WSM10: (1,1,1,1,1,1,1,0.5) | All but Cross given 1 |
* The weights in the parentheses are in the order for the datasets: MuAc, MuPref, HuAlc, HuAddChip, HuLink, Ce, Dr, Cross, respective. The description of these datasets is provided in Figure 1.
** Cross denotes the bonus score for genes who had cross-species hits except for genes from human linkage regions (see section Materials and methods).
Table 2.
Summary of the genes by their scores using 10 weighting matrices*.
| Weighting matrix |
Score | |||||||
|---|---|---|---|---|---|---|---|---|
| 0.5 | 1 | 1.5 | 2 | 2.5 | 3 | 3.5 | 4 | |
| 1 | 1722 | 535 | 40 | 38 | 102 | 21 | ||
| 2 | 1722 | 535 | 78 | 102 | 21 | |||
| 3 | 1966 | 314 | 131 | 41 | 6 | |||
| 4 | 1722 | 338 | 235 | 137 | 16 | 10 | ||
| 5 | 1966 | 220 | 137 | 112 | 15 | 8 | ||
| 6 | 244 | 1745 | 202 | 222 | 34 | 11 | ||
| 7 | 1966 | 357 | 89 | 29 | 17 | |||
| 8 | 1966 | 314 | 17 | 114 | 41 | 6 | ||
| 9 | 1966 | 314 | 149 | 29 | ||||
| 10 | 1966 | 314 | 132 | 17 | 29 | |||
* For each matrix, scores of each gene were summarized based on its evidence in each dataset (see details in Table 1). Then, the number of genes in each score range was counted and summarized in this table.
Alcohol dependence GWAS analysis for ranked genes
To assess the performance of our ranking algorithm and weighting score matrices, we explored whether the ranked genes showed non-random enrichment of significant signals in alcoholism GWAS results. Specifically, we examined enriched association signals of ranked genes in the COGA GWAS [23], one of the largest alcohol dependence GWAS datasets. To increase the effect of our analyses we filtered the data for minor allele frequency, Hardy-Weinberg equilibrium deviation, and failure rate (see section Materials and methods). This resulted in 958,380 SNPs in our follow up analysis, with an observed minimum p-value of 9.5 × 10-7. However, the minimum q-value was 0.605 after False Discovery Rate (FDR) analysis. Of those SNPs approximately 68.7% (658,008/958,380) mapped to the human non-pseudogenes in NCBI Entrez Gene database, and they were used in this study. FDR analysis was then performed on restricted subsets of markers based on gene rank score.
For each ranked gene under different weighting score matrices, we calculated the q-value of each SNP in the gene from the COGA GWAS data. The results for the ten weighting score matrices were summarized in Additional file 1. For each weighting score matrix, its gene ranking performance was expected to increase by improving the q-values of SNPs mapped in the ranked genes. To quantitatively measure performance and correct for gene size, we conducted 100 simulations, in each of which the same number of genes were randomly chosen from the whole gene set. FDR based q-value analysis was then performed on the GWAS genotyped SNPs that mapped to the randomly chosen genes. The proportion of q-values in each q-value bin (e.g., 0.1-0.2) was calculated and then compared with those from the actual ranked alcohol genes. For the simplicity of comparison, we separated q-values into different bins. The results are shown in Additional file 1.
According to our permutation results, the weighting score matrix 3 had the best performance, since it gave the lowest q-values among genes (Additional file 1). This matrix was then used to refine gene scores using 1000 permutations (Table 3). In general, the subset of SNP results restricted to the scored genes was enriched for significant effects as the gene rank score increased from 0.5 to 2.0 (see Table 4). Specifically, the minimum FDR based q-value was 0.605 for all SNPs passing QC. The minimum q-value decreased for SNPs in all scored genes, but then increased for genes with score ≥ 1 or ≥ 1.5. However, the minimum q-value became the smallest (0.357) when this analysis was applied to genes with score ≥ 2. There were 47 genes whose scores were ≥ 2, and a total of 2293 SNPs mapped to these genes. For this gene subset, we found many more SNPs having small q values, including 27 SNPs with q-value < 0.4 and 39 SNPs with q-value < 0.5, than those in other gene sets (e.g., gene subset with score ≥ 1.5 or any scored, Table 4). Although this q-value analysis was not perfect (e.g., we did not find a steady decrease of q-value by increasing gene score, Table 4), it suggests that multi-species gene ranking by optimal weighting matrix might be effective for prioritizing candidate genes for complex traits.
Table 3.
Empirical p-values estimated from 1000 permutations based on weighting score matrix 3.
| q-value | Gene score | ||||
|---|---|---|---|---|---|
| ≥0.5 | ≥1 | ≥1.5 | ≥2 | = 2.5 | |
| q < 0.9 | 0.393 (6399) | 0.367 (1863) | 0.603 (399) | 0.191 (866) | 0.052 (178) |
| q < 0.8 | 0.525 (469) | 0.292 (415) | 0.441 (163) | 0.310 (199) | 0.131 (108) |
| q < 0.7 | 0.495 (8) | 0.228 (164) | 0.627 (5) | 0.208 (117) | 0.118 (72) |
| q < 0.6 | 0.286 (5) | N/A | N/A | 0.225 (42) | 0.093 (53) |
| q < 0.5 | 0.113 (5) | N/A | N/A | 0.122 (39) | N/A |
| q < 0.4 | 0.012 (5) | N/A | N/A | 0.101 (27) | N/A |
Based on Table 2, we used weight matrix 3 to rank genes (a total of 2458 genes) and then separated them according to their q-values. The number of SNPs in each q-value and score category based on COGA dataset is shown in parentheses. N/A: not available due to absence of the real data at those categories.
Table 4.
