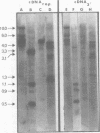
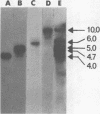
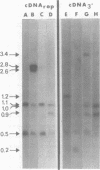
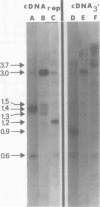
Abstract
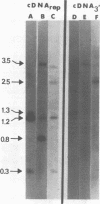
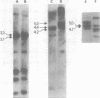
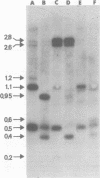
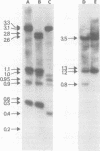
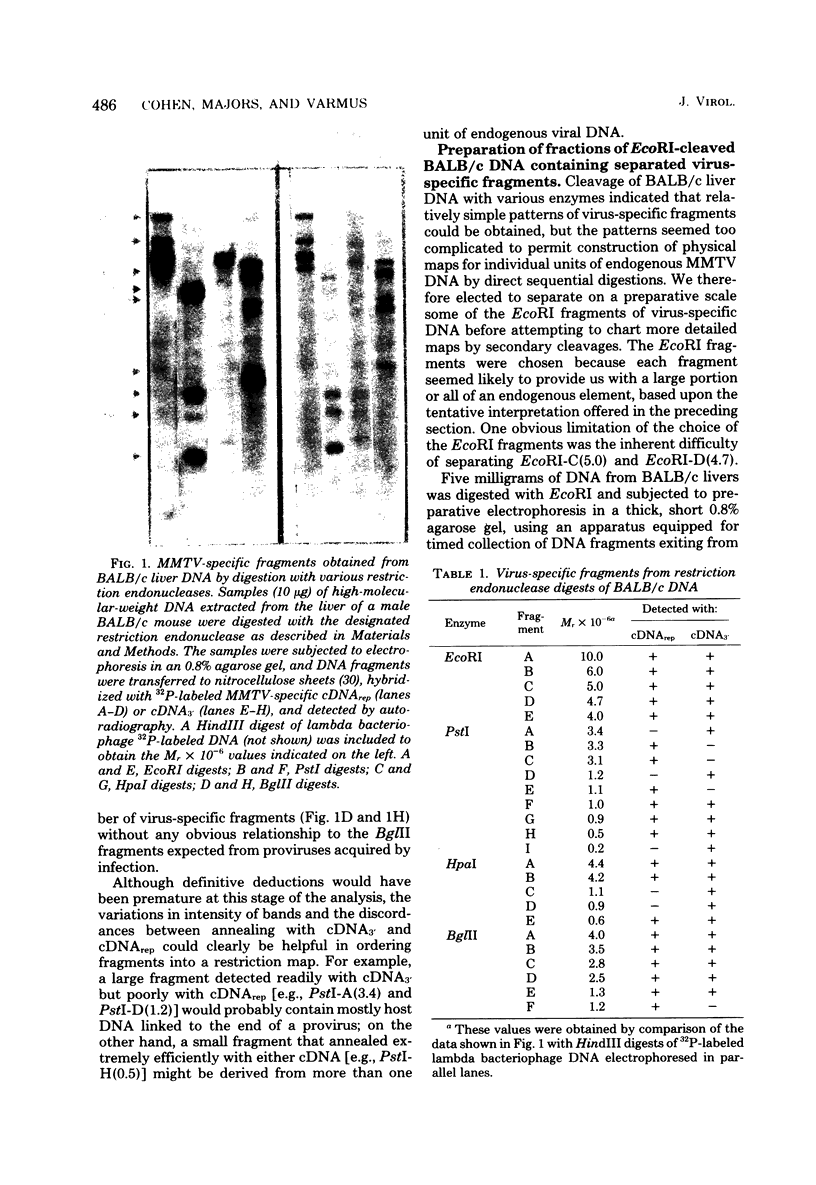
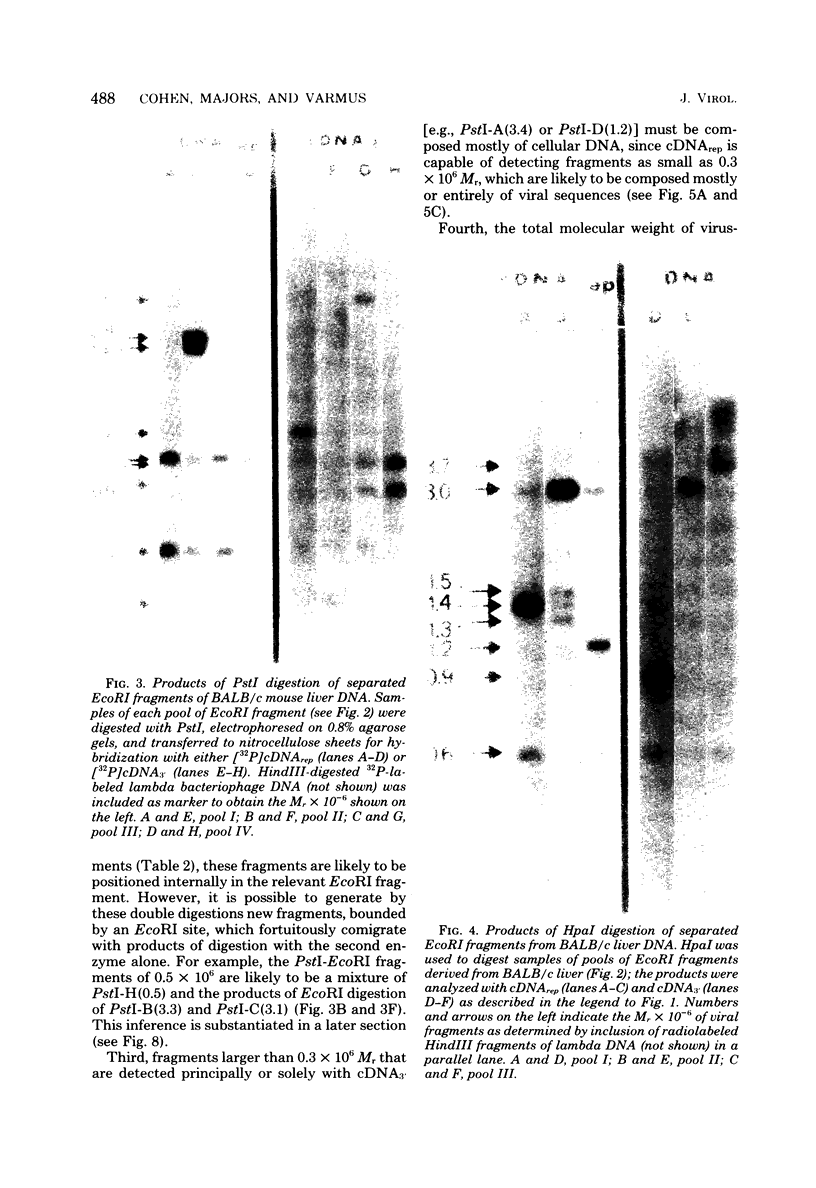
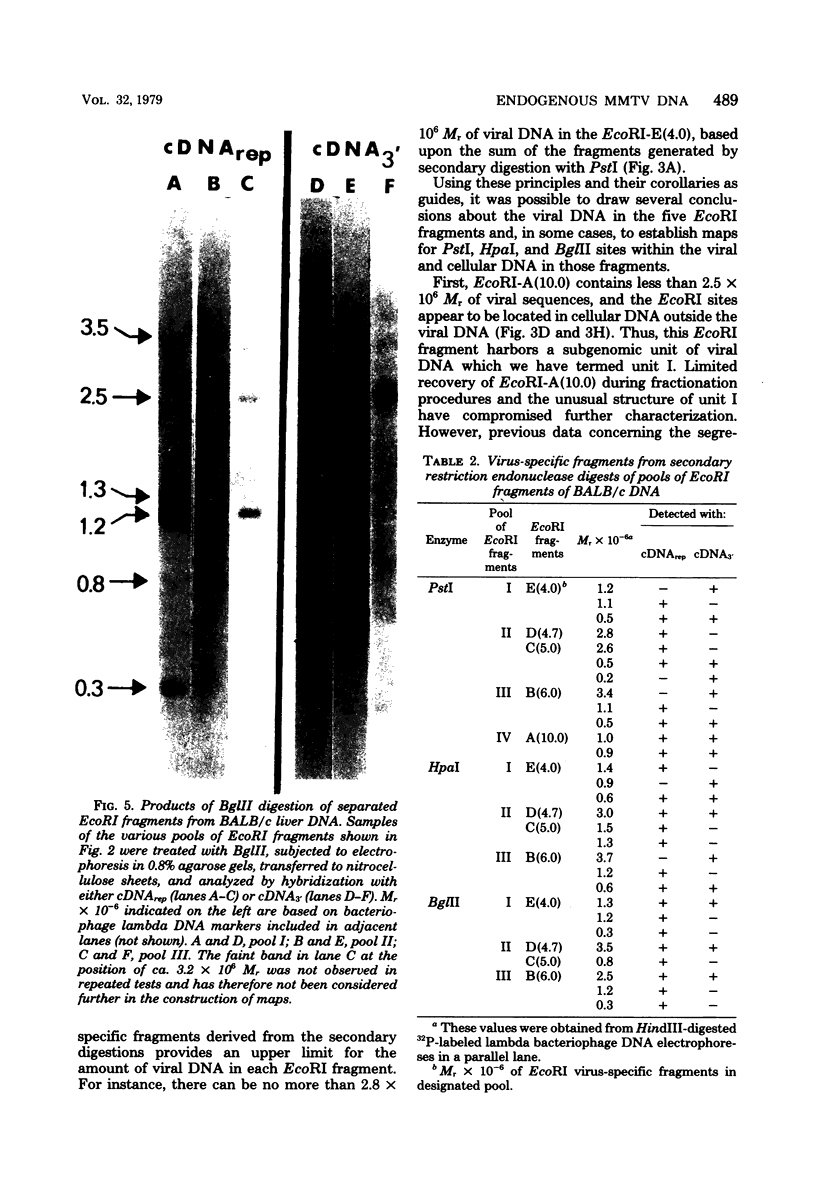
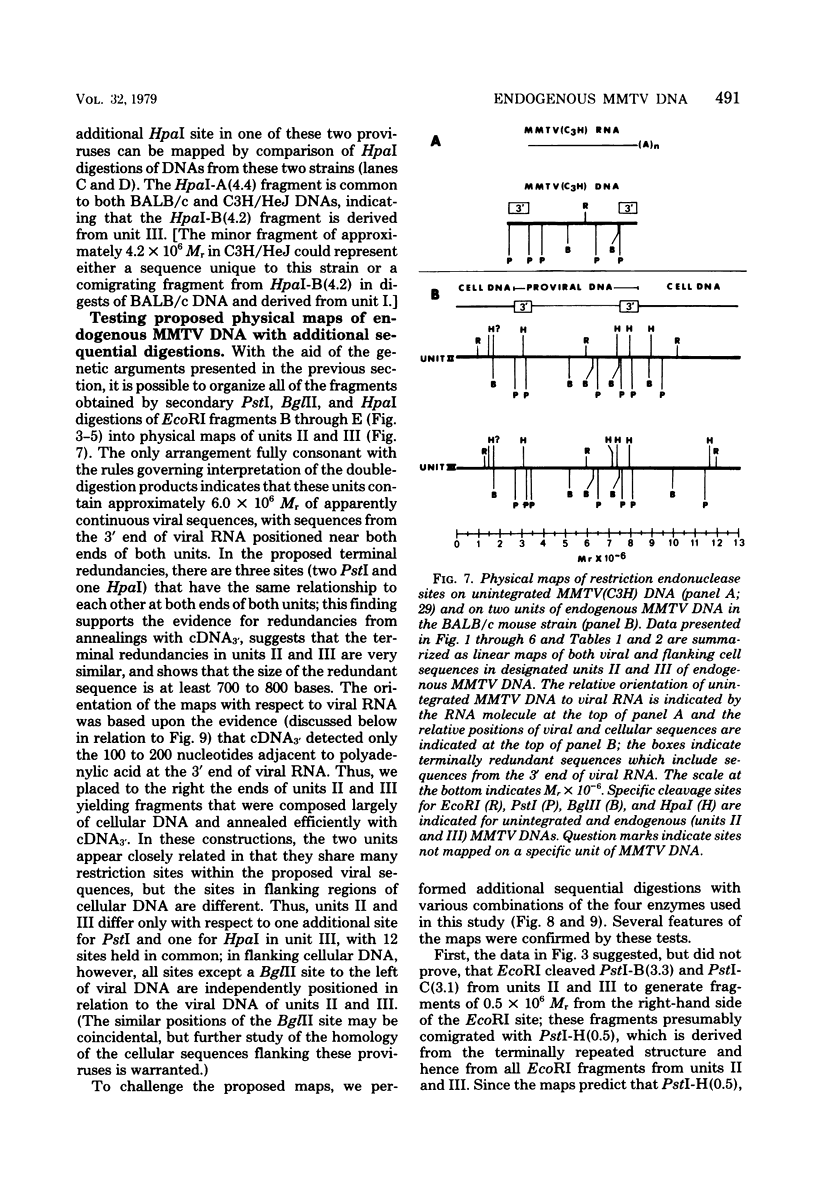
We used restriction endonucleases to prepare physical maps of the mouse mammary tumor virus (MMTV)-specific DNA endogenous to the BALB/c mouse strain. The mapping was facilitated by the DNA transfer procedure, using complementary DNAs specific for the whole and for the 3' terminus of MMTV RNA to detect fragments containing viral sequences. The strategies used for the arrangement of fragments into physical maps included sequential digestions with two or three enzymes; preparative isolation of EcoRI fragments containing viral sequences; and comparisons of virus-specific fragments derived from the DNA of several mouse strains. Most of the MMTV-related DNA in the BALB/c genome is organized into two units (II and III) which strongly resemble proviruses acquired upon horizontal infection with milk-borne strains of MMTV and other retroviruses. These units contain approximately 6.0 x 10(6) Mr of apparently uninterrupted viral sequences, they bear redundant sequences totaling at least 700 to 800 base pairs at their termini, and the terminal redundancies include sequences derived from the 3' end of MMTV RNA. Units II and III are closely related in that they share 12 of 14 recognition sites for endonucleases, but cellular sequences flanking units II and III are dissimilar by this criterion. The remainder of the MMTV-related DNA endogenous to