Abstract
Blastomyces dermatitidis is a dimorphic fungal pathogen that primarily causes blastomycosis in the midwestern and northern United States and Canada. While the genes controlling sexual development have been known for a long time, the genes controlling sexual reproduction of B. dermatitidis (teleomorph, Ajellomyces dermatitidis) are unknown. We identified the mating-type (MAT) locus in the B. dermatitidis genome by comparative genomic approaches. The B. dermatitidis MAT locus resembles those of other dimorphic fungi, containing either an alpha-box (MAT1-1) or an HMG domain (MAT1-2) gene linked to the APN2, SLA2, and COX13 genes. However, in some strains of B. dermatitidis, the MAT locus harbors transposable elements (TEs) that make it unusually large compared to the MAT locus of other dimorphic fungi. Based on the MAT locus sequences of B. dermatitidis, we designed specific primers for PCR determination of the mating type. Two B. dermatitidis isolates of opposite mating types were cocultured on mating medium. Immature sexual structures were observed starting at 3 weeks of coculture, with coiled-hyphae-containing cleistothecia developing over the next 3 to 6 weeks. Genetic recombination was detected in potential progeny by mating-type determination, PCR-restriction fragment length polymorphism (PCR-RFLP), and random amplification of polymorphic DNA (RAPD) analyses, suggesting that a meiotic sexual cycle might have been completed. The F1 progeny were sexually fertile when tested with strains of the opposite mating type. Our studies provide a model for the evolution of the MAT locus in the dimorphic and closely related fungi and open the door to classic genetic analysis and studies on the possible roles of mating and mating type in infection and virulence.
Introduction
Blastomyces dermatitidis is a thermally induced dimorphic fungus associated with severe respiratory and disseminated diseases in immunocompetent humans and other animals (1, 2). It can undergo a temperature-dependent morphological change from mycelium at environmental temperature (22°C) to pathogenic yeast at body temperature (37°C). In the lung, or in culture at 37°C, the conidia germinate into multinucleate, haploid, budding yeast cells, resulting in lung infections and, at times, systemic spread to other organs (1). B. dermatitidis is endemic to the Ohio and Mississippi River valleys, and in southeastern states, and causes pneumonia following the inhalation of conidia from the soil and subsequent conversion to budding yeast cells in animal hosts (3).
B. dermatitidis can reproduce sexually, and Ajellomyces dermatitidis is the teleomorphic form (4). While B. dermatitidis sexual structures have not been identified in nature, the teleomorph Ajellomyces dermatitidis, McDonough & Lewis, can be generated by pairing opposite mating-type cultures in vitro (4). Work by McDonough (4, 5) and by Kwon-Chung (6) in the late 1960s and early 1970s showed the formation of cleistothecia containing asci with eight ascospores each and characterized the mating system as heterothallic and bipolar. Sexual reproduction is pervasive in eukaryotes and is a major force driving the evolution of eukaryotic microbes by significantly increasing genetic diversity and, in some cases, may generate novel genotypes with altered virulence (7, 8). B. dermatitidis is generally regarded as a heterothallic species that requires isolates of both mating types to complete the sexual cycle. However, a B. dermatitidis isolate of both mating types has been reported from a patient with blastomycosis (9, 10). This suggests that patients might be infected either with two isolates of different mating types or with diploids heterozygous for mating type. Other possibilities are that B. dermatitidis also has a homothallic mating system and that some isolates are self-fertile.
Sexual reproduction may be associated with the virulence and outbreak of infectious diseases. For example, sexual reproduction of Cryptococcus gattii has been advanced as a potential contributing factor to the outbreak on Vancouver Island (11, 12). C. gattii is a sibling species of Cryptococcus neoformans and, while both species can infect both immunocompromised and immunocompetent hosts, C. neoformans causes most opportunistic infections in HIV/AIDS patients. The C. gattii VGII molecular type is causing an outbreak that began on Vancouver Island in 1999 and has since expanded to the Pacific Northwest region in both Canada and the United States (11, 13, 14). The major outbreak genotype VGIIa is highly clonal, but multilocus sequence typing (MLST) and the identification of an α/α diploid isolate of the same mating types provides evidence that the outbreak genotype may have been generated via same-sex mating to cause the outbreak and its expansion (12). Recently, studies revealed that the C. gattii VGIII molecular type is another common agent causing cryptococcosis in HIV/AIDS patients in southern California (15). The VGIII population includes two distinct sublineages, VGIIIa and VGIIIb, which differ in virulence in animal models, although both can cause infections in HIV/AIDS patients. Many VGIII isolates are fertile, and recombination has been detected in the VGIII global population (15). These findings suggest sexual reproduction may be an important force maintaining the genetic diversity and virulence of the C. gattii VGIII population that is causing disease in HIV/AIDS patients.
In this study, we used comparative genomic approaches to delineate the structure of the MAT locus of B. dermatitidis and conducted mating assays to demonstrate that the B. dermatitidis MAT locus controls sexual reproduction.
MATERIALS AND METHODS
Strains and cultivation.
B. dermatitidis strains ATCC 18187 and ATCC 18188 were obtained from the American Type Culture Collection. The yeast-phase cells were maintained at 37°C on slants of Middlebrook 7H10 agar with oleic acid-albumin-dextrose-catalase (OADC) supplement or on 3 M agar plates. Conversion to the mold form was accomplished on potato dextrose agar (PDA; 2% dry potato flakes, 1% glucose, and 1.5% agar) at 22°C, and mold-phase cells were maintained at 22°C on Sabouraud dextrose agar (SAB; BD Biosciences, Sparks, MD).
Identification of the B. dermatitidis MAT locus.
Mating in ascomycetous fungi is governed by a single MAT locus with two idiomorphs of highly divergent sequences (16). In the euascomycetes, each MAT idiomorph encodes an alpha domain or HMG domain transcription factor (16, 17). The alpha domain and HMG domain genes determine plus and minus strains, respectively, and the corresponding MAT locus is designated MAT1-1 and MAT1-2, respectively. The alpha domain and HMG domain genes that determine the mating types of dimorphic fungi are evolutionarily conserved and can be identified by homology searches using genes from the closely related species. To identify the MAT locus of B. dermatitidis, the alpha domain and HMG domain genes from the MAT locus of closely related species Histoplasma capsulatum (18, 19), Coccidioides immitis (19), and Paracoccidioides brasiliensis (20) were used as query sequences to do BLASTn searches of the B. dermatitidis genomic sequences using the Broad Institute database (http://www.broadinstitute.org/annotation/genome/blastomyces_dermatitidis/MultiHome.html). The available genome sequences are for isolates ATCC 26199, SLH14081, ER3 (ATCC MYA-2586), ATCC 18188, and ATCC 18187 (GenBank accession numbers AEII01000000, ACBU01000000, ACBT01000000, ADMK01000000, and AJJV01000000, respectively). The top hits and flanking regions were then extracted from the B. dermatitidis genomic sequences to identify the MAT locus. The alpha domain and HMG domain genes and flanking regions were aligned using the CLUSTALW program to determine the borders of the MAT locus.