Improvement in GWAS-based association FDR as multi-species gene ranking score increases.
| FDR q-value |
Total SNPs passing QC | Scored | Score ≥1 | Score ≥1.5 | Score ≥2 | Score = 2.5 |
|---|---|---|---|---|---|---|
| All SNPs | 547920 | 91774 | 18988 | 7948 | 2293 | 210 |
| < 0.9 | 48016 | 6399 | 1863 | 399 | 866 | 178 |
| < 0.8 | 643 | 469 | 415 | 163 | 199 | 108 |
| < 0.7 | 6 | 8 | 164 | 5 | 117 | 72 |
| < 0.6 | 0 | 5 | 0 | 0 | 42 | 53 |
| < 0.5 | 0 | 5 | 0 | 0 | 39 | 0 |
| < 0.4 | 0 | 5 | 0 | 0 | 27 | 0 |
| < 0.3 | 0 | 0 | 0 | 0 | 0 | 0 |
| < 0.2 | 0 | 0 | 0 | 0 | 0 | 0 |
| < 0.1 | 0 | 0 | 0 | 0 | 0 | 0 |
| < 0.05 | 0 | 0 | 0 | 0 | 0 | 0 |
| Min q | 0.605 | 0.369 | 0.667 | 0.636 | 0.357 | 0.526 |
Bioinformatics analysis of top ranked genes
As presented above and detailed in Table 5, 47 genes with score ≥ 2 had promising q-value improvement, and were used as our high priority list for follow up bioinformatics analysis. These genes also had evidence from at least 2 different species (Figure 1). We first performed functional enrichment analysis of Gene Ontology (GO) terms implemented in the WebGestalt tool. In this tool, each gene set is tested its functional enrichment with GO annotations based on the hypergeometric test. As shown in Table 6 the most significantly enriched functional terms belonged to the groupings of neurotransmitter receptor activity, ion binding, and synaptic structure. The most significantly enriched functional terms were behavior (pBH = 5.08 × 10-5), gamma-aminobutyric acid (GABA) signaling pathway (pBH = 8.51 × 10-5) and cell communication (pBH = 0.0001) in the GO category of "Biological Process"; GABA receptor activity (pBH = 3.35 × 10-9), GABA-A receptor activity (pBH = 7.38 × 10-8), and neurotransmitter binding (pBH = 7.38 × 10-8) in the category of "Molecular Function"; and postsynaptic membrane (pBH = 1.86 × 10-7), chloride channel complex (pBH = 2.76 × 10-7), and synapse part (pBH = 3.36 × 10-7) in the category of "Cellular Component". Many of these enriched functional categories are consistent with the current knowledge of alcohol dependence and ethanol response [24]. These indicate that the top ranked genes are highly enriched in functions relevant to alcoholism.
Table 5.
47 genes with ranked score ≥ 2 using weighting score matrix 3.
| Gene symbol | Gene ID | Score |
|---|---|---|
| TAC1 | 6863 | 2.5 |
| JUN | 3725 | 2.5 |
| GABRB1 | 2560 | 2.5 |
| GABRA2 | 2555 | 2.5 |
| CCKBR | 887 | 2.5 |
| CCK | 885 | 2.5 |
| TMEM165 | 55858 | 2 |
| TIMP2 | 7077 | 2 |
| TH | 7054 | 2 |
| SPP1 | 6696 | 2 |
| SLC6A11 | 6538 | 2 |
| PURB | 5814 | 2 |
| PRKCE | 5581 | 2 |
| PPP1R1B | 84152 | 2 |
| PENK | 5179 | 2 |
| PDZRN3 | 23024 | 2 |
| PDAP1 | 11333 | 2 |
| PC | 5091 | 2 |
| PBX1 | 5087 | 2 |
| NTSR2 | 23620 | 2 |
| NTRK2 | 4915 | 2 |
| NR4A3 | 8013 | 2 |
| NPY | 4852 | 2 |
| NPY2R | 4887 | 2 |
| MPDZ | 8777 | 2 |
| MAPK14 | 1432 | 2 |
| MAN1A2 | 10905 | 2 |
| LAMB1 | 3912 | 2 |
| GSK3B | 2932 | 2 |
| GPRC5B | 51704 | 2 |
| GNG12 | 55970 | 2 |
| GBA | 2629 | 2 |
| GABRG2 | 2566 | 2 |
| GABRD | 2563 | 2 |
| GABRB2 | 2561 | 2 |
| GABBR1 | 2550 | 2 |
| FOXN3 | 1112 | 2 |
| FOSL2 | 2355 | 2 |
| EIF4EBP2 | 1979 | 2 |
| DDX5 | 1655 | 2 |
| CLIC4 | 25932 | 2 |
| CHRM1 | 1128 | 2 |
| CAPZB | 832 | 2 |
| BDNF | 627 | 2 |
| ATP8A1 | 10396 | 2 |
| ASNS | 440 | 2 |
| ABHD4 | 63874 | 2 |
Table 6.
Functional enrichment test for the 47 top ranked genes using WebGestalt.
