Abstract
Candida albicans strains that are homozygous at the mating type locus can spontaneously and reversibly switch from the normal yeast morphology (white) to an elongated cell type (opaque), which is the mating-competent form of the fungus. White-opaque switching also influences the ability of C. albicans to colonize and proliferate in specific host niches and its susceptibility to host defense mechanisms. We used live imaging to observe the interaction of white and opaque cells with host phagocytic cells. For this purpose, we generated derivatives of the switching-competent strain WO-1 that express green fluorescent protein from a white-specific promoter and red fluorescent protein from an opaque-specific promoter or vice versa. When mixed populations of these differentially labeled white and opaque cells were incubated with human polymorphonuclear neutrophils (PMNs) on a glass slide, the neutrophils selectively phagocytosed and killed white cells, despite frequent physical interaction with opaque cells. White cells were attacked only after they started to form a germ tube, indicating that the suppression of filamentation in opaque cells saved them from recognition by the PMNs. In contrast to neutrophils, dendritic cells internalized white as well as opaque cells. However, when embedded in a collagen matrix, the PMNs also phagocytosed both white and opaque cells with similar efficiency. These results suggest that, depending on the environment, white-opaque switching enables C. albicans to escape from specific host defense mechanisms.
Introduction
The yeast Candida albicans is a commensal microorganism in the orogastrointestinal and urogenital tracts of most healthy people, but it can also cause superficial infections of the skin and mucosae as well as life-threatening disseminated infections, especially in immunocompromised patients. Morphological transitions play an important role in the biology of C. albicans and in the interactions of the fungus with its host. In response to various environmental signals, C. albicans changes its growth mode from the budding yeast form to filamentous growth, which facilitates tissue invasion (1). Strains that have become homozygous at the mating type locus (MTLa/a or MTLα/α), either by mitotic recombination or by loss of one copy of chromosome 5 and duplication of the homologous chromosome, can also switch from the normal white yeast form to an elongated cell form termed “opaque” (2, 3). Opaque cells are the mating-competent form of C. albicans; they can mate with other opaque cells to produce recombinant progeny (4). Switching between the white and opaque phases occurs spontaneously at a relatively low frequency, but it can also be induced by certain environmental signals (5–10).
White and opaque cells differ not only in their mating competence but also in the expression of many genes that are unrelated to mating, including adhesins and metabolic genes, suggesting that the two cell types are adapted to different environments within a mammalian host (11–13). Indeed, opaque cells are better colonizers of the skin, but they are much less virulent than white cells in a mouse model of disseminated candidiasis (14, 15). Opaque cells do not readily undergo filamentation under most conditions that promote hyphal formation in white cells, which may result in a reduced ability to invade tissues (16). In addition, opaque cells have been reported to be more susceptible than white cells to killing by polymorphonuclear neutrophils (PMNs) or cell-free oxidants, and they also stimulated more superoxide generation by PMNs (17). As neutrophils are important first-line host defense cells in the prevention of disseminated candidiasis (18), an increased susceptibility to killing by these phagocytes may contribute to the reduced virulence of opaque cells during systemic infections. On the other hand, white but not opaque cells release a chemoattractant for PMNs, indicating that opaque cells may in fact avoid being assaulted by neutrophils (19). Interestingly, white cells have also been found to be more efficiently phagocytosed than opaque cells by other types of innate immune cells, Drosophila hemocyte-derived S2 cells and mouse macrophage-derived RAW264.7 cells (20). Therefore, it remains unclear if and when white-opaque switching may enable C. albicans to escape from specific components of the host immune system.
In this work, we used live imaging by video microscopy to study the interaction of mixed populations of white and opaque cells of C. albicans with two types of phagocytic cells that play important roles in the primary host defense against microbial infections, neutrophils and dendritic cells (DCs). Our results demonstrate that neutrophils and dendritic cells differ in their ability to recognize the two types of cells and that, depending on the environment, white-opaque switching may enable C. albicans to evade attack by the host's innate immune system.
MATERIALS AND METHODS
Strains and growth conditions.