A syntenic comparison of the MAT locus and flanking regions among the four B. dermatitidis isolates was performed by using the Artemis and ACT tools (21). Briefly, the MAT locus and flanking regions of the four isolates were compared using local BLASTn and the BLAST results were then entered into the ACT tool to visualize synteny.
Isolation of genomic DNA (gDNA) and mating-type determination for B. dermatitidis by PCR.
Liquid cultures of yeast cells in histoplasma macrophage medium (HMM) were grown to mid-log phase, and ∼5 × 107 to 1 × 108 cells were washed with water two times and resuspended in 4 ml of osmotic medium (0.3 g/ml MgSO4, 10 mM NaPO4 buffer [pH 5.8]) containing 4 mg/ml Yatalase (Fisher Scientific) and 2 mg/ml lysing enzymes (Sigma, St. Louis, MO). The suspensions were incubated at 37°C, with gentle mixing. After overnight incubation, the preparations were checked for protoplasts and washed by adding 2.5 volumes (10 ml) of trapping buffer (0.6 M sorbitol, 0.1 M Tris [pH 7.0]), centrifuged at 1,000 × g for 10 min, and washed two more times with 15 ml of trapping buffer. gDNA was isolated from the protoplasts using a Qiagen genomic DNA kit, with Qiagen 20/G tips and modifications used for yeast DNA.
The available MAT1-1 and MAT1-2 sequences of four B. dermatitidis isolates were aligned using CLUSTALW to choose primers for the mating-type determination for B. dermatitidis by PCR. In addition, the selected primers were further used as query sequences in BLAST searches of the B. dermatitidis genome to avoid potential nonspecific amplification. The primers used for the mating-type determination by PCR are listed in Table 1. PCR assays were conducted in a PTC-200 automated thermal cycler (MJ Research). A total of 300 ng of each DNA preparation was amplified in a 25-μl reaction mixture containing 10 pM each primer, 2 mM each nucleotide (dATP, dCTP, dGTP, and dTTP), 2.5 μl of 10× Ex Taq buffer, 0.125 μl of Ex Taq polymerase (TaKaRa), and an appropriate volume of distilled water. The following conditions were used for the amplification: an initial 3 min of denaturation at 95°C, followed by 35 cycles of denaturation for 30 s at 94°C, an annealing time of 20 s at 54°C, and an extension cycle for 1 min at 72°C. The amplification was completed by an extension period of 5 min at 72°C. Sterile water served as a negative control in each assay. PCR products were analyzed on 1.5% agarose gels.
Table 1.
Primers used in this study
| Primer | Sequence (5′→3′) | Aim |
|---|---|---|
| BD_alpha_box_F | TTCCAAATTTACTTGACCAGCAT | Mating-type determination (MAT1-1) |
| BD_alpha_box_R | CGTTGAATTCATTCATAGCCTTC | |
| BD_HMG_F | ATCGCCTGAGGTGTGTTTATCTA | Mating-type determination (MAT1-2) |
| BD_HMG_R | GGAGCGTATTGATAGTCAGGATG | |
| PI primer 10 | AATAGAACCCATCCC | RAPD |
| PI primer 24 | CGTGCAAGGGAGCACC | RAPD |
| RFLP-1-F | CAGGTTATCTCTTTCCGTGTCC | PCR-RFLP |
| RFLP-1-R | CCCGAGACAAACTCATAGTTCC | |
| RFLP-2-F | GGGGAAATGTGAAATTGTTGTT | PCR-RFLP |
| RFLP-2-R | ACAGCTTGGCTCTCTATCAACC |
Mating assays.
The medium used for the cocultivation of paired mating strains was soil extract agar (SEA). Soil extracts were produced from 350 g of Miracle-Gro African Violet Enriched Potting Soil (obtained from a local nursery), which was placed in 2.1 liters of tap water, autoclaved for 30 min at 15 lb/in2, allowed to cool, and filtered through two layers of Miracloth. To make SEA, 1 g of yeast extract and 1 g of d-glucose were added to 1 liter of soil extract and the pH was adjusted to 6.8, prior to adding 15 g of agar. The mix was autoclaved for 10 min at 20 lb/in2, with a 15-min exhaust, and allowed to cool, and penicillin and streptomycin were added (to 100 units/ml and 100 mg/ml, respectively) before pouring either 60- or 100-mm plates. Sections of agar (∼0.5 by 1 cm) containing mold, from the leading edge of growth on PDA or SAB plates, for pairs of strains of interest, were placed ∼2 cm apart on SEA plates and incubated at 22°C for 4 to 12 weeks. Plates were examined with a low-power stereomicroscope at 45× to visualize cleistothecium formation at the junction of the spreading inoculation. The fertility of cleistothecia was analyzed in one of the following two ways: (i) cleistothecia were scraped into 10 μl of lactophenol blue on a microscope slide, squashed under a coverslip, and viewed at 400× to visualize ascospores and (ii) cleistothecia were scraped into 100 μl of water in a Microfuge tube, disrupted by vortexing, and then viewed at 400× for ascospore release. In some cases, the disrupted cleistothecia were preserved in 2% formaldehyde before microscopy.
To validate the identified MAT1-1 and MAT1-2 loci as functional mating-type determinants for B. dermatitidis, we conducted mating assays using two isolates that were analyzed in the original work by McDonough to demonstrate fruitful mating in culture: isolates ATCC 18187 and ATCC 18188. In addition, F1 progeny of crosses between the parental strains were backcrossed to the parental strains, and siblings mated with other F1 strains (see below).
F1 progeny testing.
