Abstract
Bacillus anthracis spores are the infectious form of the organism for humans and animals. However, the approved human vaccine in the United States is derived from a vegetative culture filtrate of a toxigenic, nonencapsulated B. anthracis strain that primarily contains protective antigen (PA). Immunization of mice with purified spore proteins and formalin-inactivated spores (FIS) from a nonencapsulated, nontoxigenic B. anthracis strain confers protection against B. anthracis challenge when PA is also administered. To investigate the capacity of the spore particle to act as a vaccine without PA, we immunized mice subcutaneously with FIS from nontoxigenic, nonencapsulated B. cereus strain G9241 pBCXO1−/pBC210− (dcG9241), dcG9241 ΔbclA, or 569-UM20 or with exosporium isolated from dcG9241. FIS vaccination provided significant protection of mice from intraperitoneal or intranasal challenge with spores of the virulent B. anthracis Ames or Ames ΔbclA strain. Immunization with dcG9241 ΔbclA FIS, which are devoid of the immunodominant spore protein BclA, provided greater protection from challenge with either Ames strain than did immunization with FIS from BclA-producing strains. In addition, we used prechallenge immune antisera to probe a panel of recombinant B. anthracis Sterne spore proteins to identify novel immunogenic vaccine candidates. The antisera were variably reactive with BclA and with 10 other proteins, four of which were previously tested as vaccine candidates. Overall our data show that immunization with FIS from nontoxigenic, nonencapsulated B. cereus strains provides moderate to high levels of protection of mice from B. anthracis Ames challenge and that neither PA nor BclA is required for this protection.
INTRODUCTION
Bacillus anthracis is a Gram-positive, spore-forming, rod-shaped bacterium that can cause cutaneous, inhalational, or gastrointestinal anthrax. Anthrax disease, which typically occurs in grazing mammals and incidentally in humans, develops after introduction and subsequent germination of B. anthracis spores within the host. In the United States, human anthrax cases are rare and occur predominantly after exposure to contaminated animal products such as wool or animal hides (1–3). The intentional dissemination of B. anthracis spores through the U.S. postal system in 2001 resulted in 22 cases of anthrax with five fatalities from inhalational anthrax (4). In recent years, Bacillus cereus strains that produce B. anthracis virulence factors have been isolated from humans with severe pulmonary “anthrax-like” infections (5–9). B. anthracis and B. cereus are closely related members of the B. cereus sensu lato group, but only B. anthracis is categorized by the U.S. Centers for Disease Control and Prevention as a Category A bioterrorism agent.
B. anthracis contains the two virulence plasmids, pXO1 and pXO2. Genes that encode the anthrax toxin subunits edema factor (EF), lethal factor (LF), and protective antigen (PA) are found on pXO1, and the genes needed to produce the poly-γ-d-glutamic acid capsule are encoded on pXO2. EF or LF combines with PA to form edema toxin (ET) or lethal toxin (LT), respectively. PA is essential for toxicity because PA binds to target cell receptors and mediates entry of EF or LF into the host cytosol (reviewed in reference 10). ET is a calmodulin-dependent adenylate cyclase that appears to elicit edema at the site of infection (11–13) and also has antiphagocytic effects on neutrophils (14). LT is a zinc-dependent metalloprotease that cleaves and subsequently inactivates mitogen-activated protein kinase kinases 1 and 2 (15, 16). The poly-γ-d-glutamic acid capsule protects vegetative bacilli from phagocytosis and macrophage killing (reviewed in reference 17). B. cereus G9241, which was isolated from a welder with pulmonary anthrax-like disease, contains the pXO1 homolog pBCXO1 and an unrelated megaplasmid, pBC210 (8). B. cereus G9241 produces PA, LF, and EF from the pBCXO1-carried genes pag, lef, and cya, respectively (8, 18). In addition, pBCXO1 encodes an intact and functional operon required for hyaluronic acid capsule synthesis (19), and pBC210 contains an operon that is necessary for production of a putative tetrasaccharide capsule (8, 18, 19).
In the United States, the only Food and Drug Administration-approved anthrax vaccine for human use is AVA (anthrax vaccine adsorbed) or Biothrax. AVA is derived from a vegetative culture filtrate of the attenuated B. anthracis V770-NP1-R and contains primarily PA as well as small amounts of LF and EF (20). Anti-PA antibodies generated as a result of vaccination with AVA are the main source of protection against anthrax (21). The current AVA vaccine has the following shortcomings: (i) a slightly variable composition, (ii) an 18-month/5-dose vaccination schedule with required annual boosters (22, 23), and (iii) minor to moderate local reactogenicity (24, 25). Furthermore, the effectiveness of AVA and other PA-based vaccines varies in different animal models. These vaccines provide no protection against toxigenic, encapsulated B. anthracis in mice (26–28), confer variable protection against geographically diverse B. anthracis isolates in guinea pigs (29, 30), and are highly protective in rabbits and rhesus macaques (29).
The bioterrorism threat associated with B. anthracis and the shortcomings of AVA fostered significant research support by government agencies to develop a better and more effective anthrax vaccine. While most of the newer vaccine candidates are still based on PA, additional research on other components of B. anthracis, such as the spore and spore components, has yielded promising results in animals (27, 30–35). Live, attenuated spore vaccines are used safely and effectively in humans in Russia and China and in animals worldwide (36, 37). Moreover, immunization with PA and formalin-inactivated spores (FIS) of a nontoxigenic, nonencapsulated B. anthracis strain conferred protection against subcutaneous (s.c.) and intranasal (i.n.) challenge of guinea pigs with virulent B. anthracis 9602 but protected against only s.c. challenge of mice (32, 35). Vaccination with either component by itself provided only minor protection from s.c. challenge with B. anthracis 9602 in guinea pigs and no protection in mice (32). Studies of the efficacy of spore proteins as vaccine candidates showed that immunization with PA and BclA, BxpB/ExsFA, or p5303 protected mice from challenge with attenuated B. anthracis Sterne (pXO1+/pXO2−) better than did immunization with PA alone (31, 34). In a similar vaccine study conducted in mice and guinea pigs challenged with fully virulent B. anthracis Ames (pXO1+/pXO2+), the addition of BclA, BxpB/ExsFA, and p5303 to the PA vaccine regimen enhanced protection compared to vaccination with PA alone (33). Immunization of mice with plasmids that encode PA and BclA increased survival after B. anthracis Ames challenge compared to immunization with either component separately, while vaccination with PA and live B. anthracis Sterne spores afforded complete protection (27).
As the above examples demonstrate, for B. anthracis spore- or spore component-based vaccines to protect mice from virulent B. anthracis challenge, the inclusion of PA, either exogenously or naturally, is required. However, here we report that immunization of mice with FIS from nontoxigenic, nonencapsulated B. cereus strains in the absence of PA provided moderate to full protection of BALB/c mice from challenge with highly virulent B. anthracis Ames. In addition, we showed that the presence or absence of BclA on the vaccine strain appeared to influence the capacity of the host immune system to generate an antibody response against some spore antigens. Finally, we identified several immunogenic spore antigens that merit further investigation as vaccine candidates. Our results with the B. cereus FIS vaccine indicate that antibodies against accessible components on the spore surface can protect against virulent B. anthracis in a mouse model.
(This work was presented in part at the Bacillus ACT 2011 Meeting, Bruges, Belgium, August 2011, and at the 112th General Meeting of the American Society for Microbiology, San Francisco, CA, June 2012 [38].)