BALB/c mice is found in a single subgenomic unit (unit I) with a complexity of ca. 2 x 10(6) Mr; the structure of this unit has not been further defined. These results support the hypotheses that endogenous proviruses have been acquired by infection of germinal tissues with MMTV. The physical maps are also useful for identifying the MMTV genomes endogenous to BALB/c mice in studies of the natural history of mammary tumorigenesis.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aaronson S. A., Stephenson J. R. Endogenous type-C RNA viruses of mammalian cells. Biochim Biophys Acta. 1976 Dec 23;458(4):323–354. doi: 10.1016/0304-419x(76)90006-8. [DOI] [PubMed] [Google Scholar]
- Astrin S. M. Endogenous viral genes of the White Leghorn chicken: common site of residence and sites associated with specific phenotypes of viral gene expression. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5941–5945. doi: 10.1073/pnas.75.12.5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentvelzen P. Host-virus interactions in murine mammary carcinogenesis. Biochim Biophys Acta. 1974 Dec 31;355(3-4):236–259. doi: 10.1016/0304-419x(74)90012-2. [DOI] [PubMed] [Google Scholar]
- Cohen J. C., Shank P. R., Morris V. L., Cardiff R., Varmus H. E. Integration of the DNA of mouse mammary tumor virus in virus-infected normal and neoplastic tissue of the mouse. Cell. 1979 Feb;16(2):333–345. doi: 10.1016/0092-8674(79)90010-2. [DOI] [PubMed] [Google Scholar]
- Cohen J. C., Varmus H. E. Endogenous mammary tumour virus DNA varies among wild mice and segregates during inbreeding. Nature. 1979 Mar 29;278(5703):418–423. doi: 10.1038/278418a0. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Drohan W., Kettmann R., Colcher D., Schlom J. Isolation of the mouse mammary tumor virus sequences not transmitted as germinal provirus in the C3H and RIII mouse strains. J Virol. 