The C. albicans strains used in this study are listed in Table 1. All strains were stored as frozen stocks with 15% glycerol at −80°C. The strains were subcultured separately in the white and opaque phases at room temperature on agar plates containing Lee's medium, pH 6.8 (22), and 5 μg/ml phloxine B, which selectively stains opaque colonies pink (23). Strains were routinely grown in YPD liquid medium (10 g yeast extract, 20 g peptone, 20 g glucose per liter) at 30°C in a shaking incubator. For selection of nourseothricin-resistant transformants, 200 μg/ml nourseothricin (Werner Bioagents, Jena, Germany) was added to YPD agar plates. To obtain nourseothricin-sensitive derivatives in which the SAT1 flipper cassette was excised by FLP-mediated recombination, transformants were grown overnight in YPM medium (10 g yeast extract, 20 g peptone, 20 g maltose per liter) without selective pressure to induce the MAL2 promoter controlling caFLP (Candida-adapted FLP) expression. One hundred to 200 cells were then spread on YPD plates containing 10 μg/ml nourseothricin and grown for 2 days at 30°C. Nourseothricin-sensitive clones were identified by their small colony size and confirmed by restreaking on YPD plates containing 200 μg/ml nourseothricin as described previously (24).
Table 1.
C. albicans strains used in this study
| Strain | Parent | Relevant genotypea | Reference or source |
|---|---|---|---|
| WO-1 | 21 | ||
| WOP4G41A and -B | WO-1 | OP4/OP4/op4::POP4-GFP-SAT1-FLIP | This work |
| WOP4G42A | WOP4G41A | OP4/OP4/op4::POP4-GFP-FRT | This work |
| WOP4G42B | WOP4G41B | OP4/OP4/op4::POP4-GFP-FRT | This work |
| WOP4R21A and -B | WO-1 | OP4/OP4/op4::POP4-RFP-SAT1-FLIP | This work |
| WOP4R22A | WOP4R21A | OP4/OP4/op4::POP4-RFP-FRT | This work |
| WOP4R22B | WOP4R21B | OP4/OP4/op4::POP4-RFP-FRT | This work |
| WOP4G42WH11R21A | WOP4G42A | OP4/OP4/op4::POP4-GFP-FRT WH11/wh11::PWH11-RFP-SAT1-FLIP | This work |
| WOP4G42WH11R21B | WOP4G42B | OP4/OP4/op4::POP4-GFP-FRT WH11/wh11::PWH11-RFP-SAT1-FLIP | This work |
| WOP4G42WH11R22A | WOP4G42WH11R21A | OP4/OP4/op4::POP4-GFP-FRT WH11/wh11::PWH11-RFP-FRT | This work |
| WOP4G42WH11R22B | WOP4G42WH11R21B | OP4/OP4/op4::POP4-GFP-FRT WH11/wh11::PWH11-RFP-FRT | This work |
| WOP4R22WH11G21A | WOP4R22A | OP4/OP4/op4::POP4-RFP-FRT WH11/wh11::PWH11-GFP-SAT1-FLIP | This work |
| WOP4R22WH11G21B | WOP4R22B | OP4/OP4/op4::POP4-RFP-FRT WH11/wh11::PWH11-GFP-SAT1-FLIP | This work |
| WOP4R22WH11G22A | WOP4R22WH11G21A | OP4/OP4/op4::POP4-RFP-FRT WH11/wh11::PWH11-GFP-FRT | This work |
| WOP4R22WH11G22B | WOP4R22WH11G21B | OP4/OP4/op4::POP4-RFP-FRT WH11/wh11::PWH11-GFP-FRT | This work |
SAT1-FLIP, the SAT1 flipper cassette; FRT, FLP recombination target sequence.
Plasmid constructions.
The previously described plasmid pGFP70 (25) contains a Candida-adapted green fluorescent protein reporter gene (GFP) (26) under the control of the OP4 promoter and URA3 as a selection marker. A SalI-PstI fragment from pOPT1G22 (27) was inserted between the same sites in pGFP70, thereby introducing a BglII site behind the GFP stop codon and substituting the dominant caSAT1 (Candida-adapted SAT1) selection marker for URA3 in the resulting plasmid, pOP4G2. An ApaI-BglII fragment from pOP4G2 containing the POP4-GFP fusion was then cloned together with a BglII-XhoI fragment from pNIM6 (9) containing the TEF3 transcription termination sequence in ApaI/XhoI-digested pCZF1M2 (9) to generate pOP4G3. The OP4 downstream region was amplified with the primers OPS21 (5′-CTTTAGTTAATGCCCGCGGTCAAGCTGCCTC-3′) and OPS8 (5′-TACTTGAGCTCTGCAACACTTCTTGCTCTTT-3′), and the SacII/SacI-digested PCR product was used to replace the CZF1 downstream region in pOP4G3, yielding pOP4G4 (Fig. 1A, top). A WH11 promoter fragment from pGFP68 (28) and a fragment containing the WH11 downstream region, which was amplified with the primers WHS15 (5′-GAGTGAGTAACCGCGGTTGAGTTGAAGTC-3′) and WHS16 (5′-CTTGGAGCTCAGTGTTAGGTGATACAGTC-3′), were used to replace the OP4 flanking sequences in pOP4G4 to generate pWH11G2. The red fluorescent protein gene (RFP) was amplified from plasmid pCaADH1-yEmRFP (29) with the primers RFP1 (5′-ATATGTCGACAAAATGGTTTCAAAAGGTGAAGAAG-3′) and RFP2 (5′-ATATAGATCTTATTTATATAATTCATCCATACC-3′) and used to replace GFP in pOP4G4 and pWH11G2, thereby producing pOP4R2 and pWH11R2, respectively.