| GO ID* | Term | # genes | p-value | pBH** |
|---|---|---|---|---|
| GO:0007610 (BP) | Behavior | 11 | 1.17 × 10-7 | 5.08 × 10-5 |
| GO:0007214 (BP) | Gamma-aminobutyric acid signaling pathway | 4 | 3.92 × 10-7 | 8.51 × 10-5 |
| GO:0007154 (BP) | Cell communication | 29 | 7.66 × 10-7 | 0.0001 |
| GO:0007631 (BP) | Feeding behavior | 5 | 2.16 × 10-6 | 0.0002 |
| GO:0050794 (BP) | Regulation of cellular process | 37 | 3.00 × 10-6 | 0.0002 |
| GO:0050789 (BP) | Regulation of biological process | 38 | 1.73 × 10-6 | 0.0002 |
| GO:0033555 (BP) | Multicellular organismal response to stress | 4 | 3.21 × 10-6 | 0.0002 |
| GO:0007186 (BP) | G-protein coupled receptor protein signaling pathway | 14 | 4.88 × 10-6 | 0.0002 |
| GO:0007166 (BP) | Cell surface receptor linked signal transduction | 19 | 2.53 × 10-6 | 0.0002 |
| GO:0065007 (BP) | Biological regulation | 38 | 8.70 × 10-6 | 0.0003 |
| GO:0016917 (MF) | GABA receptor activity | 6 | 3.22 × 10-11 | 3.35 × 10-9 |
| GO:0004890 (MF) | GABA-A receptor activity | 5 | 1.93 × 10-9 | 7.38 × 10-8 |
| GO:0042165 (MF) | Neurotransmitter binding | 7 | 2.13 × 10-9 | 7.38 × 10-8 |
| GO:0030594 (MF) | Neurotransmitter receptor activity | 6 | 3.56 × 10-8 | 9.26 × 10-7 |
| GO:0005254 (MF) | Chloride channel activity | 6 | 6.04 × 10-8 | 1.26 × 10-6 |
| GO:0005253 (MF) | Anion channel activity | 6 | 9.06 × 10-8 | 1.35 × 10-6 |
| GO:0031404 (MF) | Chloride ion binding | 6 | 9.06 × 10-8 | 1.35 × 10-6 |
| GO:0043168 (MF) | Anion binding | 6 | 2.64 × 10-7 | 3.43 × 10-6 |
| GO:0005230 (MF) | Extracellular ligand-gated ion channel activity | 5 | 2.22 × 10-6 | 2.57 × 10-5 |
| GO:0008509 (MF) | Anion transmembrane transporter activity | 6 | 3.99 × 10-6 | 4.15 × 10-5 |
| GO:0045211 (CC) | Postsynaptic membrane | 8 | 2.22 × 10-9 | 1.86 × 10-7 |
| GO:0034707 (CC) | Chloride channel complex | 6 | 6.57 × 10-9 | 2.76 × 10-7 |
| GO:0044456 (CC) | Synapse part | 9 | 1.20 × 10-8 | 3.36 × 10-7 |
| GO:0045202 (CC) | Synapse | 9 | 2.51 × 10-7 | 5.27 × 10-6 |
| GO:0030054 (CC) | Cell junction | 9 | 5.88 × 10-6 | 9.88 × 10-5 |
| GO:0034702 (CC) | Ion channel complex | 6 | 1.53 × 10-5 | 0.0002 |
| GO:0044459 (CC) | Plasma membrane part | 16 | 1.70 × 10-5 | 0.0002 |
| GO:0031226 (CC) | Intrinsic to plasma membrane | 11 | 0.0002 | 0.0019 |
| GO:0005887 (CC) | Integral to plasma membrane | 11 | 0.0002 | 0.0019 |
| GO:0033267 (CC) | Axon part | 3 | 0.0003 | 0.0025 |
* BP: biological process; MF: molecular function; and CC: cellular component.
** BH: p-value corrected by the Benjamini-Hochberg method (1995) [50].
To further investigate whether our approach to selecting the 47 genes is efficient, we compared the results with a similar analysis of top-ranked genes based on p values in COGA GWAS. We assigned the smallest p value of the marker mapped to a gene to represent gene-wise association significance. Then, we selected the most significant 47 genes. No functional term was significant in GO term analysis. Of note, our results were not corrected for gene length bias, a potential problem in gene-based association studies [25]. This comparison suggested that our cross-species gene ranking method may be more useful in extracting biological meaning from gene lists.
We further examined the function of the 47 genes selected by cross-species ranking by using the ToppFun online tool [26]. ToppFun provides enrichment analysis of candidate genes in many biological categories, including GO terms, biological pathways, human and mouse phenotypes, protein domains, and reference search in PubMed. We presented the results of ToppFun as complementary information for WebGestalt analysis and summarized the results of enriched pathways in Table 7 enriched mouse phenotypes in Table 8 and enriched PubMed citations in Table 9. In the pathway analysis, ToppFun uses a comprehensive collection of pathways from major databases such as KEGG, Reactome, and BioCarta [26]. The most enriched pathway is neuroactive ligand-receptor interaction (p = 5.36 × 10-5). Other significant pathways included GPCR ligand binding and G alpha signaling events; here, GPCR denotes G protein-coupled receptor (Table 7). Moreover, mouse phenotype analysis revealed that our selected genes are involved in neuron-related activity (Table 8). Overall, pathway and mouse phenotype enrichment analyses confirmed the results obtained from the GO term enrichment analysis by WebGestalt, and these analyses also revealed highlighted genes related to synaptic activity and GABA signaling as being particularly represented in significant pathways and mouse phenotypes. Finally, we queried PubMed references by ToppFun to search for publications that are overrepresented with genes from our top ranked list (Table 9). The highest scored record from this analysis was from a genetic study of gene expression associated with alcohol consumption in rats and humans [4], in which 24 of our top genes were represented in the total of 130 genes described by this study and showed significant enrichment (p < 1 × 10-6). The second highest scored record was from an association study of 182 candidate genes in anorexia nervosa (enrichment p < 1 × 10-6).
Table 7.
Pathways significantly associated with top candidate genes by ToppFun.
| Pathway ID/name | Description | Source | p-value | Terms in query | Terms in genome |
|---|---|---|---|---|---|
| hsa04080 | Neuroactive ligand-receptor interaction | KEGG pathway | 5.36 × 10-5 | 10 | 256 |
| REACTOME_GPCR_LIGAND_BINDING | Genes involved in GPCR ligand binding | MSigDB: C2.cp - Reactome | 2.66 × 10-3 | 10 | 392 |
| REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors | MSigDB: C2.cp - Reactome | 4.36 × 10-3 | 7 | 173 |
| REACTOME_DOWNSTREAM_EVENTS_IN_GPCR_SIGNALING | Genes involved in Downstream events in GPCR signaling | MSigDB: C2.cp - Reactome | 8.63 × 10-3 | 10 | 448 |
| REACTOME_CLASS_A1_RHODOPSIN_LIKE_RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) | MSigDB: C2.cp - Reactome | 1.62 × 10-2 | 8 | 292 |
| REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | Genes involved in G alpha (q) signaling events | MSigDB: C2.cp - Reactome | 2.78 × 10-2 | 6 | 157 |
Table 8.