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All bacterial strains and plasmids used in this work are listed in Table 1. Bacteria were routinely cultured in Luria-Bertani (LB) broth with shaking at 37°C (250 rpm), and selection of transformants was done on LB agar plates at 37°C unless stated otherwise. Where appropriate, antibiotics were used for selection at the following final concentrations: ampicillin (Amp), 100 μg/ml; kanamycin (Kan), 100 μg/ml; and erythromycin (Erm), 5 μg/ml.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| Top10 | High-competency cloning strain | Life Technologies |
| INV110 | dam dcm mutant strain used to produce unmethylated plasmid DNA | Life Technologies |
| B. anthracis | ||
| Ames | pXO1+/pXO2+ | 30 |
| Ames ΔbclA | pXO1+/pXO2+ ΔbclA::kan | 39 |
| Sterne 34F2 | pXO1+/pXO2− | NMRCa |
| B. cereus | ||
| 569-UM20 | Plasmidless, nonencapsulated, anthranilic acid-negative strain derived from B. cereus 569 | 40 |
| dcG9241 | pBCXO1−/pBC210−/pBClin29+ | 18 |
| dcG9241 ΔbclA | pBCXO1−/pBC210−/pBClin29+ ΔbclA::kan | This work |
| Plasmids | ||
| pGEM-T | TA-based E. coli cloning vector; Ampr | Promega |
| pUTE583 | Dual E. coli and Bacillus vector; Cmr in E. coli, Emr in Bacillus | 41 |
| pUTE618 | Source of Ωkan cassette for gene replacement; Cmr Spr | 41 |
| pBclA-U | 1-kb region upstream from bclA in pGEM-T; Ampr | This work |
| pBclA-D | 1-kb region downstream from bclA in pGEM-T; Ampr | This work |
| pBclA-UD | 1-kb regions upstream and downstream of bclA in pGEM-T; Ampr | This work |
| pBclA-UΩD | Ωkan cassette between 1-kb regions upstream and downstream of bclA in pGEM-T; Ampr | This work |
| p583BclA-UΩD | Ωkan cassette between 1-kb region upstream and downstream of bclA in pUTE583; Cmr in E. coli, Emr in Bacillus | This work |
NMRC, Naval Medical Research Center.
Spore preparation and inactivation.
B. anthracis Sterne, B. cereus, and B. anthracis Ames spores were prepared as previously reported (18, 34, 42). FIS were generated by treatment of purified spores with 4% formaldehyde in distilled water for 1 week at 4°C. The spores were washed several times with distilled water, and an aliquot was plated to ensure complete inactivation. Formalin treatment was repeated if complete loss of viability was not achieved.
Generation of dcG9241 ΔbclA.
The bclA gene (BCE_G9241_1212) in B. cereus G9241 pBCXO1−/pBC210− (dcG9241) was replaced by an omega element that contains a kanamycin resistance cassette (Ωkan), based on a previously published method (43). The 1-kb sequence upstream from bclA was amplified by PCR with bclA-up-sense (5′-GTC GAC CCA TAT ATA TAT ATC CTA TTC TTG TAC AAT TCT CTC CTC TAG GAA CAT C) and bclA-up-antisense (5′-GAA TTC AAA TTC ACC TCC ATA AAG CGT TCA TTA TAT AGT AGA TGC AAA ACC) primers; the SalI and EcoRI restriction enzyme sites are in bold and underlined, respectively. The 1-kb sequence downstream from the bclA gene was amplified by PCR with bclA-down-sense (5′-GAA TTC ACT TAG CAG TAA AAC TGA TAT CAG TTT TAC TGC TTT TTC ATT GG) and bclA-down-antisense (5′-GCG GCC GCC TAT TCT TTT CGC CAG TAA ATA CCG AAA TCA TCA ATT TGA GTC ATA GG) primers; the EcoRI and NotI restriction enzyme sites are underlined and italicized, respectively. Both upstream and downstream PCR fragments were ligated into the pGEM-T vector (Promega, Madison WI) to produce plasmids pBclA-U and pBclA-D, respectively. The correct orientation of the 1-kb upstream region cloned into pBclA-U was verified by digestion with EcoRI and NotI. The digested vector was used as the destination vector for ligation of the 1-kb downstream fragment obtained by EcoRI and NotI digestion of pBclA-D to generate pBclA-UD. The Ωkan cassette was isolated from pUTE618 by digestion with EcoRI and inserted between the upstream and downstream fragments in pBclA-UD digested with EcoRI to construct plasmid pBclA-UΩD. The plasmid pBclA-UΩD was digested with SalI to isolate the Ωkan flanked by the upstream and downstream fragments and subcloned into the pUTE583 shuttle vector to produce plasmid p583BclA-UΩD. This plasmid was transformed into Escherichia coli TOP10 cells (Life Technologies, Grand Island, NY) for maintenance and into E. coli INV110 cells (Life Technologies) to generate unmethylated DNA for electroporation. The method for electroporation of p583BclA-UΩD into dcG9241 was modified from a previously published procedure (44). Specifically, a 50-ml culture inoculated with 500 μl of an overnight (O/N) culture of dcG9241 was grown in LB broth until the optical density at 600 nm (OD600) was approximately 0.3. Glycine was added to a 3% final concentration, and the culture was incubated 1 h longer. The culture was placed on ice for 5 min and harvested by centrifugation at 8,000 × g, 4°C. The bacterial pellet was washed three times with ice-cold E buffer (272 mM sucrose, 0.5 mM MgCl2, 0.5 mM K2HPO4, 0.5 mM KH2PO4) and spun at 10,000 × g at 4°C in a microcentrifuge. The bacterial pellet was then resuspended in 500 μl E buffer. Approximately 1.5 μg of p583BclA-UΩD, isolated from E. coli INV110, was electroporated into 100 μl dcG9241 in a 2-mm electroporation cuvette at 200 Ω, 25 μF, and 1.5 kV. LB broth (1 ml) was added immediately after electroporation, and the transformation mixture was incubated for 2.5 h at 37°C. The transformation outgrowth was plated onto LB agar plates that contained Erm and Kan and incubated O/N at 37°C. Transformants were restreaked, and a single colony was used to inoculate 25 ml LB broth. After 8 h, 0.25 ml of the culture was used as the inoculum for another 25-ml culture. The subculture step was repeated three more times. Serial dilutions were plated onto LB agar plates supplemented with Kan and grown O/N. Kan-resistant colonies were patched onto LB agar plates supplemented with Erm. Erm-sensitive patches were screened by PCR with bclA-up-ex, a primer external to the cloned 1-kb upstream region (5′-GGT ACT TCC GTT GCA AGT TTA AAC CAA AAT ATC GCT TCG), and Kan-test, a primer internal to the Ωkan cassette (5′-GAC TTA CTG GGG ATC AAG CCT GAT TGG GAG). In addition, extracts of spore surface proteins (34) from dcG9241 and dcG9241 ΔbclA spores were isolated, separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with anti-BclA (45) to further verify the deletion of bclA by the absence of BclA.
Purification of dcG9241 exosporium.
The exosporium from dcG9241 spores was isolated according to previously published protocols (46, 47). Briefly, dcG9241 spores in water were filtered through a 1.6-μm glass fiber filter to remove any debris and diluted into phosphate-buffered saline (PBS) with 0.5 mM EDTA and protease inhibitor cocktail set VII (EMD Millipore Chemicals, Billerica, MA). Spores were disrupted by sonication with a Fisher Scientific model 705 Sonic Dismembrator (Thermo Fisher Scientific, Pittsburgh, PA) on ice for 5 min at 40% power with 15 s on and 30 s off. After this sonication step, the lysed spores were pelleted twice by centrifugation at 15,000 × g for 20 min at 4°C. The supernatants from both centrifugation steps were pooled and filtered through a 0.45-μm polyvinylidene difluoride (PVDF) filter to remove residual spore debris. The filtered supernatant was subjected to centrifugation at 40,000 rpm in a Ti50 rotor. The supernatant was removed, and the exosporium pellet was resuspended in PBS and stored at −80°C.
Immunization and challenge of BALB/c mice.
All B. anthracis Ames work was done under animal biosafety level 3 (ABSL3) conditions in accordance with Institutional Animal Care and Use Committee regulation at the U.S. Army Medical Research Institute of Infectious Diseases. On days 1 and 15, 6- to 8-week-old female BALB/c mice (National Cancer Institute, Frederick, MD) were immunized s.c. with 200 μl of either 1 × 108 B. cereus FIS in 0.3% alhydrogel (Sigma-Aldrich, St. Louis, MO) or 30 μg purified dcG9241 exosporium in 0.3% alhydrogel. Sera were collected from BALB/c mice on days 14 and 28 and combined into three separate pools for each vaccine group. On day 29, mice were challenged either intraperitoneally (i.p.) with 3 × 103 spores (5 to 10 times the median lethal dose [LD50]) or i.n. with 5 × 106 spores (70 to 120 times the LD50) of B. anthracis Ames or B. anthracis Ames ΔbclA (39, 48, 49). The mice were monitored for morbidity and mortality for 14 days postchallenge. Significant differences in survival after challenge were determined by Fisher's exact test, Kaplan-Meier survival analysis, and log rank tests with SAS version 8.2 (SAS Institute Inc., Cary, NC). GraphPad Prism version 5.03 (GraphPad Software Inc., La Jolla, CA) was used to calculate the median time to death (MTTD) and to test for significant differences among the groups with a nonparametric Kruskal-Wallis test and Dunn's multiple-comparison test. P values of <0.05 were considered statistically significant.