1977 Mar;21(3):986–995. doi: 10.1128/jvi.21.3.986-995.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J. P., Rosen J. M., Butel J. S. Differential expression of poly(A)-adjacent sequences of mammary tumor virus RNA in murine mammary cells. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5797–5801. doi: 10.1073/pnas.75.12.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisby D. P., Weiss R. A., Roussel M., Stehelin D. The distribution of endogenous chicken retrovirus sequences in the DNA of galliform birds does not coincide with avian phylogenetic relationships. Cell. 1979 Jul;17(3):623–634. doi: 10.1016/0092-8674(79)90270-8. [DOI] [PubMed] [Google Scholar]
- Gillespie D., Gillespie S., Gallo R. C., East J. L., Dmochowski L. Genetic origin of RD114 and other RNA tumour viruses assayed by molecular hybridization. Nat New Biol. 1973 Jul 11;244(132):51–54. doi: 10.1038/newbio244051a0. [DOI] [PubMed] [Google Scholar]
- Goulian M. Incorporation of oligodeoxynucleotides into DNA. Proc Natl Acad Sci U S A. 1968 Sep;61(1):284–291. doi: 10.1073/pnas.61.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene P. J., Heyneker H. L., Bolivar F., Rodriguez R. L., Betlach M. C., Covarrubias A. A., Backman K., Russel D. J., Tait R., Boyer H. W. A general method for the purification of restriction enzymes. Nucleic Acids Res. 1978 Jul;5(7):2373–2380. doi: 10.1093/nar/5.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. H., Payvar F., Spector D., Schimke R. T., Robinson H. L., Payne G. S., Bishop J. M., Varmus H. E. Heterogeneity of genetic loci in chickens: analysis of endogenous viral and nonviral genes by cleavage of DNA with restriction endonucleases. Cell. 1979 Oct;18(2):347–359. doi: 10.1016/0092-8674(79)90054-0. [DOI] [PubMed] [Google Scholar]