Two batches (M1 and M2) of F1 progeny from different sites on the mating plate were analyzed in our study. Individual F1 progeny were obtained by scraping several cleistothecia off the agar surface of SEA mating plates after 10 weeks or more and by placing in 100 μl of sterile water. After vortexing to release the ascospores, an aliquot was viewed with a hemocytometer at 400× to determine the presence of ascospores, their abundances, and the presence of nonascospore cells. Serial dilutions were plated on 3 M medium, and the plates were incubated at 37°C to allow the growth of independent yeast cell colonies. Individual colonies were patched onto 3 M agar and, when grown, were streaked for single colonies to ensure the clonality of individual F1 strains. An individual colony was then selected for each F1 strain and expanded on 3 M agar plates at 37°C. These recloned F1 strain cultures were used to isolate genomic DNA for molecular analyses and were converted back to mold for mating studies.
To test the fertility of F1 progeny, eight B. dermatitidis strains, including the two parental isolates ATCC 18187 (−) and ATCC 18188 (+) and six F1 progeny, M1-7 (+), M2-1 (+), M2-2 (+), M2-16 (+), M2-11 (−), and M2-18 (−), were used as tester strains for mating assays. The mating assays were conducted as described in the previous section. The production of cleistothecia with ascospores was defined as a successful mating.
Identification of SNPs between isolates ATCC 18187 and ATCC 18188.
A total of 824,355,256 reads, with a length of 101 bp, were generated for ATCC 18187 using Illumina HiSeq technology. To identify single nucleotide polymorphisms (SNPs), the ATCC 18187 reads were aligned to the ATCC 18188 assembly using BWA version 0.5.9 with default parameters. Alignments with mapping quality of at least 30 were selected using SAMtools view; these high-quality alignments have an average depth of 11 across all reference positions. Candidate variants were called from the alignments using GATK's Unified Genotyper (GenomeAnalysisTK version 1.0.3471) (22). The resulting SNPs were then filtered to require a depth of 4 or more, a maximum of 1 SNP per 10-bp window, a homopolymer run of the variant in either direction of ≤5, and strand bias values of ≤0.0 (high strand bias values denote more bias in the forward or reverse strand and are more likely to indicate false-positive calls).
The filtered set of 44,353 SNPs were then subjected to a variant recalibration process, using GATK's Variant Recalibrator. The purpose of the variant recalibration process is to assign a well-calibrated probability to each variant call in a call set.
The module evaluates variants in a two-step process. The variant recalibration procedure involves using the Variant Recalibrator walker to create a Gaussian mixture model (GMM) by looking at the different parameters for a high-quality subset of the call set and then evaluating all the variants. For the ATCC 18187 analysis, 29,727 concordant SNPs between the GATK and SAMtools pipelines (23) were used as the high-quality subset. A GMM using five statistics, QD (variant quality divided by depth), MQ (mapping quality), MQRankSum (mapping quality rank sum test), ReadPosRankSum (read position bias rank sum test), and SB (strand bias), was generated. Using the GMM tranche file generated by the previous step, the ApplyRecalibration walker examined each variant's VQS LOD value and decided which tranche it falls in (true positive or false positive). Variants in tranches that fall below the specified truth sensitivity filter level were filtered out. The variant calibration process resulted in a total of 41,892 high-quality recalibrated SNPs. These SNPs were submitted to dbSNP (handle, BROAD-GENOMEBIO; dbSNP release build identifier, B136).
Evaluation of genetic recombination in F1 progeny of B. dermatitidis.
The F1 progeny of the cross between isolates ATCC 18187 and ATCC 18188 are expected to demonstrate recombination between unlinked genetic markers. One known polymorphic locus between these lines is, of course, the MAT locus. Therefore, PCR primers were generated to yield distinct products representing one or the other mating type (see above). To find other genetic markers, random amplification of polymorphic DNA (RAPD) primers were tested and found to generate DNA products that are polymorphic between the parental strains.
We also used SNPs, as identified above, to design PCR-restriction fragment length polymorphism (PCR-RFLP) assays for genotyping strains. Genomic DNA was isolated from 22 of the F1 progeny (obtained from two different crosses of ATCC 18187 and ATCC 18188) and assayed for these PCR polymorphisms.
RESULTS
B. dermatitidis is heterothallic with two mating types, MAT1-1 and MAT1-2.
By performing BLASTn searches, an alpha domain gene that exhibits 73% DNA sequence similarity with the alpha domain gene of the MAT1-1 allele of H. capsulatum was identified in the B. dermatitidis ATCC 18188 genomic sequence. By a similar approach, an HMG domain gene that exhibits 68% DNA sequence similarity with the HMG domain gene of the MAT1-2 allele of H. capsulatum was identified in the genomic sequences of the B. dermatitidis isolates ATCC 26199, ER3, and SLH14081. The identified alpha domain and HMG domain genes were then used as query sequences to perform BLASTx searches in GenBank, and the top hits are the MAT locus genes of dimorphic fungi and the closely related dermatophyte fungi. This suggests that the alpha domain and HMG domain genes that were identified in the genomic sequences of four B. dermatitidis isolates are bona fide MAT locus genes. Analyses of the flanking regions further support the identification of the MAT locus idiomorphs of B. dermatitidis (see below). Only an alpha domain or HMG domain gene (and not both) was identified in the genomic sequence of any individual isolate, providing further support that B. dermatitidis is a heterothallic species.
B. dermatitidis has a similar MAT locus gene architecture as other dimorphic fungi and the dermatophytes.
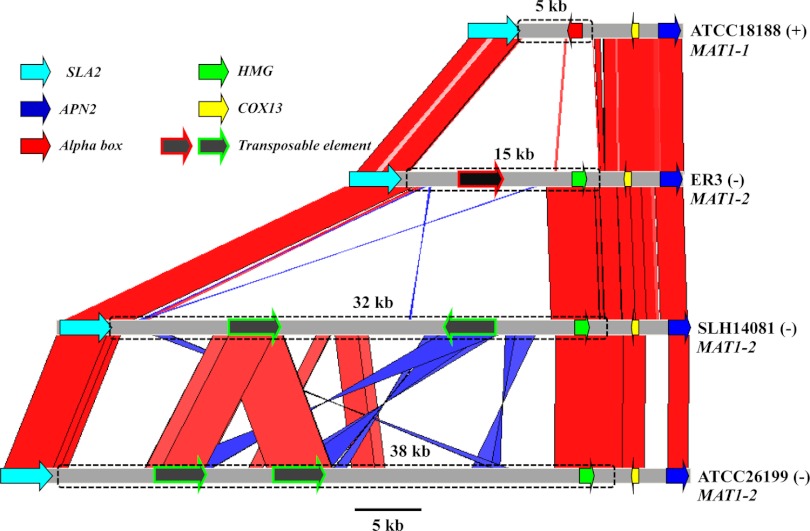
Like other dimorphic fungi, H. capsulatum, C. immitis, and P. brasiliensis (18, 19, 24), the B. dermatitidis MAT locus is tightly associated with the SLA2, APN2, and COX13 genes (Fig. 1). The SLA2 gene lies on one side, and the COX13 and APN2 genes flank the MAT locus on the other side (Fig. 1). A conserved gene order is a common feature of the MAT locus of ascomycetous fungi and is often used as a strategy to identify the MAT locus genes. However, in contrast to the dimorphic fungal pathogens, in the dermatophytes, the SLA2, COX13, and APN2 genes are instead linked on one side of the MAT locus (24).