ELISA to detect anti-spore antibodies.
The direct binding of antibodies in immune mouse sera to B. anthracis Ames or B. anthracis Ames ΔbclA spores was assayed by enzyme-linked immunosorbent assay (ELISA) as previously described (34, 50, 51). Specifically, irradiation-sterilized spores (1 × 107 spores/well) were added to an Immulon II HB microplate (Thermo Fisher Scientific) and allowed to incubate O/N at 4°C. The plate was washed three times with PBS containing 0.1% Tween 20 (PBS-T) and then blocked with PBS containing 0.5% Tween 20 and 5% milk for 24 to 48 h at 4°C. The three serum pools from each vaccine group were diluted into the blocking buffer and serially diluted in triplicate. After incubation at 37°C for 1 h, the plate was washed as before and goat anti-mouse horseradish peroxidase-conjugated IgG (KPL, Inc., Gaithersburg, MD) diluted 1:1,000 was added. After 1 h of incubation at 37°C, the plate was washed with PBS-T six times and the horseradish peroxidase substrate was added [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS) 2-component microwell peroxidase substrate kit; KPL, Inc.]. The plate was incubated at 37°C for 30 min and the absorbance at 405 nm (A405) read on a microplate reader (BioTek Instruments, Winooski, VT). The reported average A405 value was the mean A405 value from triplicate samples from each pool. ELISA data were analyzed with SPSS version 20.0.0 (IBM Inc., Armonk, NY). Overall differences in antispore antibody titers among immunization groups were compared by two-way analysis of variance (ANOVA), adjusting for differences among dilutions, followed by Tukey pairwise post hoc comparisons. Differences at a given dilution were analyzed by one-way ANOVA followed by Tukey pairwise post hoc comparisons.
Expression and purification of exosporium proteins.
All recombinant exosporium proteins used in this study (Table 2) were described previously (34). The proteins were expressed in E. coli BL21(DE3)pLysS cells grown overnight at 30°C in autoinducing LB medium (ForMedium; Hunstanton, Norfolk, United Kingdom) with Amp. Cultures were harvested by centrifugation and frozen at −20°C until needed. Frozen cell pellets were resuspended in 1× Bugbuster (EMD Millipore Chemicals) with Benzonase nuclease (EMD Millipore Chemicals) and protease inhibitor cocktail set VII (EMD Millipore Chemicals) and incubated for 15 min at room temperature (RT) with gentle rocking. The lysates were diluted approximately 7-fold into 9 M urea, 20 mM Tris (pH 7.5), and 500 mM NaCl and incubated for an additional 15 min at RT with gentle rocking. The denatured cell lysates were clarified by centrifugation at 13,000 × g and 10°C for 15 min. The supernatants were transferred to clean tubes and gently rocked with Ni-nitrilotriacetic acid (NTA) resin (Qiagen Inc., Valencia, CA) for 30 min at RT. The resin was washed with 9 resin volumes of 7 M urea, 20 mM Tris (pH 7.5), 500 mM NaCl, and 10 mM imidazole. The protein was eluted from the resin in 4 resin volumes of 7 M urea, 20 mM Tris (pH 7.5), 500 mM NaCl, and 500 mM imidazole and then stored at −20°C.
Table 2.
B. anthracis genes expressed in this studya
| Ames locus tag | Sterne locus tag | Protein name |
|---|---|---|
| BA0108 | BAS0108 | Translation elongation factor Tu |
| BA0252 | BAS0238 | Alanine racemase |
| BA0355 | BAS0340 | CotB homolog |
| BA0803 | BAS0766 | CotJC |
| BA0804 | BAS0767 | CotJB |
| BA0805 | BAS0768 | CotJA |
| BA1222 | BAS1130 | BclA |
| BA1234 | BAS1141 | CotZ1/ExsY |
| BA1237 | BAS1144 | BxpB/ExsFA |
| BA1238 | BAS1145 | CotZ2/CotY |
| BA1489 | BAS1378 | Fe-Mn superoxide dismutase (SOD15) |
| BA1786 | BAS1655 | ExsE |
| BA2150 | NAb | ExsG |
| BA2162 | BAS2008 | BxpA |
| BA2292 | BAS2138 | Hypothetical protein (p2138) |
| BA2332 | BAS2174 | BxpC |
| BA2554 | BAS2377 | ExsK |
| BA2617 | BAS2439 | ExsD |
| BA2888 | BAS2693 | Inosine-uridine-preferring nucleoside hydrolase |
| BA3211 | BAS2986 | Hypothetical protein (p2986) |
| BA3668 | BAS3402 | Glycosyl hydrolase, family 18 |
| BA3906 | BAS3619 | CotE |
| BA4266 | BAS3957 | Hypothetical protein (p3957) |
| BA4499 | BAS4177 | Mn superoxide dismutase (SODA1) |
| BA4722 | BAS4383 | ThiJ/PfpI family protein |
| BA4898 | BAS4544 | Small, acid-soluble spore protein B |
| BA5640 | BAS5241 | Cell wall hydrolase |
| BA5641 | BAS5242 | YwdL |
| BA5699 | BAS5303 | Hypothetical protein (p5303) |
See reference 34.
NA, not applicable.
Immuno-dot blots.
All dot blots were done on a 96-well Minifold I Dot-Blot apparatus (Whatman, Piscataway, NJ) with a 0.45-μm nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA). Each well of the dot blot apparatus was prefilled with 200 μl of PBS to which 10 μl of each sample was added. Samples were loaded in triplicate across each row; wells A1 to A3 contained only buffer (i.e., negative control), wells A4 to A9 contained diluted normal mouse sera, and the remainder of the wells contained the purified protein samples. Vacuum was applied to the dot blot apparatus to bind the samples to the membrane. Once each well emptied, the vacuum was removed, the wells were washed with 200 μl PBS, and the vacuum was reapplied. The membrane was removed from the apparatus and blocked in PBS-T and 5% milk for 2 h at RT. The day 28 mouse antisera (pooled from each vaccine group) were added to the membranes at a final concentration of 40 μg/ml in PBS-T and 5% milk and incubated O/N at 4°C (the protein concentration in each pooled antiserum sample was calculated from the absorbance at 280 nm measured on a NanoDrop 1000 spectrophotometer [Thermo Fisher Scientific] with the estimate that a 0.1% protein solution has an absorbance at 280 nm of 1.0 in a 1-cm path length). The membrane was washed several times with PBS-T and incubated for 2 h at RT with 0.125 μg/ml goat anti-mouse IgG–Alexa Fluor 488 conjugate (Life Technologies). The membrane was washed once with PBS-T and several times with PBS before visualization on an ImageQuant LAS 4000 (GE Healthcare Biosciences, Piscataway, NJ). All membranes were imaged with identical settings. Integrated spot intensities were determined with ImageQuant TL array version 7.0 software (GE Healthcare Biosciences) with the background for each spot calculated from the average value of the baseline surrounding the spot. Analyses of the data were done in GraphPad Prism version 5.03 (Graph Pad Software Inc.); the triplicate integrated spot intensities for each sample were averaged and then normalized between 0 and 100%, the average values for the PBS/no-protein blank and BA_5610, respectively. Baseline correction was calculated by subtraction of the PBS-serum group normalized value from each sample. Comparisons among the normalized pooled antisera responses to a given protein for each vaccine group were done by two-way ANOVA followed by Bonferroni posttests with GraphPad Prism version 5.03 (Graph Pad Software Inc.).
RESULTS
Immunization with inactivated B. cereus spores protects BALB/c mice from B. anthracis Ames or Ames ΔbclA challenge.