- Hughes S. H., Shank P. R., Spector D. H., Kung H. J., Bishop J. M., Varmus H. E., Vogt P. K., Breitman M. L. Proviruses of avian sarcoma virus are terminally redundant, co-extensive with unintegrated linear DNA and integrated at many sites. Cell. 1978 Dec;15(4):1397–1410. doi: 10.1016/0092-8674(78)90064-8. [DOI] [PubMed] [Google Scholar]
- Kelly R. B., Cozzarelli N. R., Deutscher M. P., Lehman I. R., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XXXII. Replication of duplex deoxyribonucleic acid by polymerase at a single strand break. J Biol Chem. 1970 Jan 10;245(1):39–45. [PubMed] [Google Scholar]
- Keshet E., Temin H. M. Sites of integration of reticuloendotheliosis virus DNA in chicken DNA. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3372–3376. doi: 10.1073/pnas.75.7.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder P., Tilghman S. M., Tiemeier D. C., Polsky F. I., Seidman J. G., Edgell M. H., Enquist L. W., Leder A., Norman B. The cloning of mouse globin and surrounding gene sequences in bacteriophage lambda. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):915–920. doi: 10.1101/sqb.1978.042.01.093. [DOI] [PubMed] [Google Scholar]
- Michalides R., Schlom J. Relationship in nucleic acid sequences between mouse mammary tumor virus variants. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4635–4639. doi: 10.1073/pnas.72.11.4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalides R., van Deemter L., Nusse R., Röpcke G., Boot L. Involvement of mouse mammary tumor virus in spontaneous and hormone-induced mammary tumors in low-mammary-tumor mouse strains. J Virol. 1978 Sep;27(3):551–559. doi: 10.1128/jvi.27.3.551-559.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris V. L., Kozak C., Cohen J. C., Shank P. R., Jolicoeur P., Ruddle F., Varmus H. E. Endogenous mouse mammary tumor virus DNA is distributed among multiple mouse chromosomes. Virology. 1979 Jan 15;92(1):46–55. doi: 10.1016/0042-6822(79)90213-7. [DOI] [PubMed] [Google Scholar]