Mouse phenotypes significantly associated with top candidate genes by ToppFun.
| Phenotype ID | Phenotype name | p-value | Terms in query | Terms in genome |
|---|---|---|---|---|
| MP:0009745 | Abnormal behavioral response to xenobiotic | 6.77 × 10-7 | 12 | 215 |
| MP:0002206 | Abnormal CNS synaptic transmission | 4.31 × 10-6 | 14 | 382 |
| MP:0003635 | Abnormal synaptic transmission | 3.57 × 10-5 | 14 | 450 |
| MP:0002062 | Abnormal conditioning behavior | 6.87 × 10-5 | 10 | 199 |
| MP:0002063 | Abnormal learning/memory/conditioning | 9.26 × 10-5 | 13 | 405 |
| MP:0002065 | Abnormal fear/anxiety-related behavior | 1.05 × 10-4 | 10 | 208 |
| MP:0001362 | Abnormal anxiety-related response | 3.59 × 10-4 | 9 | 179 |
| MP:0002572 | Abnormal emotion/affect behavior | 8.34 × 10-4 | 11 | 329 |
| MP:0001454 | Abnormal cued conditioning behavior | 1.41 × 10-3 | 6 | 66 |
| MP:0003633 | Abnormal nervous system physiology | 2.44 × 10-3 | 20 | 1333 |
| MP:0001399 | Hyperactivity | 2.57 × 10-3 | 9 | 226 |
| MP:0001363 | Increased anxiety-related response | 2.86 × 10-3 | 7 | 117 |
| MP:0009357 | Abnormal seizure response to inducing agent | 9.04 × 10-3 | 7 | 139 |
| MP:0001449 | Abnormal learning/memory | 1.22 × 10-2 | 10 | 349 |
| MP:0003088 | Abnormal prepulse inhibition | 1.45 × 10-2 | 5 | 57 |
| MP:0003313 | Abnormal locomotor activation | 1.46 × 10-2 | 13 | 630 |
| MP:0000950 | abnormal seizure response to pharmacological agent | 1.53 × 10-2 | 6 | 99 |
| MP:0002945 | Abnormal inhibitory postsynaptic currents | 1.58 × 10-2 | 5 | 58 |
| MP:0004008 | Abnormal GABA-mediated receptor currents | 2.85 × 10-2 | 3 | 11 |
| MP:0009747 | Impaired behavioral response to xenobiotic | 3.45 × 10-2 | 5 | 68 |
| MP:0004747 | Abnormal cochlear OHC afferent innervation pattern | 3.48 × 10-2 | 2 | 2 |
Table 9.
PubMed citations significantly over-represented with top candidate genes by ToppFun.
| PubMed ID | Description | p-value | Terms in query | Terms in publication |
|---|---|---|---|---|
| 19874574 | Genetical genomic determinants of alcohol consumption in rats and humans. | < 1 × 10-6 | 24 | 130 |
| 20468064 | Association study of 182 candidate genes in anorexia nervosa. | < 1 × 10-6 | 15 | 182 |
| 18985723 | GABA neurotransmitter signaling in the developing mouse lens: dynamic regulation of components and functionality. | < 1 × 10-6 | 7 | 18 |
| 21205893 | TrkB receptor controls striatal formation by regulating the number of newborn striatal neurons. | < 1 × 10-6 | 6 | 12 |
| 16987237 | Reduced expression of neuropeptide genes in a genome-wide screen of a secretion-deficient mouse. | < 1 × 10-6 | 8 | 67 |
Discussion
In this work, we applied a unique cross-species, evidence-based gene prioritization strategy for genes involved in alcoholism. We started with a set of genes with prior microarray expression evidence of involvement in ethanol response, representing approximately 10% of the human protein-coding genes. These genes were ranked using additional sources of evidence across multiple species, including humans, mice, C. elegans and Drosophila. We used the COGA GWAS dataset and applied permutation analysis to evaluate the best weighting score matrix for gene ranking. Based on these results, we selected the top 47 genes with the best evidence for follow up bioinformatics analysis. Our functional enrichment test of these 47 genes suggested that this ranking algorithm identifies gene sets with coherent biological functions relevant to brain responses to ethanol and neural adaptations occurring with alcoholism. Remarkably, higher ranking scores were predictive of genes containing an enrichment of significant SNP associations in the context of COGA alcohol dependence GWAS results. These results provide initial evidence that a cross-species analysis of gene networks correlated with molecular or behavioral responses to ethanol may provide a powerful strategy to identify candidate genes that contribute to alcoholism.
The identification of genes mediating biological responses to ethanol, including the modification of risk profiles for alcoholism, is an area of intense research interest due to the possibility of pinpointing targets for future alcoholism therapies. Recent advances in behavioral genetics and genomics have identified large numbers of genes that potentially contribute to phenotypic responses to ethanol in both human and animal models. However, little progress has been made in narrowing or organizing these large lists of genes into a tractable scheme for understanding the neurobiology and genetics of alcoholism. One approach that has been used for large collections of microarray data has been the performance of a meta-analysis across data on rodent models of divergent ethanol drinking collected from multiple centers and strains [3]. However, this analysis identified 3,800 genes associated with variation in ethanol intake, making downstream hypothesis-driven studies difficult to formulate.
As discussed in the Background, in our research approach, we pursued a gene ranking algorithm constructed to integrate data on ethanol-related genes across species. We recognized that direct behavioral parallels with ethanol response across humans, mice, Drosophila and C. elegans were likely to be tenuous or non-existent. However, molecular commonalities underlying ethanol responses across species, if they could be identified, should provide a powerful validation mechanism for candidate genes involved in ethanol behavioral responses, even if those particular behavioral components differ across species.
Our ranking algorithm, while largely empirical at this stage, identified a ranked list of genes with obvious coherence in terms of functional gene networks. A remarkably large number of genes already validated as altering behavioral responses to ethanol were contained in the higher ranks. In addition, bioinformatics analysis showed several interesting biological functions that were over-represented among the ranked genes (Tables 6, 7, 8, 9), which is largely consistent with our previous analysis based on a network approach [27]. Again, a number of individual gene members from the constructed networks have strong prior validation as candidate genes that influence alcoholism traits in humans or behavioral responses to ethanol in animal models. These validated genes serve to increase the probability for the entire gene network playing a role in ethanol responses.
Although gene targeting approaches in animal models might ultimately be the most robust method for validating the role of individual genes in ethanol response behaviors, such studies are complex and time-consuming. We chose, as an initial approach to validate our cross-species ranking algorithm, a study of the association of the gene ranking score with alcoholism traits in a GWAS analysis. We found a reduction in the minimum FDR q-value as the ranking score increased to 2. It is important to note that this effect is not due to the progressive limiting of markers examined. In this study, FDR is not dependent on the number of tests performed.