We previously reported that live spores of dcG9241, a plasmid-cured derivative of B. cereus G9241, is avirulent in mice (18). In a preliminary study, we found that vaccination with live dcG9241 spores completely protected A/J mice against i.n. and s.c. challenge with 10 times the LD50 of B. anthracis Sterne spores (data not shown). Since dcG9241 does not produce toxins or capsule, we concluded that antibodies against spore components or vegetative antigens were sufficient to protect the mice from infection with the attenuated B. anthracis strain. Here we asked whether spores from nontoxigenic, nonencapsulated B. cereus strains could protect mice from challenge with the fully virulent B. anthracis Ames strain. We used formalin to inactivate spores of dcG9241 and 569-UM20, a derivative of the laboratory isolate B. cereus 569 (52) produced by UV mutagenesis (40); spores were inactivated to prevent outgrowth of the vegetative form and thus to allow attribution of any protection to the spore components themselves. We vaccinated BALB/c mice twice, 2 weeks apart, with 569-UM20 or dcG9241 FIS or with exosporium isolated from dcG9241 and then challenged the mice with a lethal dose of B. anthracis Ames spores on day 29. As shown in Table 3, s.c. immunization with 569-UM20 or dcG9241 FIS protected 40 to 60% of mice challenged by i.n. or i.p. inoculation. In addition, the MTTD for those mice that succumbed to infection after immunization with FIS from either strain was significantly longer (14 and 11 days for dcG9241 and 569-UM20, respectively) than that for mice given the adjuvant control (3 days) (Table 3). In contrast, vaccination with the isolated exosporium conferred no protection against B. anthracis Ames challenge. These results demonstrate that immunization with FIS alone from two nontoxigenic, nonencapsulated B. cereus strains can partially protect mice from challenge with the virulent B. anthracis Ames.
Table 3.
Survival and times to death for vaccination studies
| B. anthracis challenge strain | Challenge route | Vaccine | Survivala,c | MTTD (days)b,c |
|---|---|---|---|---|
| Ames | Intranasal | PBS/alhydrogel | 0/20 (0) | 3 |
| dcG9241 exosporium | 0/10 (0) | 3.5 | ||
| 569-UM20 FIS | 10/20 (50)*** | 11** | ||
| dcG9241 FIS | 11/20 (55)*** | 14*** | ||
| dcG9241 ΔbclA FIS | 8/10 (80)**** | 14*** | ||
| Intraperitoneal | PBS/alhydrogel | 2/20 (10) | 2 | |
| dcG9241 exosporium | 1/10 (10) | 3 | ||
| 569-UM20 FIS | 8/20 (40) | 3.5 | ||
| dcG9241 FIS | 12/20 (60)** | 14*** | ||
| dcG9241 ΔbclA FIS | 10/10 (100)**** | 14*** | ||
| Ames ΔbclA | Intranasal | PBS/alhydrogel | 0/10 (0) | 3 |
| dcG9241 exosporium | 0/10 (0) | 3 | ||
| 569-UM20 FIS | 4/10 (40) | 7.5* | ||
| dcG9241 FIS | 5/10 (50)* | 9.5* | ||
| dcG9241 ΔbclA FIS | 8/10 (80)** | 14*** | ||
| Intraperitoneal | PBS/alhydrogel | 0/10 (0) | 2 | |
| dcG9241 exosporium | 0/10 (0) | 2.5 | ||
| 569-UM20 FIS | 7/10 (70)** | 14** | ||
| dcG9241 FIS | 9/10 (90)*** | 14*** | ||
| dcG9241 ΔbclA FIS | 9/10 (90)*** | 14*** |
Number of survivors/number challenged (percent survival).
MTTD, median time to death.
Values that are statistically different from that for the PBS/alhydrogel group are indicated by asterisks (****, P < 0.0001; ***, P < 0.001; **, P < 0.01; and *, P < 0.05).
Previous reports showed that immunization with the immunodominant spore protein BclA can enhance protection from challenge with B. anthracis, especially when BclA is administered as part of a vaccine regimen that includes PA (27, 31, 33). To determine whether BclA contributed to the protection we observed after immunization with intact spores, we immunized BALB/c mice on days 1 and 15 with dcG9241 ΔbclA FIS and challenged them on day 29 with a lethal dose of B. anthracis Ames. We observed 80% and 100% survival after i.n. and i.p. challenge, respectively, with B. anthracis Ames (Table 3). The percent survival afforded by immunization with dcG9241 ΔbclA FIS compared to immunization with dcG9241 FIS (P < 0.029) or 569-UM20 FIS (P < 0.0016) was statistically significant for the i.p. challenge route but only suggestive for the i.n. challenge route. The increased survival trend for the dcG9241 ΔbclA FIS-immunized mice suggests that removal of the immunodominant protein BclA from the vaccine may provide a beneficial effect. These findings parallel previous observations that the presence of BclA on the spore surface occludes immunogenic antigens and that the removal of BclA makes these antigens more available to the host immune system (33, 34, 45, 53, 54).
To determine whether the presence of BclA on the spore surface of the challenge strain hinders the protective capacity of antibodies generated against other, less abundant or less accessible spore proteins in response to the vaccination regimen, we challenged immunized mice with B. anthracis Ames ΔbclA spores. Deletion of bclA from B. anthracis Sterne or Ames does not reduce the virulence of either strain in mice (39, 55). Immunization with FIS from any of the strains tested provided significant protection from lethal i.n. or i.p. challenge with B. anthracis Ames ΔbclA; however, purified exosporium failed to confer protection (Table 3). As with B. anthracis Ames challenge, we observed a trend toward increased protection from B. anthracis Ames ΔbclA challenge in mice that were immunized with dcG9241 ΔbclA FIS compared to mice immunized with dcG9241 FIS or 569-UM20 FIS. Overall, these results show that immunization with B. cereus FIS partially or completely protects mice from B. anthracis Ames and Ames ΔbclA i.p. and i.n. challenge and that BclA is not a major contributor to the protection afforded by FIS; in fact, the presence of BclA may actually impede the capacity of the host immune system to recognize and respond to other immunogenic spore proteins.
Antisera from B. cereus FIS-immunized mice are reactive with intact B. anthracis spores.
To assess the serum antibody response against inactivated B. anthracis Ames or B. anthracis Ames ΔbclA spores, we used an ELISA to analyze pooled antisera collected from each vaccination group on days 14 and 28. The day 14 antisera reacted only minimally with spores (data not shown), but the day 28 antisera exhibited a strong antispore response (Fig. 1). The day 28 antisera from mice immunized with dcG9241 or 569-UM20 FIS were significantly more reactive with irradiated B. anthracis Ames and B. anthracis Ames ΔbclA spores (P < 0.001) than were antisera from the PBS/alhydrogel group. Despite the strong protection from B. anthracis Ames and B. anthracis Ames ΔbclA challenge that we observed in dcG9241 ΔbclA FIS-immunized mice, the overall antispore titers in the antisera from mice immunized with dcG9241 ΔbclA FIS differed significantly (P < 0.001) only from those for the PBS/alhydrogel group when tested against B. anthracis Ames ΔbclA spores. However, the reactivity of antisera from mice immunized with dcG9241 ΔbclA FIS against both B. anthracis Ames spores and B. anthracis Ames ΔbclA spores differed significantly from the antibody response generated by mice immunized with dcG9241 FIS or 569-UM20 FIS (P < 0.001). Furthermore, immunization with dcG9241 ΔbclA FIS elicited a significantly lower antibody response against Ames spores and a higher response against Ames ΔbclA spores than did immunization with dcG9241 FIS or 569-UM20 FIS. Consistent with the survival data, the antispore titers generated in response to vaccination with isolated exosporium were similar to those in response to the PBS/alhydrogel control. Taken together, these data demonstrate that vaccination followed by one boost with FIS elicits a robust antispore antibody response in mice at 28 days after the initial immunization. In addition, these findings strongly suggest that the presence of BclA alters the immune response to other spore surface antigens and that other spore proteins contribute to protection.
Fig 1.
Antisera from B. cereus FIS-immunized mice reacted with B. anthracis Ames or B. anthracis Ames ΔbclA spores. The spore-binding activity of the day 28 prechallenge antisera from each of the vaccine groups, i.e., dcG9241 FIS (♦), dcG9241 ΔbclA FIS (▲), 569-UM20 FIS (●), dcG9241 exosporium (■), or PBS/alhydrogel alone (×), was determined by ELISA. (A) Postvaccination, prechallenge antiserum reactivity with inactivated wild-type B. anthracis Ames spores; (B) postvaccination, prechallenge antiserum reactivity with inactivated B. anthracis Ames ΔbclA spores. Asterisks indicate dilutions that were significantly different (P < 0.05) from the same dcG9241 ΔbclA FIS dilution.