Fig 1.
Syntenic comparison of the MAT locus of B. dermatitidis. Red bars/lines represent highly syntenic and similar DNA regions, and blue bars/lines represent highly syntenic and similar DNA regions with reverse orientation. The B. dermatitidis MAT locus is flanked by the SLA2, COX13, and APN2 genes, which share high sequence identity among these isolates. Isolate ATCC 18188 is designated plus (+) mating type due to the presence of the MAT1-1 locus, which is determined by an alpha-box gene. Isolates ER3, SLH-14081, and ATCC 26199 are designated minus (−) mating type, based on the identification of an HMG gene in the MAT1-2 locus. The genomic regions depicted with boxes in dashed outlines are the MAT locus. These TEs result in expansion of the MAT locus of B. dermatitidis. TEs are present in the MAT locus of three minus isolates ER3, SLH-14081, and ATCC 26199.
B. dermatitidis MAT locus size varies significantly due to the presence of transposable elements.
Typically, the fungal MAT locus is highly diverged and exhibits significant DNA sequence dissimilarity compared to other genomic regions. The MAT locus boundaries and size are determined by DNA sequence alignment and comparison between isolates of different mating types (24–26). The MAT locus sizes of B. dermatitidis determined by DNA sequence alignment between isolates ATCC 18188 (+) and ER3 (−) are 5 and 15 kb, respectively. However, the MAT locus (−) idiomorph sizes as determined by DNA sequence alignment between the plus isolate ATCC 18188 and the minus isolates SLH14081 and ATCC 26199 are 32 and 38 kb, respectively. The size variation of the B. dermatitidis MAT locus is mainly the consequence of the presence of transposable elements within the MAT locus. One retrotransposon is identified in the minus isolate ER3, and two different retrotransposons are found in the minus isolates SLH14081 and ATCC 26199 (Fig. 1). These retrotransposons are flanked by terminal repeats and encode a DEE superfamily endonuclease domain and a PRK00247 60-kDa IMP superfamily domain and are also found in other genomic regions. However, no transposable element was identified in the plus isolate ATCC 18188. It is not known if other minus strains lack transposable elements or if other plus strains harbor them.
Sexual reproduction of B. dermatitidis isolates of opposite mating types.
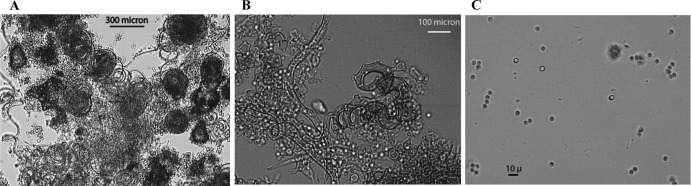
The sexual development of B. dermatitidis has been reported previously (4, 5). However, the biochemical and molecular basis of the control of B. dermatitidis sexual reproduction by the MAT locus was not clear at that time. In our study, two heterothallic B. dermatitidis isolates of the opposite mating type, ATCC 18188 and ATCC 18187, were cocultured on appropriate mating medium to study the sexual development of B. dermatitidis and to confirm that the MAT locus controls the sexual cycle. Several types of agar medium were tested to find a medium that would support mating, and SEA (see Materials and Methods section) was found to reproducibly support mating in fertile strains of the opposite mating type. After 3 or 4 weeks at room temperature (22°C), spherical structures representing immature cleistothecia could be seen by low-power light microscopy. With longer incubations, the cleistothecia grew larger and acquired a tan pigmentation. Putative cleistothecia were scraped from the agar surface, squashed on a microscope slide, and viewed under high power, where we observed sexual structures, including coiled hyphae with greatly thickened hyphal structures, asci, and spores, as reported in previous studies (Fig. 2) (4, 5). Multiple attempts to find cleistothecial structures from self-pairing were unsuccessful.
Fig 2.
Sexual development of B. dermatitidis. After coculture of B. dermatitidis isolates ATCC 18188 (+) and ATCC 18187 (−) on SEA medium for 10 weeks, cleistothecia were scraped off the agar, disrupted in water, and fixed in formaldehyde prior to microscopic examination. (A) 20× light microscopy shows multiple disrupted cleistothecia, with highly spiraled hyphae. (B) 40× image showing thickened, coiled hyphae and specialized vegetative cells attached to the spiral hyphae. (C) 60× image shows multiple ascospores released from asci. The ascospores are characterized by a light halo around the cell.
F1 progeny are fertile.
The F1 progeny of this mating were isolated and analyzed for two reasons: (i) to confirm that a sexual cycle had been completed and (ii) to demonstrate that the sequence structures, identified as MAT1-1 and MAT1-2, correspond to the functional mating-type determinants. An F1 progeny population allows us to analyze genetic marker recombination to confirm meiosis, and it allows analysis of the linkage between the functional MAT1 mating phenotype and the PCR-based identity of the MAT1 idiomorph for each strain.
The fertility of F1 progeny was tested by crossing each back to the two parental isolates (ATCC 18187 and ATCC 18188) and to six sibling F1 progeny. Fertility was monitored microscopically by the production of cleistothecia that release ascospores (Fig. 2C). All of the F1 progeny are fertile (Table 2). They can mate with one or more strains of opposite mating type, but all fail to mate with any strain of the same mating type (Table 2). However, F1 progeny were not sexually fertile in combination with all of the potentially compatible tester strains. We found ∼56% (111/198) successful matings of the F1 progeny with strains of the opposite mating type (Table 2). This finding suggests that other genetic modifiers of fertility unlinked to the MAT locus remain to be identified.
Table 2.