- Morris V. L., Medeiros E., Ringold G. M., Bishop J. M., Varmus H. E. Comparison of mouse mammary tumor virus-specific DNA in inbred, wild and Asian mice, and in tumors and normal organs from inbred mice. J Mol Biol. 1977 Jul;114(1):73–91. doi: 10.1016/0022-2836(77)90284-4. [DOI] [PubMed] [Google Scholar]
- Parks W. P., Scolnick E. M. Murine mammary tumor cell clones with varying degrees of virus expression. Virology. 1973 Sep;55(1):163–173. doi: 10.1016/s0042-6822(73)81018-9. [DOI] [PubMed] [Google Scholar]
- Ringold G. M., Blair P. B., Bishop J. M., Varmus H. E. Nucleotide sequence homologies among mouse mammary tumor viruses. Virology. 1976 Apr;70(2):550–553. doi: 10.1016/0042-6822(76)90297-x. [DOI] [PubMed] [Google Scholar]
- Ringold G. M., Shank P. R., Varmus H. E., Ring J., Yamamoto K. R. Integration and transcription of mouse mammary tumor virus DNA in rat hepatoma cells. Proc Natl Acad Sci U S A. 1979 Feb;76(2):665–669. doi: 10.1073/pnas.76.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D. L., Varmus H. E. Structural analysis of the intracellular RNAs of murine mammary tumor virus. J Virol. 1979 May;30(2):576–589. doi: 10.1128/jvi.30.2.576-589.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabran J. L., Hsu T. W., Yeater C., Kaji A., Mason W. S., Taylor J. M. Analysis of integrated avian RNA tumor virus DNA in transformed chicken, duck and quail fibroblasts. J Virol. 1979 Jan;29(1):170–178. doi: 10.1128/jvi.29.1.170-178.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank P. R., Cohen J. C., Varmus H. E., Yamamoto K. R., Ringold G. M. Mapping of linear and circular forms of mouse mammary tumor virus DNA with restriction endonucleases: evidence for a large specific deletion occurring at high frequency during circularization. Proc Natl Acad Sci U S A. 1978 May;75(5):2112–2116. doi: 10.1073/pnas.75.5.