Antisera from B. cereus FIS-immunized mice are reactive with potentially novel spore surface immunogens.
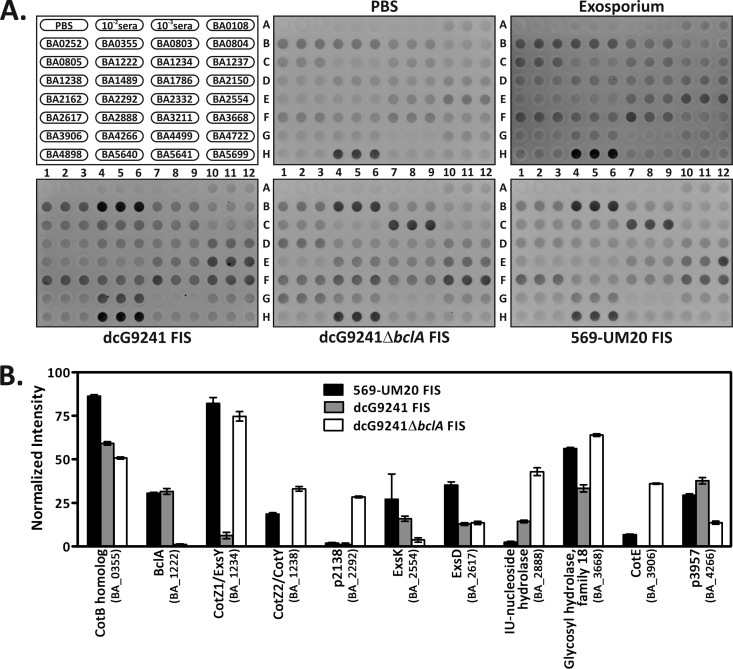
To identify specific B. anthracis proteins to which antibodies were raised in response to vaccination with B. cereus FIS, we tested the reactivity of the day 28 mouse antisera pooled from each vaccination group with a previously reported panel of recombinant spore surface proteins (34) (Fig. 2A). We observed the highest reactivity of the antisera to the cell wall hydrolase (BA_5640) for all of the vaccine groups, including the PBS/alhydrogel group, so we used the cell wall hydrolase signal intensity for the 100% data normalization value. As predicted by the survival and ELISA data, the reactivity of the exosporium vaccine group antisera did not differ significantly from that of the PBS/alhydrogel control group antisera toward any of the proteins in the panel. Compared to the PBS/alhydrogel control group antisera, the antisera from at least one of the FIS-immunized groups reacted significantly with 11 spore proteins (P < 0.001) (Fig. 2B). A comparison of the reactivities of antisera against dcG9241 FIS and dcG9241 ΔbclA FIS with the spore protein panel further illustrated the role of BclA in spore antigen availability; immunization with dcG9241 ΔbclA FIS reduced the antibody response against BclA, ExsK, and p3957 and increased the response against CotZ1/ExsY, CotZ2/CotY, p2138, inosine-uridine-preferring nucleoside hydrolase, glycosyl hydrolase family 18, and CotE compared to immunization with dcG9241 FIS. We conclude that FIS-immunized mice generate antibodies against several proteins that are localized to the accessible surface of the spore and that reactivity of the antisera with some of these proteins is directly related to the presence or absence of BclA on the spore surface.
Fig 2.
Antisera from FIS-immunized mice react with purified spore proteins. (A) Twenty-nine spore proteins were purified, blotted to a nitrocellulose membrane in triplicate, and probed with the day 28 prechallenge antisera from the different immunization groups. The format for each dot blot is shown at the top left. (B) Normalized signal intensities for the 569-UM20 FIS, dcG9241 FIS, or dcG9241 ΔbclA FIS antisera that reacted with proteins from the spore protein panel. The error bars represent one standard error of the mean.
DISCUSSION
The consensus from the literature is that immunization with B. anthracis live spores, inactivated spores, or spore components requires PA to protect mice against challenge with virulent B. anthracis (27, 31–35, 56, 57). Despite this requirement for PA, all of these examples demonstrate the capacity of spore antigens to contribute to vaccine efficacy. In this study, we not only provided further evidence for the contribution of spore components toward vaccine efficacy but also showed that immunization with FIS from nontoxigenic, nonencapsulated B. cereus strains 569-UM20, dcG9241, and dcG9241 ΔbclA protected mice from the highly virulent B. anthracis Ames strain without the addition of exogenous PA to the vaccine regimen. Furthermore, to our knowledge, inactivated B. cereus spores have never been tested as an anthrax vaccine in mice, animals that are inherently difficult to protect from a virulent B. anthracis challenge (28, 58). Thus, the novelty of our protection experiments is in the use B. cereus inactivated spores and in the absence of any form of PA.
Brossier et al. showed that administration of FIS in conjunction with PA elicits a response that completely protects mice and guinea pigs from s.c. challenge with virulent B. anthracis 9602, an encapsulated, toxigenic strain of B. anthracis with virulence similar to that of Ames; either FIS or PA alone provide only minor protection in guinea pigs and no protection in mice (32). A follow-up study by Gauthier et al. showed that vaccination with FIS plus PA protects guinea pigs from i.n. challenge with B. anthracis 9602 but fails to protect mice (35). In those experiments, the FIS vaccine strain was B. anthracis RPLC2, a nontoxigenic Sterne derivative (32, 35). We hypothesize that differences in B. cereus spore composition or antigen presentation compared to B. anthracis RPLC2 spores contributed to the efficacy of B. cereus FIS in our experiments. However, we cannot rule out that experimental differences between our FIS vaccination studies and those previously reported could have contributed to our increased FIS vaccine efficacy. These experimental differences include (i) the use of Swiss outbred mice versus BALB/c mice, (ii) challenge with B. anthracis 9602 versus B. anthracis Ames, (iii) the number of spores administered, and (iv) the time between vaccination and challenge. The use of different mouse strains probably makes little difference from a virulence viewpoint because the LD50 values for the related strain B. anthracis Vollum 1B vary by at most only 10-fold between Swiss outbred and BALB/c mice (58). The LD50 values for B. anthracis Ames and 9602 are similar for i.p. inoculation (32, 49), but B. anthracis 9602 is 10- to 100-fold less virulent than Ames by i.n. administration (35, 48, 59). Since the challenge doses in each experiment were based on LD50 values for each bacterial challenge strain in a given mouse strain, the actual number of spores administered also should not contribute to the different FIS vaccination outcomes between our study and previous reports. Lastly, in our experiments, we challenged the mice 2 weeks after the booster injection, while the challenge occurred 3 weeks after the boost in the previously reported FIS studies. Since our mice were challenged 1 week earlier than mice in the other study, our mice actually had less time to mount an immune response. Therefore, the time between vaccination and challenge most likely did not contribute to the different outcomes. We contend that the most significant difference between our experiments and those of others was the choice of vaccine strain. The increased survival rates in the dcG9241 and dcG9241 ΔbclA FIS-vaccinated mice were all statistically significant compared to those for the PBS/alhydrogel controls, while the increased protection afforded by 569-UM20 FIS vaccination was only variably significant (Table 3). This trend suggests that vaccination with either dcG9241 or dcG9241 ΔbclA FIS is more protective, a finding that highlights the importance of the strain chosen for the FIS vaccine.
BclA is highly immunogenic and is the major immunogen on the spore surface (47). However, here we demonstrated that the presence of BclA on FIS did not contribute to better protection against Ames challenge; in fact, the increased survival trend observed in the dcG9241 ΔbclA FIS vaccination group suggests that removal of BclA from the spore surface is beneficial to the FIS vaccine. Thus, mice vaccinated with dcG9241 ΔbclA FIS were better protected than those vaccinated with the BclA-positive strain dcG2941 or 569-UM20 FIS, although this enhanced protection was statistically significant only for the i.p. challenge route. The relative lack of importance of BclA in protection is supported by the observations that B. anthracis Sterne FIS and Sterne ΔbclA FIS induce similar gamma interferon responses in mouse splenocytes (60) and that BclA can occlude other spore surface antigens from the host (34, 39, 53). Thus, removal of BclA from the spore surface, as we did genetically, would allow these potential antigens to be better recognized and presented to the host immune system. The B. anthracis Ames and Ames ΔbclA ELISAs further support this hypothesis. When Ames spores were probed with antisera from FIS-immunized mice, the BclA-positive FIS groups displayed the highest reactivity and were statistically significantly different from the PBS/alhydrogel and dcG9241 ΔbclA FIS groups; however, when Ames ΔbclA spores were probed, the antiserum pool from the dcG9241 ΔbclA FIS group displayed the highest reactivity and was statistically significantly different from the PBS/alhydrogel and the BclA-positive groups. The survival data combined with ELISA data suggest that protective antigens other than BclA are present on the spore surface and that the lack of BclA on the dcG9241 ΔbclA FIS surface is the reason that this group was protected most effectively from B. anthracis Ames challenge.