Summary results for mating F1 strains with testers of the same or opposite mating typea
| Strain | Crossed with tester strains of: |
|
|---|---|---|
| Same MAT1 | Opposite MAT1 | |
| MAT1-2 strains | ||
| 18187 | 0/13 | 23/34 |
| M2-11 | 0/8 | 6/17 |
| M2-18 | 0/13 | 16/30 |
| M1-4 | 0/5 | 1/7 |
| M1-5 | 0/5 | 6/7 |
| M2-3 | 0/5 | 1/7 |
| M2-17 | 0/5 | 2/7 |
| MAT1-1 strains | ||
| 18188 | 0/29 | 6/13 |
| M1-7 | 0/16 | 5/9 |
| M2-1 | 0/30 | 10/16 |
| M2-2 | 0/16 | 7/9 |
| M2-16 | 0/16 | 7/9 |
| M1-2 | 0/7 | 3/5 |
| M2-4 | 0/7 | 1/5 |
| M2-6 | 0/7 | 1/5 |
| M2-20 | 0/7 | 4/5 |
| Nonrecombinant MAT1-1 strains | ||
| M1-1 | 0/7 | 3/5 |
| M1-3 | 0/7 | 2/5 |
| M1-6 | 0/7 | 1/5 |
| M1-8 | 0/7 | 2/5 |
| M1-9 | 0/7 | 3/5 |
| M1-10 | 0/7 | 1/5 |
| Total for both MAT1 mating types | 0/184 | 111/198 |
Results are presented as the number of ascospore-producing crosses/total number of crosses attempted with 8 mating tester strains.
Evidence of genetic recombination of the F1 progeny of B. dermatitidis.
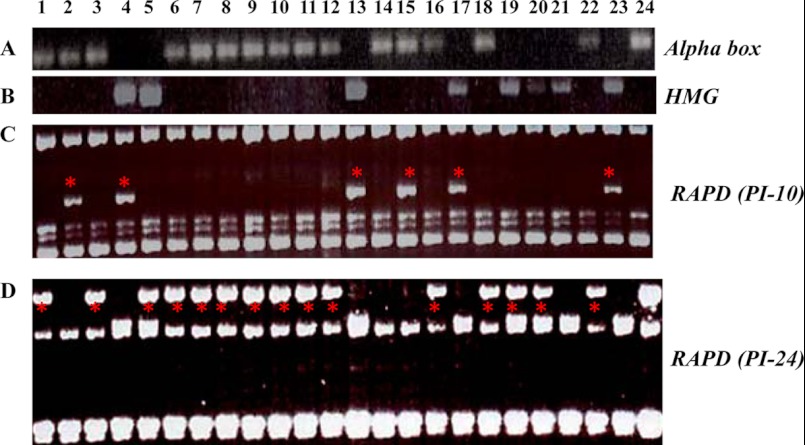
Meiosis, the hallmark of sexual reproduction, is characterized by chromosome segregation and genetic recombination. To confirm that meiosis had indeed occurred, progeny from two mating experiments (M1 and M2) of ATCC 18188 (+) and ATCC 18187 (−) were isolated and their DNA was tested for recombination using five genetic markers: two RAPD markers, two PCR-RFLP markers, and also the mating-type locus itself (as determined by PCR with primers specific for MAT1-1 and MAT1-2). None of the F1 progeny was positive for both of the MAT1 markers, suggesting that each strain does represent a single-cell clone (Fig. 3A and B). Based on a combination of the five genetic markers, we detected genetic recombination in 16 progeny out of 22 tested (Fig. 3 and 4; Table 3). All of the progeny in the M2 mating experiment show recombinant genotypes, indicating that they are the products of successful sexual reproduction. In contrast, genetic recombination was only detected in three of the M1 mating assay progeny. The lack of genetic recombination in some M1 progeny may be explained by the limited genetic markers used or by some of the progeny strains representing mitotic cells of the parental culture, because these progeny were not microdissected from asci to ensure that only ascospores were cultured.
Fig 3.
(A and B) Mating-type determination of B. dermatitidis parental isolates (ATCC 18187 and ATCC 18188) and 22 potential progeny by PCR. Primers were designed to specifically amplify the alpha-box (plus mating type) and HMG (minus mating type) genes, which are characteristic of the B. dermatitidis MAT locus (see Table 1). (C and D) RAPD analysis of the progeny of isolates ATCC 18188 and ATCC 18187. Two RAPD primers, PI-10 (C) and PI-24 (D), reveal genetic variations (indicated by an asterisk) between these two parental isolates and the potential progeny. Lane 1, M1-1; lane 2, M1-2; lane 3, M1-3; lane 4, M1-4; lane 5, M1-5, lane 6, M1-6; lane 7, M1-7; lane 8, M1-8; lane 9, M1-9; lane 10, M1-10; lane 11, M2-1; lane 12, M2-2; lane 13, M2-3; lane 14, M2-4; lane 15, M2-6; lane 16, M2-7; lane 17, M2-11; lane 18, M2-16; lane 19, M2-17; lane 20, M2-18; lane 21, M2-19; lane 22, M2-20; lane 23, ATCC 18187; and lane 24, ATCC 18188.
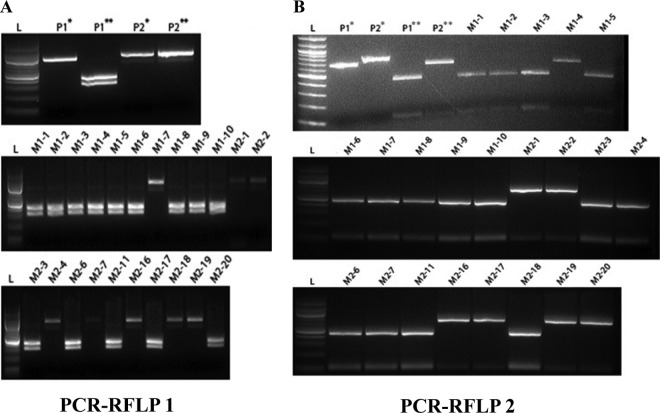
Fig 4.