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steffen D., Weinberg R. A. The integrated genome of murine leukemia virus. Cell. 1978 Nov;15(3):1003–1010. doi: 10.1016/0092-8674(78)90284-2. [DOI] [PubMed] [Google Scholar]
- Swanstrom R., Shank P. R. X-Ray Intensifying Screens Greatly Enhance the Detection by Autoradiography of the Radioactive Isotopes 32P and 125I. Anal Biochem. 1978 May;86(1):184–192. doi: 10.1016/0003-2697(78)90333-0. [DOI] [PubMed] [Google Scholar]
- Tal J., Kung H. J., Varmus H. E., Bishop J. M. Characterization of DNA complementary to nucleotide sequences adjacent to poly(A) at the 3'-terminus of the avian sarcoma virus genome. Virology. 1977 Jun 1;79(1):183–197. doi: 10.1016/0042-6822(77)90344-0. [DOI] [PubMed] [Google Scholar]
- Todaro G. J., Benveniste R. E., Callahan R., Lieber M. M., Sherr C. J. Endogenous primate and feline type C viruses. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):1159–1168. doi: 10.1101/sqb.1974.039.01.133. [DOI] [PubMed] [Google Scholar]
- Todaro G. J., Huebner R. J. N.A.S. symposium: new evidence as the basis for increased efforts in cancer research. Proc Natl Acad Sci U S A. 1972 Apr;69(4):1009–1015. doi: 10.1073/pnas.69.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E., Bishop J. M., Nowinski R. C., Sarker N. H. Mammary tumour virus specific nucleotide sequences in mouse DNA. Nat New Biol. 1972 Aug 9;238(84):189–191. doi: 10.1038/newbio238189a0. [DOI] [PubMed] [Google Scholar]
- Varmus H. E., Quintrell N., Medeiros E., Bishop J. M., Nowinski R. C., Sarkar N. H. Transcription of mouse mammary tumor virus genes in tissues from high and low tumor incidence mouse strains. J Mol Biol. 1973 Oct 5;79(4):663–679. doi: 10.1016/0022-2836(73)90070-3. [DOI] [PubMed] [Google Scholar]