Although the results are encouraging, the limitations of the current analysis and possible improvements must be noted. We noted that when the gene rank score cutoff increased from 2.0 to 2.5, the size of the q-values reversed. This observation might be attributed to overly restricted gene selection given that number of SNPs in genes dropped from 2293 in 47 genes to 210 in only 6 genes. Another limitation is that the use of genes from the addiction/alcoholism array represents hypotheses about important genes, as selected by expert review, rather than selection from empirical association data. We could improve the current approach in the following ways. First, although we included seven datasets in the gene ranking, many additional datasets now exist or will be released in the near future that may be used in multi-species data integration. Additionally, this single GWAS dataset is likely to be underpowered given the recent evidence showing many loci of small effect influence most complex human traits. However, a network or pathway analysis approach to analyze a set of genes might improve power [12].
While there are undoubtedly numerous ways to score or weight genes, we have shown that this simple empirical approach is effective. Our results demonstrate the utility of gene ranking after cross-species data integration. Since this initial study demonstrated the utility of this approach, we are continuing to expand the number of data sets and improve the scoring scheme through a more sophisticated optimization of weighting parameters. As more data is included, additional alcohol GWAS results become available, and more sophisticated gene ranking algorithms are developed, we expect improvement in specificity and sensitivity. For example, there are many gene expression studies in rat brain from animals evaluated for alcohol-preference behavior [2,28-31], and they will be integrated in future gene ranking. However, our initial gene targeting experiments in animal models, using the ranked gene lists derived in this study, have already identified several novel genes that alter ethanol response behaviors in mice, Drosophila or C. elegans (unpublished data). This provides direct support of our cross-species gene ranking.
Conclusion
In this study, we proposed a cross-species, evidence-based gene ranking strategy and demonstrated it in the eight alcoholism or ethanol response related datasets from four species (human, mouse, fly, and worm). Through the use of permutation and FDR analyses, we evaluated 10 weighting score matrices and found that one of them had the best performance. Using this optimal weighting matrix, we selected 47 genes whose scores were greater than 2 for follow up bioinformatics analysis. Functional enrichment tests revealed that these 47 genes are involved in brain responses to ethanol and neural adaptations occurring with alcoholism. Our results, with further experimental validation in three animal models, suggest that our approach is useful for cross-species gene prioritization.
Materials and methods
Cross species gene ranking
In an effort to populate an inclusive gene list with non-biased data from at least two species, we used published microarray gene expression data from our own and other laboratories. As shown in Figure 1, microarray gene expression data was used from three sources: acute responses to ethanol in C57BL/6 and DBA2/J mice (whole genome analysis of samples from reference [14]) that had been supplemented with additional microarray studies (U74B and U74C arrays, Affymetrix) on the same samples, a meta-analysis of genes involved in ethanol preference drinking across multiple mouse strains [3], and analysis of prefrontal cortex from autopsied samples of alcoholic and non-alcoholic brains [15]. We then merged these datasets by utilizing gene homology mapping features within the WebGestalt [32]. This produced a list of 2458 genes. These genes were ranked by scores resulting from the following algorithm:
The symbols refer to sources diagramed in Figure 1. MuAc, MuPref and HuAlc refer to presence in the microarray studies mentioned above. HuAddChip are selected genes from human association studies on alcohol dependence using the "addiction chip" designed by David Goldman and colleagues at the National Institute on Alcohol Abuse and Alcoholism (NIAAA) [19]. HuLink refers to genes contained within linkage regions that have been implicated multiple times across human studies of alcohol-related phenotypes on chromosomes 1, 4, and 7 [16-18]. The region on chromosome 1 ranges from D1S1613 at 64,007,000 bp to D1S2624 at 154,898,000 bp (according to HapMap build 36) and encompasses a variety of overlapping linkage signals to alcohol-related phenotypes, including alcohol dependence, heavy drinking, sensitivity to alcohol, and tolerance, across a number of samples [17,18,33-38]. The chromosome 4 region ranges from D4S2382 at 39,727,200 bp to D4S1615 at 128,429,200 bp and encompasses linkage peaks from four independent samples [16,18,39-42]. The chromosome 7 region ranged from D7S691 at 41,996,200 bp to D7S1817 at 109,026,000 bp and constitutes the strongest linkage region in the Collaborative Study of the Genetics of Alcoholism (COGA) sample [18,43-45]. The invertebrate gene sets are from published studies in C. elegans [22] and Drosophila [20,21]. Finally, the "Cross" term is a bonus score added for cross-species hits for a given gene except for genes from human linkage regions. The weighting terms wi (i = 1, 2, 3, ... 8) were empirically chosen with 0.5 or 1.0 in 10 different weighting score matrices (Table 1). After a permutation test with COGA GWAS data, we found the weighting score matrix 3 could provide the best performance.
Analysis of COGA alcohol dependence GWAS dataset
The COGA GWAS dataset was used to evaluate the gene rankings. It contains 1205 cases and 700 controls [23]. All cases met DSM-IV criteria for alcohol dependence. Controls were defined as individuals who have consumed alcohol, but did not meet any definition of alcohol dependence or alcohol abuse, nor did they meet any DSM-IIIR or DSM-IV definition of abuse or dependence for other drugs (except nicotine). The Illumina human 1M chipset was used for genotyping. Only DNA samples achieving a call rate of > 95% were included. A total of 1,041,465 SNP markers were used for case-control analyses. We conducted population stratification and association analyses using PLINK, a highly flexible, fast, and user-friendly package for GWAS analysis [46]. In our analyses we included only SNPs if their genome-wide failure rate did not exceed 0.05. SNPs were further excluded if minor allele frequency was less than 0.01. After these data filtering processes, 958,380 SNPs were used for further analyses. Then, we mapped these SNPs to non-pseudogenes in the NCBI Entrez Gene database. Specifically, a SNP belongs to a gene if it locates in the region within 10 kb upstream to 10 kb downstream of the gene.
FDR control
To control the risk of false discoveries in GWAS studies, for each p-value, we calculated a q-value [47,48]. A q-value is an estimate of the proportion of false discoveries among all significant markers (i.e., q-values are FDRs) when the corresponding p-value is used as the threshold to declare significance. As argued previously [49], we preferred this approach to more traditional multiple testing methods that control the probability of producing one or more false discoveries for a set of tests [50]. Our approach was preferred because these q-values 1) provide a better balance between the competing goals of finding true positives versus controlling false discoveries, 2) allow the use of more similar standards in terms of the proportion of false discoveries produced across studies due to much less dependence on the number of tests performed, 3) are relatively robust against the effects of correlated tests [47,49,51-56], and 4) rather than an all-or-nothing conclusion about whether a study produces significant results, instead provide a more subtle picture about the possible relevance of the tested markers. The FDR procedure is performed in the R statistical package.