PCR-RFLP analysis of the progeny of isolates ATCC 18188 (+) and ATCC 18187 (−). SNP and insertion/deletion between isolates ATCC 18188 and ATCC 18187 were used to design a PCR-RFLP analysis to detect genetic variation between these two isolates and the potential progeny. An SNP was identified between isolates ATCC 18188 and ATCC 18187 that abolished a single BamHI restriction site in ATCC 18188 (A). Note that for ATCC 18188, digestion results in two distinct bands, while digestion of ATCC 18187 fails to produce any novel fragments. Progeny were scored as either genotype 1 or 2, denoting their relationship to ATCC 18188 or ATCC 18187, respectively (see Table 3). A second SNP identified within a different region abolished an NdeI restriction site in ATCC 18187. However, sequences of the amplified region predicted an additional NdeI restriction site shared by both parents, which accounts for the faint low-molecular-weight band appearing after digestion of ATCC 18187 DNA (B). For panels A and B, parental DNAs representing the amplified region around the SNP prior to and after digestion are indicated by one asterisk and two asterisks, respectively. L, DNA ladder.
Table 3.
Evidence of genetic recombination of the progeny of B. dermatitidis ATCC18187 and ATCC18188a
| Isolate | PCR-RFLP 1 | PCR-RFLP 2 | Mating type | RAPD 1 | RAPD 2 | Recombination |
|---|---|---|---|---|---|---|
| 18188 (P1) | 1 | 1 | 1 | 1 | 1 | Not applicable |
| 18187 (P2) | 2 | 2 | 2 | 2 | 2 | Not applicable |
| M1-1 | 1 | 1 | 1 | 1 | 1 | Not detected |
| M1-2 | 1 | 1 | 1 | 2 | 2 | Yes |
| M1-3 | 1 | 1 | 1 | 1 | 1 | Not detected |
| M1-4 | 1 | 2 | 2 | 2 | 2 | Yes |
| M1-5 | 1 | 1 | 2 | 1 | 1 | Yes |
| M1-6 | 1 | 1 | 1 | 1 | 1 | Not detected |
| M1-7 | 2 | 1 | 1 | 1 | 1 | Yes |
| M1-8 | 1 | 1 | 1 | 1 | 1 | Not detected |
| M1-9 | 1 | 1 | 1 | 1 | 1 | Not detected |
| M1-10 | 1 | 1 | 1 | 1 | 1 | Not detected |
| M2-1 | 2 | 2 | 1 | 1 | 1 | Yes |
| M2-2 | 2 | 2 | 1 | 1 | 1 | Yes |
| M2-3 | 1 | 1 | 2 | 2 | 2 | Yes |
| M2-4 | 2 | 1 | 1 | 1 | 2 | Yes |
| M2-6 | 1 | 1 | 1 | 2 | 2 | Yes |
| M2-7 | 2 | 1 | 1 | 1 | 1 | Yes |
| M2-11 | 1 | 1 | 2 | 2 | 2 | Yes |
| M2-16 | 2 | 2 | 1 | 1 | 1 | Yes |
| M2-17 | 1 | 2 | 2 | 1 | 1 | Yes |
| M2-18 | 2 | 1 | 2 | 1 | 1 | Yes |
| M2-19 | 2 | 2 | 2 | 1 | 2 | Yes |
| M2-20 | 1 | 2 | 1 | 1 | 1 | Yes |
The mating types were determined by PCR assay (see Fig. 3). Amplification of an alpha-box gene is determined as mating type 1 (MAT1-1), and amplification of an HMG gene is designated as mating type 2 (MAT1-2).
Based on the shared conservation of the alpha domain and HMG domain genes of the MAT1 idiomorphs in related ascomycetous fungi, it is expected that the control of mating in B. dermatitidis will be governed by the MAT1 locus. The results for 16 different F1 strains, for which both mating tests and marker analysis were preformed, indicate complete congruence between the functional mating test phenotype and molecular identification of the MAT1 idiomorph (Table 3).
DISCUSSION
The teleomorphic form of B. dermatitidis is Ajellomyces dermatitidis. The sexual structures—such as thick-walled structures (cleistothecia) and spiraling hyphae surrounding numerous asci, each containing eight ascospores—were previously observed in B. dermatitidis (4, 5, 27); however the molecular characteristics of the MAT locus and its control of sexual reproduction were unknown. In this study, we defined the DNA-sequence structure of the MAT locus of B. dermatitidis and conducted mating assays by coculturing two B. dermatitidis isolates of opposite mating types on appropriate mating medium. Our genetic studies reveal a complete sexual cycle of B. dermatitidis based on the detection of genetic recombination.
Both homothallic and heterothallic mating systems have been identified throughout the Ascomycota. Some studies have hypothesized that the ancestral mating system in the Ascomycota was heterothallic (19, 28, 29), but other studies have derived the opposite conclusion, namely, that the ancestral mating system was homothallic (30, 31). This controversy may in part result from a limited number of fungal species with identified MAT loci and defined sexual cycles, as well as the challenge in assigning ancestral states to characters subject to recent and ongoing transitions. Our mating assays suggest that B. dermatitidis is a heterothallic species on the basis of the observation of successful mating of B. dermatitidis strains of opposite mating types rather than the same mating types or during solo culture (Tables 2 and 3). In addition, either a single MAT1-1 or a single MAT1-2 idiomorph, and not both, was identified for each of the four sequenced B. dermatitidis genomes (Fig. 1). Only a heterothallic mating system has been identified in dimorphic fungi and the dermatophytes (18–20, 24). It is therefore likely that the ancestral mating system of the dimorphic fungi and dermatophytes was heterothallic.
In three minus strains of B. dermatitidis, we identified two different retrotransposons that are not found in the MAT locus of the one plus isolate analyzed in our study (Fig. 1). Further genomic analyses will need to be done to determine the frequency of retrotransposons among B. dermatitidis strains. The transposable elements and additional nonsyntenic DNA in the MAT locus of some strains significantly and variably increase the MAT locus size in the four B. dermatitidis strains studied, 5 kb, 15 kb, 32 kb, and 38 kb. In other fungi, transposons have been found within or flanking MAT loci, such as in the homothallic fungus Neosartorya fischeri (29) and in the MAT locus of C. neoformans (26) and P. brasiliensis (24), but the influence of transposon transposition on MAT locus genes or mating ability is unknown. There is evidence that insertion of a transposon in an avirulence gene caused a gain of virulence in the rice blast fungus Magnaporthe grisea (32). Transposition of transposons into or around the MAT locus may significantly increase the sequence diversity between the two idiomorphs and decrease recombination of this genomic region if sexual reproduction is still possible. Insertion and local transposition likely also provide homologous sequences driving recombination events at the MAT locus.