The immuno-dot blots show the postvaccination, prechallenge antiserum reactivity with 11 spore proteins from the recombinant spore protein panel and also demonstrate the contribution of BclA toward antigen availability. In addition to BclA (BA_1222), the proteins ExsK (BA_2554) and p3957 (BA_4266) reacted less with the dcG9241 ΔbclA FIS group's antisera than with the dcG9241 FIS group's antisera. BclA is required for ExsK to form high-molecular-weight complexes on the surface of the exosporium (54), and therefore, removal of BclA from the spore would inhibit localization of ExsK on the spore surface. The possible requirement of BclA to localize the hypothetical protein p3957 to the spore surface is currently unknown. Six proteins were more reactive with antisera raised by immunization with BclA-negative FIS: CotZ1/ExsY (BA_1234), CotZ2/CotY (BA_1238), p2138 (BA_2292), inosine-uridine-preferring nucleoside hydrolase (BA_2888), glycosyl hydrolase family 18 (BA_3668), and CotE (BA_3906). Both CotZ1/ExsY and CotZ2/CotY form multimeric complexes with BclA and BxpB/ExsFA (46, 61), and this complex may prevent recognition of CotZ1/ExsY and CotZ2/CotY by the host immune system. There are no reports of direct interactions between BclA and any of the remaining four proteins. The increased signal of the dcG9241 ΔbclA FIS antisera versus dcG9241 FIS antisera with the remaining four proteins may be due to the proximity of these proteins to BclA on the spore surface. Of note, the 569-UM20 FIS antisera and dcG9241 ΔbclA FIS antisera had similar reactivities with three of these six proteins, CotZ1/ExsY, CotZ2/CotY, and glycosyl hydrolase, an observation that could suggest that the accessible spore surface of 569-UM20 is sufficiently different from the dcG9241 spore surface to permit generation of antibodies toward these proteins. A BLAST query (62, 63) of the B. cereus G9241 BclA amino acid sequence against the nonredundant protein sequences database limited to B. cereus 569 (B. cereus ATCC 10876) yielded two possible BclA homologs present in 569-UM20, BCERE0002_43750 and BCERE0002_21830. The 569-UM20 homologs are 69 and 77 amino acids larger than the G9241 BclA homolog, and they have a relatively low amino acid sequence identity (∼37% as calculated by a global alignment with ALIGN [64, 65]) (data not shown). Regardless of which gene product is the true BclA homolog, both proteins are sufficiently different from the BclA protein encoded by dcG9241 to alter spore surface accessibility of 569-UM20 toward CotZ1/ExsY, CotZ2/CotY, and glycosyl hydrolase. The increased reactivity of the 569-UM20 FIS antisera toward the two remaining proteins, the CotB homolog (BA_0355) and ExsD (BA_2617), may also be explained by the probability of a different spore surface morphology between these two bacterial strains. The differences in reactivity between dcG9241 and 569-UM20 FIS antisera could also be a result of amino acid sequence variations between the spore protein homologs of these two species, which, in turn, could provide them with different antigenic properties.
Of the 11 antiserum-reactive proteins, six have been previously identified as vaccine candidates. Rabbit antiserum generated against live B. anthracis Sterne spores was previously found to be reactive with glycosyl hydrolase in a screen to find novel vaccine candidates (66). BclA, CotZ1/ExsY, ExsK, ExsD, and p3957 were detected by rabbit polyclonal antiserum 311001-01 generated against inactivated whole spores (34). Of these six proteins, all but ExsD and glycosyl hydrolase were previously evaluated as vaccine candidates in conjunction with PA (27, 31, 33, 34); only vaccination with BclA provided some protection to mice from either B. anthracis Sterne (31) or Ames (27) challenge. It should be noted that all of the previously tested vaccine proteins were cloned from B. anthracis Sterne and that amino acid sequence differences exist between B. anthracis Sterne and the B. cereus G9241 protein homologs. The amino acid sequence identities between the G9241 and Sterne proteins are as follows: BclA, 66.5%; CotZ1/ExsY, 82.2%; ExsK, 54.8%; and p3957, 85.6% (global alignment and percent identity for each pair of protein sequences was calculated with ALIGN [64, 65]) (data not shown). It is possible that the G9241 variants of these previously tested proteins may perform better as vaccine components than the Sterne counterparts. In addition, there are likely other spore proteins that have yet to be recognized.
The complete lack of efficacy of vaccination with purified dcG9241 exosporium was unexpected. There are several possible explanations for this finding. First, the lack of a significant response to exosporium as indicated by the ELISA data suggests that insufficient amounts of the exosporium were used for immunization. Steichen et al. reported the generation of antibodies directed toward purified exosporium in which they injected BALB/c mice with 50 μg of purified exosporium initially in complete Freund's adjuvant and then 4 more times in saline every 3 days (47). In our experiments we used 30 μg of exosporium injected with an alhydrogel adjuvant only twice, 2 weeks apart. The difference between 30 μg and 50 μg is probably not significant and most likely would not explain the lack of an immune response. However, the increased time between boosters, the decreased injection frequency, and the choice of adjuvant used in our experiments all could have contributed to the lack of a robust host immune response from purified exosporium. Furthermore, it is also possible that purified exosporium has a short half-life in the mouse, which could reduce its immunogenicity. While we could have increased the frequency of the immunizations, we wanted the immunization schedule to be the same for all groups. It is also possible that the method we used to isolate exosporium resulted in the loss of protective spore antigens. The procedure incorporated sonication to disrupt the spore particle, a low-speed centrifugation step to remove nonlysed spores and debris, and a high-speed ultracentrifugation step to pellet the exosporium-containing outer spore membrane fraction. With such a procedure, any proteins not tightly associated with or integral to the outer spore membrane could have been lost during the purification process. Another possible explanation for the ineffectiveness of the exosporium as a vaccine is that the spore particle itself may act as a scaffold for antigen presentation, may protect the antigens from degradation, or may act as an additional adjuvant. The approximate size of spores may alter the immune response or antigen presentation, as observed with the use of synthetic microparticles in other vaccine platforms (67). A requirement for a scaffold could explain the lack of protection afforded by immunization with recombinant spore proteins alone in previous studies; however, it is also possible that antibodies against these spore proteins simply were not protective in the absence of PA and that an effective vaccine comprised solely of spore proteins remains to be discovered.
ACKNOWLEDGMENTS
We thank Diana Fisher for her expert statistical analyses of the survival data, Cara Olsen for her advice and assistance with the ELISA statistical analysis, and Steven Tobery and Avesta Ebrahimi for their technical assistance.
This work was funded by the Biological Defense Research Directorate, Naval Medical Research Center, U.S. Navy (to A.D.O.), and JSTO-CBD/DTRA projects 1.1A0010-070RDB and CMB VAXBT-03-10-RDP 004 (to S.L.W.).
The opinions, interpretations, conclusions, and recommendations expressed are those of the authors and do not reflect an official policy or position of the Department of Army, Department of Navy, Department of Defense, or U.S. Government. Research was conducted under an IACUC-approved protocol in compliance with the Animal Welfare Act, PHS policy, and other Federal statutes and regulations relating to animals and experiments involving animals. The facility where this research was conducted is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International, and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011.
We have no conflicts of interest.