Random permutation for different score matrix
To test the significance of the gene ranking enrichment result for each weighting score matrix, we did 100 random permutations for the q-value enrichment. Since longer genes tend to have more SNPs in GWAS data, to reduce this gene length bias, we restricted the gene length of the random selections in each permutation within ± 50 kb of the average length of our ranked genes. We set the permutation p-value as the proportion of permutation times in which there are higher q-value proportions in randomly selected genes than in our ranked genes in the corresponding score region. For example, there were n genes with a score s under a specific weighting score matrix. Then, in each permutation, n genes were selected from all human genes whose length is ± 50 kb of the average length of the n ranked genes. The q-values for SNPs in the randomly selected genes were calculated based on the GWAS data. For simplicity of comparison, we compared the number of SNPs in each q-value range (e.g., < 0.9, < 0.8, etc.). The proportion of the q-value number for each q-value range in randomly selected genes was then calculated. If the proportion was larger than our ranked alcohol genes at the same q-value range, we counted this permutation as a "significant permutation" for the specific score range s and q-value range. After 100 permutations, the proportion of "significant permutation" was set as the p-value of our permutation result at the corresponding score and q-value range. For the weighting score matrix with the best performance, permutation testing was repeated 1000 times again to check the significance.
Bioinformatics analysis of cross-species ranked gene list
The 47 top ranked genes with a score ≥ 2 were examined for enriched GO terms using the WebGestalt online tool (version 2) [32]. This tool examines the over-representation of genes of interest in GO terms based on the hypergeometric test followed by the Benjamini-Hochberg (1995) adjustment of p-values [50]. We then used the ToppFun online tool [57], which is an integrated over-representation analysis tool, to interrogate databases for biological pathways, mouse phenotypes, and PubMed citations.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AYG FA HJE DMD MAS AA JK and JIN collected and prepared data for this study. AYG, FA, BTW, ZZ, PJ, and MFM conducted data analysis. ZZ, EJO, BPR, DMD, JCB, AGD, MSG, KSK, BTW and MFM conceived and designed the study. ZZ, AYG, BTW, MFM, PJ, HJE, MAS, AA, JK, and JIN contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Summary of the number and proportion of q-values, and p-value of 100 permutation results for different ranked score and q-value range under each of the 10 weighting score matrices.
Contributor Information
Zhongming Zhao, Email: zhongming.zhao@vanderbilt.edu.
An-Yuan Guo, Email: guoay@hust.edu.cn.
Edwin JCG van den Oord, Email: ejvandenoord@vcu.edu.
Fazil Aliev, Email: faliev@vcu.edu.
Peilin Jia, Email: peilin.jia@vanderbilt.edu.
Howard J Edenberg, Email: edenberg@iupui.edu.
Brien P Riley, Email: bpriley@vcu.edu.
Danielle M Dick, Email: ddick@vcu.edu.
Jill C Bettinger, Email: jcbettinger@vcu.edu.
Andrew G Davies, Email: agdavies@vcu.edu.
Michael S Grotewiel, Email: msgrotewiel@vcu.edu.
Marc A Schuckit, Email: mschuckit@ucsd.edu.
Arpana Agrawal, Email: agrawala@psychiatry.wustl.edu.
John Kramer, Email: john-kramer@uiowa.edu.
John I Nurnberger, Jr, Email: jnurnber@iupui.edu.
Kenneth S Kendler, Email: kendler@vcu.edu.
Bradley T Webb, Email: btwebb@vcu.edu.
Michael F Miles, Email: mfmiles@vcu.edu.
Acknowledgements
This work was supported by NIAAA grants P20AA017828 (MFM, KSK), U01AA0116667 (MFM), U01AA016662 (MFM), P30AA019372 (KSK), and R21AA017437 (ZZ).
The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes ten different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, A. Goate, J. Rice, K. Bucholz); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield); Southwest Foundation (L. Almasy), Howard University (R. Taylor) and Virginia Commonwealth University (D. Dick). Other COGA collaborators include: L. Bauer (University of Connecticut); D. Koller, S. O'Connor, L. Wetherill, X. Xuei (Indiana University); Grace Chan (University of Iowa); N. Manz, M. Rangaswamy (SUNY Downstate); A. Hinrichs, J. Rohrbaugh, J-C Wang (Washington University in St. Louis); A. Brooks (Rutgers University); and F. Aliev (Virginia Commonwealth University). A. Parsian and M. Reilly are the NIAAA Staff Collaborators.
We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, currently a consultant with COGA, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). Funding support for GWAS genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the National Institute on Alcohol Abuse and Alcoholism, the NIH GEI (U01HG004438), and the NIH contract "High throughput genotyping for studying the genetic contributions to human disease" (HHSN268200782096C). The authors thank Kim Doheny and Elizabeth Pugh from CIDR and Justin Paschall from the NCBI dbGaP staff for valuable assistance with genotyping and quality control in developing the dataset available at dbGaP.
This article has been published as part of BMC Genomics Volume 13 Supplement 8, 2012: Proceedings of The International Conference on Intelligent Biology and Medicine (ICIBM): Genomics. The full contents of the supplement are available online at http://www.biomedcentral.com/bmcgenomics/supplements/13/S8.