Compared to closely related fungal species (including the other dimorphic fungi H. capsulatum, C. immitis, Coccidioides posadasii, and P. brasiliensis and the dermatophytes Microsporum gypseum, M. canis, Trichophyton equinum, T. rubrum, and T. tonsurans), B. dermatitidis has a similar structure of the MAT locus that harbors either an alpha-box gene or an HMG domain gene and is flanked by the SLA2, COX13, and APN2 genes. Previous studies suggested that the MAT locus of dimorphic fungi is expanding based on the comparison of the MAT locus between dimorphic fungi and the dermatophytes (24). In the Cryptococcus species complex, the MAT locus has also expanded by capturing adjacent genes into the MAT locus (25, 26, 33). This fungal MAT locus expansion is mainly caused by the capture of adjacent genes into the MAT locus and, as a result, increases the DNA sequence similarity between isolates of the opposite mating type (7). Herein, we present another mechanism of fungal MAT locus expansion involving the incorporation of transposable elements.
Both mating types were found in the natural population of B. dermatitidis, and they have a similar frequency in nature (10, 34). In two other dimorphic fungi, H. capsulatum and Coccidioides, both have a similar ratio of the two mating types in nature (35, 36), and a recent study performed by Torres et al. reveals an ∼1:1 distribution of the MAT1-1 and MAT1-2 strains of P. brasiliensis in South America (20). The approximately equal proportion of the two mating types in nature argues that dimorphic fungi may have a sexual cycle in nature. Studies of population genetics further support that these dimorphic fungi reproduce sexually in nature (21, 37–40). Strain typing results based on 27 microsatellite markers suggest genetic recombination and sexual reproduction of B. dermatitidis in nature (10). The identification of the B. dermatitidis MAT locus that exhibits a highly similar structure with other fungi and the population genetic studies indicate that B. dermatitidis may reproduce sexually in the environment. Sexual reproduction is linked with virulence in some pathogenic fungi. For example, the α mating type dominates the Cryptococcus neoformans population and is more virulent than a mating-type isolates in some genetic backgrounds (41, 42). Moreover, spores produced by sexual reproduction of C. neoformans are infectious propagules providing another link between sex and virulence in this species (43–45). Many pathogenic fungi have the genes encoding sexual machinery in their genomes, but no known sexual cycle has been observed in nature or the laboratory (7). It is not clear whether B. dermatitidis reproduces sexually in nature and, if so, whether the sexual spores are infectious propagules. Animal experiments using sexual spores dissected from the sexual reproduction of B. dermatitidis in the laboratory and virulence comparisons of strains of the opposite mating type could be useful to understand the possible associations of sex and virulence. In addition, comparison of virulence and mating ability between strains with and without transposon insertions in the MAT locus can help to understand the function or consequences of transposons in sexual reproduction and pathogenesis.
ACKNOWLEDGMENTS
We thank Sheng Sun and Sujal Phadke for valuable discussions and for reading the manuscript.
This study was supported by NIH/NIAID R37 grant AI39115 to J.H. and NIH grant AI35681 to B.S.K. and has been funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract number HHSN272200900018C to the Broad Institute.
Footnotes
Published ahead of print 9 November 2012
REFERENCES
- 1. Klein BS, Tebbets B. 2007. Dimorphism and virulence in fungi. Curr. Opin. Microbiol. 10:314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tenenbaum MJ, Greenspan J, Kerkering TM. 1982. Blastomycosis. Crit. Rev. Microbiol. 9:139–163 [DOI] [PubMed] [Google Scholar]
- 3. Klein BS, Vergeront JM, Davis JP. 1986. Epidemiologic aspects of blastomycosis, the enigmatic systemic mycosis. Semin. Respir. Infect. 1:29–39 [PubMed] [Google Scholar]
- 4. McDonough ES, Lewis AL. 1967. Blastomyces dermatitidis: production of the sexual stage. Science 156:528–529 [DOI] [PubMed] [Google Scholar]
- 5. McDonough ES, Lewis AL. 1968. The ascigerous stage of Blastomyces dermatitidis. Mycologia 60:76–83 [PubMed] [Google Scholar]
- 6. Kwon-Chung KJ. 1971. Genetic analysis on the incompatibility system of Ajellomyces dermatitidis. Sabouraudia 9:231–238 [PubMed] [Google Scholar]
- 7. Lee SC, Ni M, Li W, Shertz C, Heitman J. 2010. The evolution of sex: a perspective from the fungal kingdom. Microbiol. Mol. Biol. Rev. 74:298–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li W, Savelkoul E, Heitman J, Logsdon JM. 2012. Evolution of meiosis, recombination, and sexual reproduction in eukaryotic microbes, p 17–43 In Sibley LD, Howlett BJ, Heitman J. (ed), Evolution of virulence of eukaryotic microbes, 1st ed. Wiley-Blackwell, Hoboken, NJ [Google Scholar]
- 9. McDonough ES, Chan DM, McNamara WJ. 1977. Dual infection by “+” and “−” mating types of Ajellomyces (Blastomyces) dermatitidis. Am. J. Epidemiol. 106:67–71 [DOI] [PubMed] [Google Scholar]
- 10. Meece JK, Anderson JL, Fisher MC, Henk DA, Sloss BL, Reed KD. 2011. Population genetic structure of clinical and environmental isolates of Blastomyces dermatitidis, based on 27 polymorphic microsatellite markers. Appl. Environ. Microbiol. 77:5123–5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Byrnes EJ, III, Li W, Lewit Y, Ma H, Voelz K, Ren P, Carter DA, Chaturvedi V, Bildfell RJ, May RC, Heitman J. 2010. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 6:e1000850 doi:10.1371/journal.ppat.1000850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, Allen A, Stajich JE, Dietrich FS, Perfect JR, Heitman J. 2005. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437:1360–1364 [DOI] [PubMed] [Google Scholar]