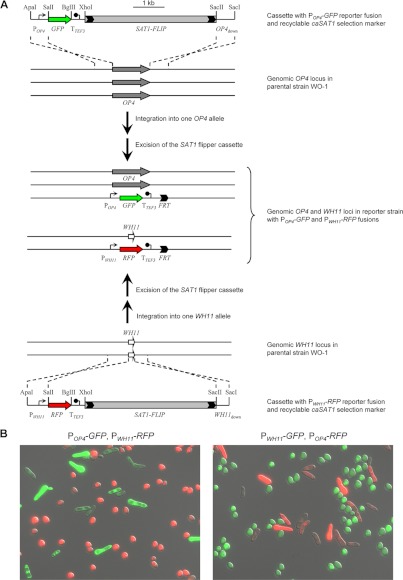
Fig 1.
Generation of phase-specifically labeled C. albicans strains. (A) Schematic of the sequential integration of POP4-GFP (top) and PWH11-RFP (bottom) reporter gene fusions into the genomic OP4 and WH11 loci of the parental wild-type strain WO-1 with the help of the recyclable SAT1 flipper cassette (SAT1-FLIP) to generate strains WOP4G42WH11R22A and -B (see also Table 1). Strains WOP4R22WH11G22A and -B, which contain POP4-RFP and PWH11-GFP reporter fusions, were generated in an analogous fashion. The parental strain WO-1 is trisomic for chromosome 1 and contains three copies of OP4, which is located on this chromosome (9). The genomic OP4 and WH11 coding sequences are indicated by the gray and white arrows, respectively; the cloned upstream (POP4 and PWH11) and downstream (OP4down and WH11down) sequences served for integration of the reporter cassettes into the respective genomic loci by homologous recombination. TTEF3, transcription termination sequence of the TEF3 gene; FRT, FLP recombination target sequence. Relevant restriction sites used for the generation of the reporter cassettes (see Materials and Methods) are indicated. (B) Phenotype of white and opaque cells of the reporter strains. Cells from white and opaque colonies of strains containing the indicated reporter fusions were mixed and observed by fluorescence microscopy. The pictures show an overlay of transmission and epifluorescence micrographs with GFP and RFP filter settings and demonstrate the phase-specific expression of GFP and RFP in white and opaque cells of the reporter strains. (Left) Strain WOP4G42WH11R22B; (right) strain WOP4R22WH11G22A.
Strain construction and analysis.
C. albicans strains were transformed by electroporation (24) with the gel-purified inserts from plasmids pOP4G4, pWH11G2, pOP4R2, and pWH11R2 described above. The correct integration of each construct and subsequent excision of the SAT1 flipper cassette were confirmed by Southern hybridization using the flanking sequences as probes. In each case, two independent series of strains (A and B) were generated.
Isolation of genomic DNA and Southern hybridization.
Genomic DNA from C. albicans strains was isolated as described previously (24). The DNA was digested with appropriate restriction enzymes, separated on a 1% agarose gel, transferred by vacuum blotting onto a nylon membrane, and fixed by UV cross-linking. Southern hybridization with enhanced chemiluminescence-labeled probes was performed with an Amersham ECL direct nucleic acid labeling and detection system (GE Healthcare UK Limited, Little Chalfont, Buckinghamshire, United Kingdom) according to the instructions of the manufacturer.
Isolation of neutrophils and dendritic cells.