References
- Guo AY, Webb BT, Miles MF, Zimmerman MP, Kendler KS, Zhao Z. ERGR: An ethanol-related gene resource. Nucleic Acids Res. 2009;37:D840–845. doi: 10.1093/nar/gkn816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpel MW, Strother WN, McClintick JN, Carr LG, Liang T, Edenberg HJ, McBride WJ. Functional gene expression differences between inbred alcohol-preferring and -non-preferring rats in five brain regions. Alcohol. 2007;41:95–132. doi: 10.1016/j.alcohol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ. et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci USA. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Printz M, Flodman P, Hodgkinson C, Goldman D, Koob G, Richardson HN, Kechris K, Bell RL. et al. Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol. 2009;7:70. doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Han L, Zhao Z. Gene- and evidence-based candidate gene selection for schizophrenia and gene feature analysis. Artif Intell Med. 2010;48:99–106. doi: 10.1016/j.artmed.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Jia P, Fanous AH, Webb BT, van den Oord EJ, Chen X, Bukszar J, Kendler KS, Zhao Z. A multi-dimensional evidence-based candidate gene prioritization approach for complex diseases-schizophrenia as a case. Bioinformatics. 2009;25:2595–2602. doi: 10.1093/bioinformatics/btp428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd ZA, Bertsch BA, Strother WN, Le-Niculescu H, Balaraman Y, Hayden E, Jerome RE, Lumeng L, Nurnberger JI, Edenberg HJ. et al. Candidate genes, pathways and mechanisms for alcoholism: an expanded convergent functional genomics approach. Pharmacogenomics J. 2007;7:222–256. doi: 10.1038/sj.tpj.6500420. [DOI] [PubMed] [Google Scholar]
- Huang HC, Zheng S, VanBuren V, Zhao Z. Discovering disease-specific biomarker genes for cancer diagnosis and prognosis. Technol Cancer Res Treat. 2010;9:219–230. doi: 10.1177/153303461000900301. [DOI] [PubMed] [Google Scholar]
- Jia P, Sun J, Guo A, Zhao Z. SZGR: a comprehensive schizophrenia gene resource. Mol Psychiatry. 2010;15:453–462. doi: 10.1038/mp.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Kuo PH, Riley BP, Kendler KS, Zhao Z. Candidate genes for schizophrenia: a survey of association studies and gene ranking. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1173–1181. doi: 10.1002/ajmg.b.30743. [DOI] [PubMed] [Google Scholar]
- Kendler KS. Schizophrenia genetics and dysbindin: a corner turned? Am J Psychiatry. 2004;161:1533–1536. doi: 10.1176/appi.ajp.161.9.1533. [DOI] [PubMed] [Google Scholar]
- Jia P, Wang L, Meltzer HY, Zhao Z. Pathway-based analysis of GWAS datasets: effective but caution required. Int J Neuropsychopharmacol. 2011;14:567–572. doi: 10.1017/S1461145710001446. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. Low level of response to alcohol as a predictor of future alcoholism. Am J Psychiatry. 1994;151:184–189. doi: 10.1176/ajp.151.2.184. [DOI] [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW, Miles MF. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25:2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002;81:802–813. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- Prescott CA, Sullivan PF, Kuo PH, Webb BT, Vittum J, Patterson DG, Thiselton DL, Myers JM, Devitt M, Halberstadt LJ. et al. Genomewide linkage study in the Irish affected sib pair study of alcohol dependence: evidence for a susceptibility region for symptoms of alcohol dependence on chromosome 4. Mol Psychiatry. 2006;11:603–611. doi: 10.1038/sj.mp.4001811. [DOI] [PubMed] [Google Scholar]
- Kuo PH, Neale MC, Riley BP, Webb BT, Sullivan PF, Vittum J, Patterson DG, Thiselton DL, van den Oord EJ, Walsh D. et al. Identification of susceptibility loci for alcohol-related traits in the Irish Affected Sib Pair Study of Alcohol Dependence. Alcohol Clin Exp Res. 2006;30:1807–1816. doi: 10.1111/j.1530-0277.2006.00217.x. [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K. et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. doi: 10.1002/(SICI)1096-8628(19980508)81:3<207::AID-AJMG1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Yuan Q, Xu K, Shen PH, Heinz E, Lobos EA, Binder EB, Cubells J, Ehlers CL, Gelernter J. et al. Addictions biology: haplotype-based analysis for 130 candidate genes on a single array. Alcohol Alcohol. 2008;43:505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova TV, Anholt RR, Mackay TF. Phenotypic and transcriptional response to selection for alcohol sensitivity in Drosophila melanogaster. Genome Biol. 2007;8:R231. doi: 10.1186/gb-2007-8-10-r231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova TV, Anholt RR, Mackay TF. Transcriptional response to alcohol exposure in Drosophila melanogaster. Genome Biol. 2006;7:R95. doi: 10.1186/gb-2006-7-10-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon JY, Hong M, Choi MS, Kang S, Duke K, Kim S, Lee S, Lee J. Ethanol-response genes and their regulation analyzed by a microarray and comparative genomic approach in the nematode Caenorhabditis elegans. Genomics. 2004;83:600–614. doi: 10.1016/j.ygeno.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F. et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Rose RJ, Kaprio J. The next challenge for psychiatric genetics: characterizing the risk associated with identified genes. Ann Clin Psychiatry. 2006;18:223–231. doi: 10.1080/10401230600948407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, Tian J, Zhao Z. Assessing gene length biases in gene set analysis of Genome-Wide Association Studies. Int J Comput Biol Drug Des. 2010;3:297–310. doi: 10.1504/IJCBDD.2010.038394. [DOI] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo AY, Sun J, Jia P, Zhao Z. Network analysis of ethanol-related candidate genes. Chem Biodiv. 2010;7:1142–1152. doi: 10.1002/cbdv.200900318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Strother WN, McClintick JN, Tian H, Stephens M, Jerome RE, Lumeng L, Li TK, McBride WJ. Gene expression in the hippocampus of inbred alcohol-preferring and -nonpreferring rats. Genes Brain Behav. 2005;4:20–30. doi: 10.1111/j.1601-183X.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Kimpel MW, Edenberg HJ, Bell RL, Strother WN, McClintick JN, Carr LG, Liang T, McBride WJ. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats. Pharmacol Biochem Behav. 2008;89:481–498. doi: 10.1016/j.pbb.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr LG, Kimpel MW, Liang T, McClintick JN, McCall K, Morse M, Edenberg HJ. Identification of candidate genes for alcohol preference by expression profiling of congenic rat strains. Alcohol Clin Exp Res. 2007;31:1089–1098. doi: 10.1111/j.1530-0277.2007.00397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T, Kimpel MW, McClintick JN, Skillman AR, McCall K, Edenberg HJ, Carr LG. Candidate genes for alcohol preference identified by expression profiling in alcohol-preferring and -nonpreferring reciprocal congenic rats. Genome Biol. 2010;11:R11. doi: 10.1186/gb-2010-11-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WebGestal. http://bioinfo.vanderbilt.edu/webgestalt/