Footnotes
Published ahead of print 31 October 2012
REFERENCES
- 1. Centers for Disease Control and Prevention 2008. Cutaneous anthrax associated with drum making using goat hides from West Africa—Connecticut, 2007. 57:628–631 [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention 2010. Gastrointestinal anthrax after an animal-hide drumming event—New Hampshire and Massachusetts, 2009. 59:872–877 [PubMed] [Google Scholar]
- 3. Guh A, Heyman ML, Barden D, Fontana J, Hadler JL. 2010. Lessons learned from the investigation of a cluster of cutaneous anthrax cases in Connecticut. J. Public Health Manag. Pract. 16:201–210 [DOI] [PubMed] [Google Scholar]
- 4. Jernigan DB, Raghunathan PL, Bell BP, Brechner R, Bresnitz EA, Butler JC, Cetron M, Cohen M, Doyle T, Fischer M, Greene C, Griffith KS, Guarner J, Hadler JL, Hayslett JA, Meyer R, Petersen LR, Phillips M, Pinner R, Popovic T, Quinn CP, Reefhuis J, Reissman D, Rosenstein N, Schuchat A, Shieh WJ, Siegal L, Swerdlow DL, Tenover FC, Traeger M, Ward JW, Weisfuse I, Wiersma S, Yeskey K, Zaki S, Ashford DA, Perkins BA, Ostroff S, Hughes J, Fleming D, Koplan JP, Gerberding JL, National Anthrax Epidemiologic Investigation Team. 2002. Investigation of bioterrorism-related anthrax, United States, 2001: epidemiologic findings. Emerg. Infect. Dis. 8:1019–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Avashia SB, Riggins WS, Lindley C, Hoffmaster A, Drumgoole R, Nekomoto T, Jackson PJ, Hill KK, Williams K, Lehman L, Libal MC, Wilkins PP, Alexander J, Tvaryanas A, Betz T. 2007. Fatal pneumonia among metalworkers due to inhalation exposure to Bacillus cereus containing Bacillus anthracis toxin genes. Clin. Infect. Dis. 44:414–416 [DOI] [PubMed] [Google Scholar]
- 6. Bottone EJ. 2010. Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 23:382–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hoffmaster AR, Hill KK, Gee JE, Marston CK, De BK, Popovic T, Sue D, Wilkins PP, Avashia SB, Drumgoole R, Helma CH, Ticknor LO, Okinaka RT, Jackson PJ. 2006. Characterization of Bacillus cereus isolates associated with fatal pneumonias: strains are closely related to Bacillus anthracis and harbor B. anthracis virulence genes. J. Clin. Microbiol. 44:3352–3360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoffmaster AR, Ravel J, Rasko DA, Chapman GD, Chute MD, Marston CK, De BK, Sacchi CT, Fitzgerald C, Mayer LW, Maiden MC, Priest FG, Barker M, Jiang L, Cer RZ, Rilstone J, Peterson SN, Weyant RS, Galloway DR, Read TD, Popovic T, Fraser CM. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl. Acad. Sci. U. S. A. 101:8449–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller JM, Hair JG, Hebert M, Hebert L, Roberts FJ, Weyant RS. 1997. Fulminating bacteremia and pneumonia due to Bacillus cereus. J. Clin. Microbiol. 35:504–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collier RJ, Young JAT. 2003. Anthrax toxin. Annu. Rev. Cell Dev. Biol. 19:45–70 [DOI] [PubMed] [Google Scholar]
- 11. Firoved AM, Miller GF, Moayeri M, Kakkar R, Shen Y, Wiggins JF, McNally EM, Tang W-J, Leppla SH. 2005. Bacillus anthracis edema toxin causes extensive tissue lesions and rapid lethality in mice. Am. J. Pathol. 167:1309–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leppla SH. 1982. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc. Natl. Acad. Sci. U. S. A. 79:3162–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pezard C, Berche P, Mock M. 1991. Contribution of individual toxin components to virulence of Bacillus anthracis. Infect. Immun. 59:3472–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Brien J, Friedlander A, Dreier T, Ezzell J, Leppla S. 1985. Effects of anthrax toxin components on human neutrophils. Infect. Immun. 47:306–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duesbery NS, Webb CP, Leppla SH, Gordon VM, Klimpel KR, Copeland TD, Ahn NG, Oskarsson MK, Fukasawa K, Paull KD, Vande Woude GF. 1998. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science 280:734–737 [DOI] [PubMed] [Google Scholar]
- 16. Vitale G, Pellizzari R, Recchi C, Napolitani G, Mock M, Montecucco C. 1999. Anthrax lethal factor cleaves the N-terminus of MAPKKS and induces tyrosine/threonine phosphorylation of MAPKS in cultured macrophages. J. Appl. Microbiol. 87:288. [DOI] [PubMed] [Google Scholar]
- 17. Fouet A. 2009. The surface of Bacillus anthracis. Mol. Aspects Med. 30:374–385 [DOI] [PubMed] [Google Scholar]
- 18. Wilson MK, Vergis JM, Alem F, Palmer JR, Keane-Myers AM, Brahmbhatt TN, Ventura CL, O'Brien AD. 2011. Bacillus cereus G9241 makes anthrax toxin and capsule like highly virulent B. anthracis Ames but behaves like attenuated toxigenic nonencapsulated B. anthracis Sterne in rabbits and mice. Infect. Immun. 79:3012–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oh SY, Budzik JM, Garufi G, Schneewind O. 2011. Two capsular polysaccharides enable Bacillus cereus G9241 to cause anthrax-like disease. Mol. Microbiol. 80:455–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Turnbull PC. 1991. Anthrax vaccines: past, present and future. Vaccine 9:533–539 [DOI] [PubMed] [Google Scholar]
- 21. Little SF, Ivins BE, Fellows PF, Friedlander AM. 1997. Passive protection by polyclonal antibodies against Bacillus anthracis infection in guinea pigs. Infect. Immun. 65:5171–5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Friedlander AM, Little SF. 2009. Advances in the development of next-generation anthrax vaccines. Vaccine 27:D28–D32 [DOI] [PubMed] [Google Scholar]
- 23. Wright JG, Quinn CP, Shadomy S, Messonnier N, Centers for Disease Control and Prevention. 2010. Use of anthrax vaccine in the United States: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2009. MMWR Recomm. Rep. 59:1–30 [PubMed] [Google Scholar]
- 24. Marano N, Plikaytis BD, Martin SW, Rose C, Semenova VA, Martin SK, Freeman AE, Li H, Mulligan MJ, Parker SD, Babcock J, Keitel W, El Sahly H, Poland GA, Jacobson RM, Keyserling HL, Soroka SD, Fox SP, Stamper JL, McNeil MM, Perkins BA, Messonnier N, Quinn CP, Anthrax Vaccine Research Program Working Group. 2008. Effects of a reduced dose schedule and intramuscular administration of anthrax vaccine adsorbed on immunogenicity and safety at 7 months: a randomized trial. JAMA 300:1532–1543 [DOI] [PubMed] [Google Scholar]
- 25. Niu M, Ball R. 2009. 2009. Adverse events after anthrax vaccination reported to the Vaccine Adverse Event Reporting System (VAERS), 1990–2007. Vaccine 27:290–297 [DOI] [PubMed] [Google Scholar]
- 26. Flick-Smith HC, Waters EL, Walker NJ, Miller J, Stagg AJ, Green M, Williamson ED. 2005. Mouse model characterisation for anthrax vaccine development: comparison of one inbred and one outbred mouse strain. Microb. Pathog. 38:33–40 [DOI] [PubMed] [Google Scholar]
- 27. Hahn UK, Boehm R, Beyer W. 2006. DNA vaccination against anthrax in mice—combination of anti-spore and anti-toxin components. Vaccine 24:4569–4571 [DOI] [PubMed] [Google Scholar]
- 28. Welkos SL, Friedlander AM. 1988. Comparative safety and efficacy against Bacillus anthracis of protective antigen and live vaccines in mice. Microb. Pathog. 5:127–139 [DOI] [PubMed] [Google Scholar]
- 29. Fellows PF, Linscott MK, Ivins BE, Pitt MLM, Rossi CA, Gibbs PH, Friedlander AM. 2001. Efficacy of a human anthrax vaccine in guinea pigs, rabbits, and rhesus macaques against challenge by Bacillus anthracis isolates of diverse geographical origin. Vaccine 19:3241–3247 [DOI] [PubMed] [Google Scholar]