Neutrophils were isolated from human blood samples by using polymorphPrep solution from Progen. The isolation was carried out according to the instructions of the manufacturer. Viability was assessed by trypan blue staining. The cells were suspended in RPMI 1640 medium containing 5% fetal calf serum (FCS) and stored on ice until usage. Bone marrow-derived dendritic cells (BMDCs) were generated in 7- to 9-day cultures as described previously (30). Briefly, bone marrow cells were flushed out of both hind legs of 10- to 12-week-old female BALB/c mice. After erythrocyte lysis, the cell suspension was precultured in a 10-cm cell culture dish for 2 h at 37°C in RPMI 1640 medium (supplemented with nonessential amino acids [NEAAs], 5% FCS, 2 mM l-glutamine, 50 μM β-mercaptoethanol, 50 μg/ml gentamicin, 250 U/ml granulocyte-macrophage colony-stimulating factor [GM-CSF], 100 U/ml interleukin-4 [IL-4]) to eliminate adherent macrophage-like cells. Subsequently, the nonadherent cells were transferred to 6-well cell culture plates at a concentration of 3 × 106 cells per well and cultured at 37°C in supplemented BMDC medium (see above) for 7 to 9 days (exchange of medium at day 3; transfer to new culture plates at day 6). Cell lines secreting murine GM-CSF or IL-4 were kindly provided by Thomas Blankenstein.
Time-lapse video microscopy.
Experiments were carried out as described by Behnsen et al. (31), with some modifications. Fresh colonies of C. albicans were washed once in phosphate-buffered saline and once in RPMI 1640 medium containing 5% FCS. For the experiments on glass slides, 1.25 × 106 cells of each phase (opaque and white) were mixed with 5 × 105 purified viable neutrophils in 200 μl RPMI 1640 medium containing 5% FCS. The suspension was poured in a glass chamber, and video microscopy was performed for 5 h at 37°C with two frames per minute using an Olympus BX61 microscope with a ×60 LUMPLFL W/IR (numerical aperture, 0.9) lens, together with the cellR software (version 2.1) from Olympus Biosystems. For the experiments within a collagen matrix, 1 × 107 yeast cells of each phase (opaque and white) were mixed with 5 × 105 purified viable neutrophils in 66 μl minimal essential medium and 133 μl type I collagen stock solution (Vitrogen-100; Nutacon) to a final collagen concentration of 1.7 mg/ml and poured in a glass chamber. The chamber was incubated for 20 min at 37°C to allow polymerization of the matrix and subsequently used for video microscopy. The experiment with DCs was performed as described above for the neutrophils on glass slides, except that the coincubation was for 6 h with one frame per minute. All experiments were performed with two independent cultures of the C. albicans strains, and each coincubation was observed over time at four different positions. Scale bars in all videos are 20 μm.
RESULTS
Generation of phase-specifically labeled C. albicans strains.
To uncover differences in the interaction of neutrophils and dendritic cells with white and opaque cells of C. albicans, we performed coincubation experiments with the phagocytes and mixed white/opaque cell populations instead of studying the interaction with white and opaque cells separately, thus allowing the immune cells to encounter the two different forms of the pathogen simultaneously. Although white and opaque cells usually can be well distinguished under the microscope by their different morphologies, we decided to use differentially labeled white and opaque cells to facilitate the recognition of the two yeast forms by video microscopy during phagocytosis. For this purpose, we sequentially introduced the GFP reporter gene under the control of the opaque-phase-specific OP4 promoter and the RFP reporter gene under the control of the white-phase-specific WH11 promoter into the corresponding endogenous genomic loci using a recyclable nourseothricin resistance marker for the selection of transformants (Fig. 1A). In addition, we also generated complementary strains expressing GFP from the WH11 promoter and RFP from the OP4 promoter to exclude the (unlikely) possibility that any differences between the two cell types were caused by the expression of GFP or RFP. This strategy had the advantage that white and opaque cells of the same reporter strain rather than different derivatives of a particular strain could be compared. The MTLα/α wild-type strain WO-1, in which white-opaque switching was originally discovered and which has been widely used to study this developmental program, was used as the parent for the construction of the reporter strains. Two independent series of strains were generated (Table 1) and used in subsequent experiments. After verifying the correct genomic configurations of the reporter strains, cells in the white and opaque phases were isolated, mixed, and observed by fluorescence microscopy. As can be seen in Fig. 1B, white and opaque cells of the reporter strains exhibited the expected phenotypes and appeared green or red in the respective phases.
Interaction of white and opaque cells with human neutrophils.