- Dick DM, Nurnberger J, Edenberg HJ, Goate A, Crowe R, Rice J, Bucholz KK, Kramer J, Schuckit MA, Smith TL. et al. Suggestive linkage on chromosome 1 for a quantitative alcohol-related phenotype. Alcohol Clin Exp Res. 2002;26:1453–1460. doi: 10.1111/j.1530-0277.2002.tb02443.x. [DOI] [PubMed] [Google Scholar]
- Guerrini I, Cook CC, Kest W, Devitgh A, McQuillin A, Curtis D, Gurling HM. Genetic linkage analysis supports the presence of two susceptibility loci for alcoholism and heavy drinking on chromosome 1p22.1-11.2 and 1q21.3-24.2. BMC Genet. 2005;6:11. doi: 10.1186/1471-2156-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SY, Shen S, Zezza N, Hoffman EK, Perlin M, Allan W. A genome wide search for alcoholism susceptibility genes. Am J Med Genet B Neuropsychiatr Genet. 2004;128B:102–113. doi: 10.1002/ajmg.b.30013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen J, Long JC, Eggert M, Ozaki N, Robin RW, Brown GL, Naukkarinen H, Virkkunen M, Linnoila M, Goldman D. Linkage of antisocial alcoholism to the serotonin 5-HT1B receptor gene in 2 populations. Arch Gen Psychiatry. 1998;55:989–994. doi: 10.1001/archpsyc.55.11.989. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Foroud T, Flury L, Su J, Meyer ET, Hu K, Crowe R, Edenberg H, Goate A, Bierut L. et al. Evidence for a locus on chromosome 1 that influences vulnerability to alcoholism and affective disorder. Am J Psychiatry. 2001;158:718–724. doi: 10.1176/appi.ajp.158.5.718. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Edenberg HJ, Kalmijn J, Flury L, Smith TL, Reich T, Bierut L, Goate A, Foroud T. A genome-wide search for genes that relate to a low level of response to alcohol. Alcohol Clin Exp Res. 2001;25:323–329. doi: 10.1111/j.1530-0277.2001.tb02217.x. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Wall TL, Phillips E, Feiler H, Wilhelmsen KC. Genomic screen for loci associated with alcohol dependence in Mission Indians. Am J Med Genet B Neuropsychiatr Genet. 2004;129B:110–115. doi: 10.1002/ajmg.b.30057. [DOI] [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, Goldman D. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet. 1998;81:216–221. doi: 10.1002/(SICI)1096-8628(19980508)81:3<216::AID-AJMG2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, Foroud T, Li TK, Begleiter H, Reich T, Rice JP. A genome screen of maximum number of drinks as an alcoholism phenotype. Am J Med Genet. 2000;96:632–637. doi: 10.1002/1096-8628(20001009)96:5<632::AID-AJMG8>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Williams JT, Begleiter H, Porjesz B, Edenberg HJ, Foroud T, Reich T, Goate A, Van Eerdewegh P, Almasy L, Blangero J. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. II. Alcoholism and event-related potentials. Am J Hum Genet. 1999;65:1148–1160. doi: 10.1086/302571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn G, Hinrichs AL, Bertelsen S, Jin CH, Kauwe JS, Suarez BK, Bierut LJ. Microsatellites versus single-nucleotide polymorphisms in linkage analysis for quantitative and qualitative measures. BMC Genet. 2005;6(Suppl 1):S122. doi: 10.1186/1471-2156-6-S1-S122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, Bierut LJ, Conneally PM, Nurnberger JI, Bucholz KK. et al. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res. 2000;24:933–945. doi: 10.1111/j.1530-0277.2000.tb04634.x. [DOI] [PubMed] [Google Scholar]
- Wang JC, Hinrichs AL, Stock H, Budde J, Allen R, Bertelsen S, Kwon JM, Wu W, Dick DM, Rice J. et al. Evidence of common and specific genetic effects: association of the muscarinic acetylcholine receptor M2 (CHRM2) gene with alcohol dependence and major depressive syndrome. Hum Mol Genet. 2004;13:1903–1911. doi: 10.1093/hmg/ddh194. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. The positive false discovery rate: A Bayesian interpretation and the q-value. Annals Stat. 2003;31:2013–2035. doi: 10.1214/aos/1074290335. [DOI] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Oord EJ, Sullivan PF. False discoveries and models for gene discovery. Trends Genet. 2003;19:537–542. doi: 10.1016/j.tig.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995. pp. 289–300.
- Brown BW, Russell K. Methods of correcting for multiple testing: operating characteristics. Stat Med. 1997;16:2511–2528. doi: 10.1002/(SICI)1097-0258(19971130)16:22<2511::AID-SIM693>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Fernando RL, Nettleton D, Southey BR, Dekkers JC, Rothschild MF, Soller M. Controlling the proportion of false positives in multiple dependent tests. Genetics. 2004;166:611–619. doi: 10.1534/genetics.166.1.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn EL, Troendle JF, McShane LM, Simon R. Controlling the number of false discoveries: application to high-dimensional genomic data. J Stat Plan Infer. 2004;124:379–398. doi: 10.1016/S0378-3758(03)00211-8. [DOI] [Google Scholar]
- Sabatti C, Service S, Freimer N. False discovery rate in linkage and association genome screens for complex disorders. Genetics. 2003;164:829–833. doi: 10.1093/genetics/164.2.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CA, Hsueh HM, Chen JJ. Estimation of false discovery rates in multiple testing: application to gene microarray data. Biometrics. 2003;59:1071–1081. doi: 10.1111/j.0006-341X.2003.00123.x. [DOI] [PubMed] [Google Scholar]
- van den Oord EJ. Controlling false discoveries in candidate gene studies. Mol Psychiatry. 2005;10:230–231. doi: 10.1038/sj.mp.4001581. [DOI] [PubMed] [Google Scholar]
- ToppFun. http://toppgene.cchmc.org/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of the number and proportion of q-values, and p-value of 100 permutation results for different ranked score and q-value range under each of the 10 weighting score matrices.