- 13. Byrnes EJ, III, Bildfell RJ, Frank SA, Mitchell TG, Marr KA, Heitman J. 2009. Molecular evidence that the range of the Vancouver Island outbreak of Cryptococcus gattii infection has expanded into the Pacific Northwest in the United States. J. Infect. Dis. 199:1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, Macdougall L, Boekhout T, Kwon-Chung KJ, Meyer W. 2004. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc. Natl. Acad. Sci. U. S. A. 101:17258–17263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Byrnes EJ, III, Li W, Ren P, Lewit Y, Voelz K, Fraser JA, Dietrich FS, May RC, Chaturvedi S, Chaturvedi V, Heitman J. 2011. A diverse population of Cryptococcus gattii molecular type VGIII in southern Californian HIV/AIDS patients. PLoS Pathog. 7:e1002205 doi:10.1371/journal.ppat.1002205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coppin E, Debuchy R, Arnaise S, Picard M. 1997. Mating types and sexual development in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 61:411–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Turgeon BG, Yoder OC. 2000. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet. Biol. 31:1–5 [DOI] [PubMed] [Google Scholar]
- 18. Bubnick M, Smulian AG. 2007. The MAT1 locus of Histoplasma capsulatum is responsive in a mating type-specific manner. Eukaryot. Cell 6:616–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fraser JA, Stajich JE, Tarcha EJ, Cole GT, Inglis DO, Sil A, Heitman J. 2007. Evolution of the mating type locus: insights gained from the dimorphic primary fungal pathogens Histoplasma capsulatum, Coccidioides immitis, and Coccidioides posadasii. Eukaryot. Cell 6:622–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Torres I, Garcia AM, Hernandez O, Gonzalez A, McEwen JG, Restrepo A, Arango M. 2010. Presence and expression of the mating type locus in Paracoccidioides brasiliensis isolates. Fungal Genet. Biol. 47:373–380 [DOI] [PubMed] [Google Scholar]
- 21. Carter DA, Taylor JW, Dechairo B, Burt A, Koenig GL, White TJ. 2001. Amplified single-nucleotide polymorphisms and a (GA)(n) microsatellite marker reveal genetic differentiation between populations of Histoplasma capsulatum from the Americas. Fungal Genet. Biol. 34:37–48 [DOI] [PubMed] [Google Scholar]
- 22. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li W, Metin B, White TC, Heitman J. 2010. Organization and evolutionary trajectory of the mating type (MAT) locus in dermatophyte and dimorphic fungal pathogens. Eukaryot. Cell 9:46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fraser JA, Diezmann S, Subaran RL, Allen A, Lengeler KB, Dietrich FS, Heitman J. 2004. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2:e384 doi:10.1371/journal.pbio.0020384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lengeler KB, Fox DS, Fraser JA, Allen A, Forrester K, Dietrich FS, Heitman J. 2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1:704–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Glick AD, Kwon-Chung KJ. 1973. Ultrastructural comparison of coils and ascospores of Emmonsiella capsulata and Ajellomyces dermatitidis. Mycologia 65:216–220 [PubMed] [Google Scholar]
- 28. Inderbitzin P, Harkness J, Turgeon BG, Berbee ML. 2005. Lateral transfer of mating system in Stemphylium. Proc. Natl. Acad. Sci. U. S. A. 102:11390–11395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rydholm C, Dyer PS, Lutzoni F. 2007. DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri. Eukaryot. Cell 6:868–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D'Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115 [DOI] [PubMed] [Google Scholar]
- 31. Woo PC, Chong KT, Tse H, Cai JJ, Lau CC, Zhou AC, Lau SK, Yuen KY. 2006. Genomic and experimental evidence for a potential sexual cycle in the pathogenic thermal dimorphic fungus Penicillium marneffei. FEBS Lett. 580:3409–3416 [DOI] [PubMed] [Google Scholar]
- 32. Kang S, Lebrun MH, Farrall L, Valent B. 2001. Gain of virulence caused by insertion of a Pot3 transposon in a Magnaporthe grisea avirulence gene. Mol. Plant Microbe Interact. 14:671–674 [DOI] [PubMed] [Google Scholar]
- 33. Findley K, Sun S, Fraser JA, Hsueh YP, Averette AF, Li W, Dietrich FS, Heitman J. 2012. Discovery of a modified tetrapolar sexual cycle in Cryptococcus amylolentus and the evolution of MAT in the Cryptococcus species complex. PLoS Genet. 8:e1002528 doi:10.1371/journal.pgen.1002528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McDonough ES, McNamara WJ, Chan DM, Wallenfang MA. 1973. Geographic distribution of “plus” and “minus” isolates of Blastomyces (Ajellomyces) dermatitidis in North America. Am. J. Epidemiol. 98:63–67 [DOI] [PubMed] [Google Scholar]
- 35. Kwon-Chung KJ, Bartlett MS, Wheat LJ. 1984. Distribution of the two mating types among Histoplasma capsulatum isolates obtained from an urban histoplasmosis outbreak. Sabouraudia 22:155–157 [PubMed] [Google Scholar]
- 36. Mandel MA, Barker BM, Kroken S, Rounsley SD, Orbach MJ. 2007. Genomic and population analyses of the mating type loci in Coccidioides species reveal evidence for sexual reproduction and gene acquisition. Eukaryot. Cell 6:1189–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burt A, Carter DA, Koenig GL, White TJ, Taylor JW. 1996. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc. Natl. Acad. Sci. U. S. A. 93:770–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carter DA, Burt A, Taylor JW, Koenig GL, White TJ. 1996. Clinical isolates of Histoplasma capsulatum from Indianapolis, Indiana, have a recombining population structure. J. Clin. Microbiol. 34:2577–2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koufopanou V, Burt A, Szaro T, Taylor JW. 2001. Gene genealogies, cryptic species, and molecular evolution in the human pathogen Coccidioides immitis and relatives (Ascomycota, Onygenales). Mol. Biol. Evol. 18:1246–1258 [DOI] [PubMed] [Google Scholar]
- 40. Koufopanou V, Burt A, Taylor JW. 1997. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc. Natl. Acad. Sci. U. S. A. 94:5478–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kwon-Chung KJ, Edman JC, Wickes BL. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:602–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Litvintseva AP, Marra RE, Nielsen K, Heitman J, Vilgalys R, Mitchell TG. 2003. Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in sub-Saharan Africa. Eukaryot. Cell 2:1162–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Botts MR, Giles SS, Gates MA, Kozel TR, Hull CM. 2009. Isolation and characterization of Cryptococcus neoformans spores reveal a critical role for capsule biosynthesis genes in spore biogenesis. Eukaryot. Cell 8:595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Giles SS, Dagenais TR, Botts MR, Keller NP, Hull CM. 2009. Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infect. Immun. 77:3491–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Velagapudi R, Hsueh YP, Geunes-Boyer S, Wright JR, Heitman J. 2009. Spores as infectious propagules of Cryptococcus neoformans. Infect. Immun. 77:4345–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]