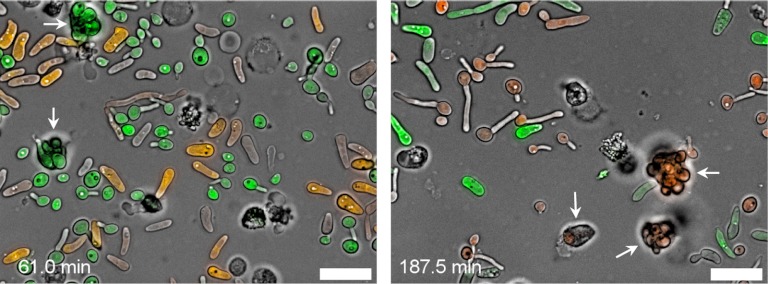
To study the interaction of white and opaque cells with PMNs, freshly isolated human neutrophils were incubated with mixed white/opaque cell populations of the labeled C. albicans strains on glass slides and observed by video microscopy. Under these conditions, the neutrophils selectively phagocytosed cells in the white phase, whereas internalization of opaque cells was rarely observed, despite frequent physical contact (Fig. 2; see Videos S1 and S2 in the supplemental material). Therefore, the fact that opaque cells do not secrete a chemoattractant for neutrophils cannot be the sole explanation for their ability to prevent phagocytosis by these host defense cells. Another possible reason could be the larger size and elongated morphology of opaque yeast cells compared with white yeast cells. However, the culture conditions employed in these experiments promoted the yeast-hyphal switch in white cells, which formed germ tubes that became more and more elongated over time. Even white cells with germ tubes that were considerably longer than whole opaque cells were efficiently phagocytosed by the neutrophils (Fig. 3; see Video S3 in the supplemental material), indicating that the size of opaque cells is not the limiting factor that precludes phagocytosis.
Fig 2.
Interaction of white and opaque cells with human neutrophils on a glass slide. (Left) Neutrophils that were coincubated with a mixture of GFP-expressing white cells and RFP-expressing opaque cells of strain WOP4R22WH11G22B. Two neutrophils that have exclusively phagocytosed white-phase cells are labeled with arrows. (Right) Neutrophils that were coincubated with a mixture of RFP-expressing white cells and GFP-expressing opaque cells of strain WOP4G42WH11R22B. Three neutrophils that have exclusively phagocytosed white-phase cells are labeled with arrows. Pictures were taken from Videos S1 (left) and S2 (right) in the supplemental material; time points are indicated. Bars, 20 μm.
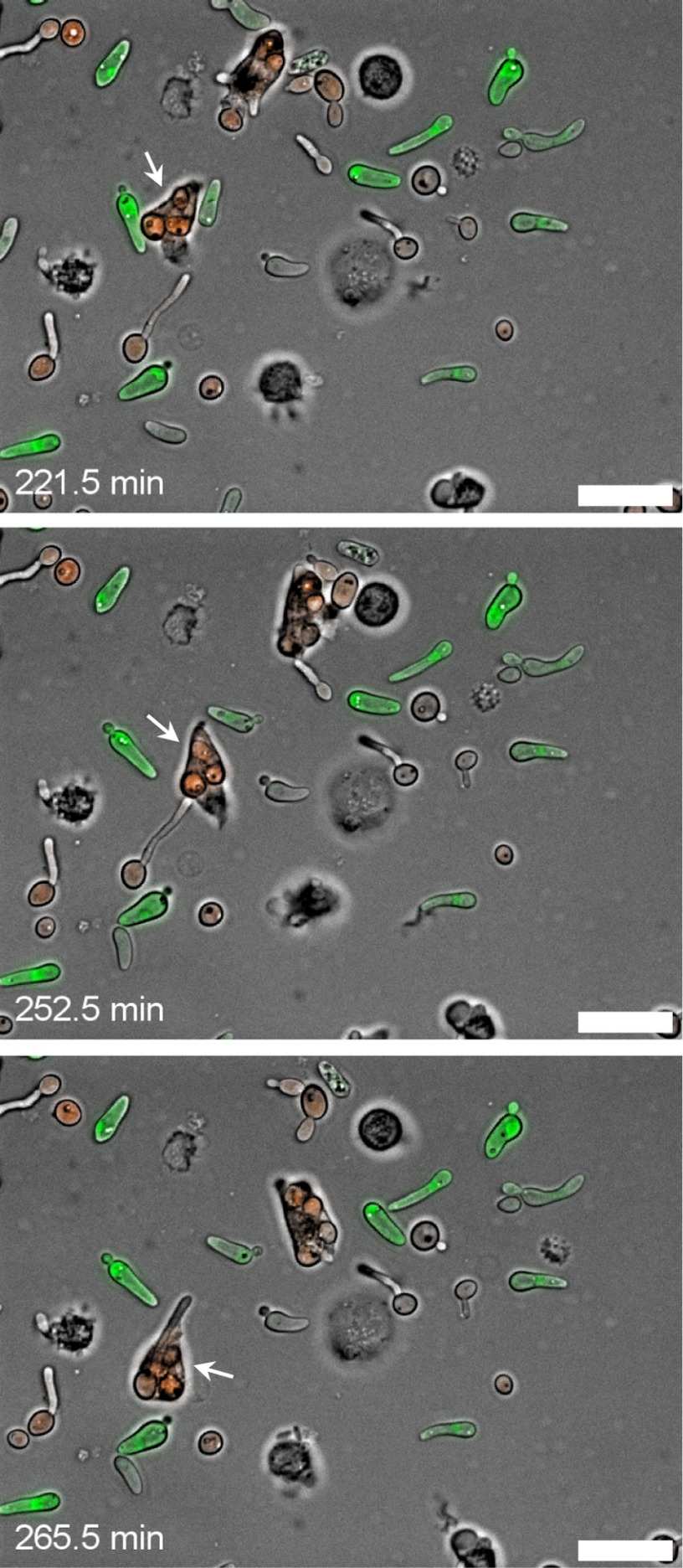
Fig 3.
Neutrophils can phagocytose large cells. (Top) A neutrophil (marked with a white arrow) that has taken up several RFP-expressing white cells but does not phagocytose opaque cells despite close physical contact; (middle) the same neutrophil attacking a white cell with a large germ tube, which is followed by phagocytosis of the complete cell (bottom). Strain WOP4G42WH11R22A was used for this experiment. For more details, see Video S3 in the supplemental material, from which the pictures were taken at the indicated time points. Bars, 20 μm.