- 30. Little SF, Knudson GB. 1986. Comparative efficacy of Bacillus anthracis live spore vaccine and protective antigen vaccine against anthrax in the guinea pig. Infect. Immun. 52:509–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brahmbhatt TN, Darnell SC, Carvalho HM, Sanz P, Kang TJ, Bull RL, Rasmussen SB, Cross AS, O'Brien AD. 2007. Recombinant exosporium protein BclA of Bacillus anthracis is effective as a booster for mice primed with suboptimal amounts of protective antigen. Infect. Immun. 75:5240–5247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brossier F, Levy M, Mock M. 2002. Anthrax spores make an essential contribution to vaccine efficacy. Infect. Immun. 70:661–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cote CK, Kaatz L, Reinhardt J, Bozue J, Tobery S, Bassett A, Sanz P, Darnell SC, Alem F, O'Brien AD, Welkos SL. 2012. Characterization of a multi-component anthrax vaccine designed to target the initial stages of infection as well as toxemia. J. Med. Microbiol. 61:1380–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cybulski RJ, Jr, Sanz P, McDaniel D, Darnell S, Bull RL, O'Brien AD. 2008. Recombinant Bacillus anthracis spore proteins enhance protection of mice primed with suboptimal amounts of protective antigen. Vaccine 26:4927–4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gauthier YP, Tournier JN, Paucod JC, Corre JP, Mock M, Goossens PL, Vidal DR. 2009. Efficacy of a vaccine based on protective antigen and killed spores against experimental inhalational anthrax. Infect. Immun. 77:1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shlyakhov EN, Rubinstein E. 1994. Human live anthrax vaccine in the former USSR. Vaccine 12:727–730 [DOI] [PubMed] [Google Scholar]
- 37. Turnbull PC. (ed). 2008. Anthrax in humans and animals, 4th ed World Health Organization Press, Geneva, Switzwerland [Google Scholar]
- 38. Vergis JM, Cote CK, Bozue J, Alem F, Ventura CL, Welkos SL, O'Brien AD. 2012. Abstr. 112th Gen. Meet. Am. Soc. Microbiol., San Francisco, CA, June 2012, abstr 812 [Google Scholar]
- 39. Bozue J, Cote CK, Moody KL, Welkos SL. 2007. Fully virulent Bacillus anthracis does not require the immunodominant protein BclA for pathogenesis. Infect. Immun. 75:508–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Green BD, Battisti L, Koehler TM, Thorne CB, Ivins BE. 1985. Demonstration of a capsule plasmid in Bacillus anthracis. Infect. Immun. 49:291–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen Y, Tenover FC, Koehler TM. 2004. Beta-lactamase gene expression in a penicillin-resistant Bacillus anthracis strain. Antimicrob. Agents Chemother. 48:4873–4877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cote CK, Van Rooijen N, Welkos SL. 2006. Roles of macrophages and neutrophils in the early host response to Bacillus anthracis spores in a mouse model of infection. Infect. Immun. 74:469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dai Z, Sirard J-C, Mock M, Koehler TM. 1995. The atxA gene product activates transcription of the anthrax toxin genes and is essential for virulence. Mol. Microbiol. 16:1171–1181 [DOI] [PubMed] [Google Scholar]
- 44. Peng D, Luo Y, Guo S, Zeng H, Ju S, Yu Z, Sun M. 2009. Elaboration of an electroporation protocol for large plasmids and wild-type strains of Bacillus thuringiensis. J. Appl. Microbiol. 106:1849–1858 [DOI] [PubMed] [Google Scholar]
- 45. Brahmbhatt TN, Janes BK, Stibitz ES, Darnell SC, Sanz P, Rasmussen SB, O'Brien AD. 2007. Bacillus anthracis exosporium protein BclA affects spore germination, interaction with extracellular matrix proteins, and hydrophobicity. Infect. Immun. 75:5233–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Redmond C, Baillie LWJ, Hibbs S, Moir AJG, Moir A. 2004. Identification of proteins in the exosporium of Bacillus anthracis. Microbiology 150:355–363 [DOI] [PubMed] [Google Scholar]
- 47. Steichen C, Chen P, Kearney JF, Turnbough J, Charles L. 2003. Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J. Bacteriol. 185:1903–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lyons CR, Lovchik J, Hutt J, Lipscomb MF, Wang E, Heninger S, Berliba L, Garrison K. 2004. Murine model of pulmonary anthrax: kinetics of dissemination, histopathology, and mouse strain susceptibility. Infect. Immun. 72:4801–4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Popov SG, Popova TG, Grene E, Klotz F, Cardwell J, Bradburne C, Jama Y, Maland M, Wells J, Nalca A, Voss T, Bailey C, Alibek K. 2004. Systemic cytokine response in murine anthrax. Cell. Microbiol. 6:225–233 [DOI] [PubMed] [Google Scholar]
- 50. Cote CK, Rossi CA, Kang AS, Morrow PR, Lee JS, Welkos SL. 2005. The detection of protective antigen (PA) associated with spores of Bacillus anthracis and the effects of anti-PA antibodies on spore germination and macrophage interactions. Microb. Pathog. 38:209–225 [DOI] [PubMed] [Google Scholar]
- 51. Welkos SL, Cote CK, Rea KM, Gibbs PH. 2004. A microtiter fluorometric assay to detect the germination of Bacillus anthracis spores and the germination inhibitory effects of antibodies. J. Microbiol. Methods 56:253–265 [DOI] [PubMed] [Google Scholar]
- 52. LePage GA, Morgan JF, Campbell ME. 1946. Production and purification of penicillinase. J. Biol. Chem. 166:465–472 [PubMed] [Google Scholar]
- 53. Basu S, Kang TJ, Chen WH, Fenton MJ, Baillie L, Hibbs S, Cross AS. 2007. Role of Bacillus anthracis spore structures in macrophage cytokine responses. Infect. Immun. 75:2351–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Severson KM, Mallozzi M, Bozue J, Welkos SL, Cote CK, Knight KL, Driks A. 2009. Roles of the Bacillus anthracis spore protein ExsK in exosporium maturation and germination. J. Bacteriol. 191:7587–7596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sylvestre P, Couture-Tosi E, Mock M. 2002. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol. 45:169–178 [DOI] [PubMed] [Google Scholar]
- 56. Barnard JP, Friedlander AM. 1999. Vaccination against anthrax with attenuated recombinant strains of Bacillus anthracis that produce protective antigen. Infect. Immun. 67:562–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cohen S, Mendelson I, Altboum Z, Kobiler D, Elhanany E, Bino T, Leitner M, Inbar I, Rosenberg H, Gozes Y, Barak R, Fisher M, Kronman C, Velan B, Shafferman A. 2000. Attenuated nontoxinogenic and nonencapsulated recombinant Bacillus anthracis spore vaccines protect against anthrax. Infect. Immun. 68:4549–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Welkos SL, Keener TJ, Gibbs PH. 1986. Differences in susceptibility of inbred mice to Bacillus anthracis. Infect. Immun. 51:795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Steward J, Lever MS, Simpson AJH, Sefton AM, Brooks TJG. 2004. Post-exposure prophylaxis of systemic anthrax in mice and treatment with fluoroquinolones. J. Antimicrob. Chemother. 54:95–99 [DOI] [PubMed] [Google Scholar]
- 60. Glomski IJ, Fritz JH, Keppler SJ, Balloy V, Chignard M, Mock M, Goossens PL. 2007. Murine splenocytes produce inflammatory cytokines in a MyD88-dependent response to Bacillus anthracis spores. Cell. Microbiol. 9:502–513 [DOI] [PubMed] [Google Scholar]
- 61. Steichen CT, Kearney JF, Turnbough CL. 2005. Characterization of the exosporium basal layer protein BxpB of Bacillus anthracis. J. Bacteriol. 187:5868–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schäffer AA, Yu Y-K. 2005. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272:5101–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pearson WR. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63–98 [DOI] [PubMed] [Google Scholar]
- 65. Pearson WR, Lipman DJ. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U. S. A. 85:2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gat O, Grosfeld H, Ariel N, Inbar I, Zaide G, Broder Y, Zvi A, Chitlaru T, Altboum Z, Stein D, Cohen S, Shafferman A. 2006. Search for Bacillus anthracis potential vaccine candidates by a functional genomic-serologic screen. Infect. Immun. 74:3987–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Oyewumi MO, Kumar A, Cui Z. 2010. Nano-microparticles as immune adjuvants: correlating particle sizes and the resultant immune responses. Expert Rev. Vaccines 9:1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]