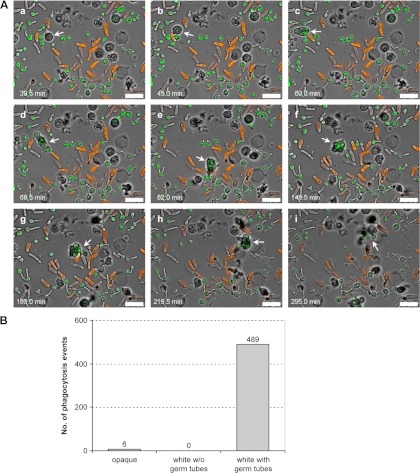
Close inspection of all phagocytosis events observed in videos from independent experiments revealed that all white cells that were taken up by the neutrophils had started to germinate, whereas white yeast cells without a recognizable germ tube were not taken up, even after repeated contact with a neutrophil. An illustrative example is provided in Video S4 in the supplemental material, representative pictures of which are shown in Fig. 4A. The neutrophil that is highlighted in Fig. 4A could be followed over the whole time course of the experiment. It frequently and repeatedly interacted with all types of C. albicans cells, often forming extensions that seemed to probe the surface of white and opaque cells, but phagocytosed only germinating white cells, while finally leaving opaque cells and white cells without germ tubes alone. It can also be seen in Video S4 in the supplemental material that when the neutrophil had reached the end of its life span and disintegrated after many phagocytosis events (Fig. 4Ai), most of the internalized C. albicans cells appeared to have been destroyed, indicating efficient killing by the neutrophil. From a total of 495 phagocytosis events analyzed in independent experiments with all four differentially labeled C. albicans strains, uptake of an opaque cell was observed only six times (1%), whereas the remaining 489 internalized fungal cells (99%) were germinating white cells (Fig. 4B). Therefore, under the experimental conditions of these assays, neutrophils appear to predominantly recognize fungal structures that are selectively expressed on germ tubes/hyphae of C. albicans but not on the surface of opaque cells.
Fig 4.
Neutrophils selectively phagocytose germ tube-forming white cells. (A) A series of pictures from Video S4 in the supplemental material (time points are indicated) in which the behavior of a single neutrophil (marked by the white arrow) can be followed over time. After the neutrophil becomes active and gets in physical contact with surrounding GFP-expressing white and RFP-expressing opaque cells (a), it takes up a germinating white cell (b), which is followed by phagocytosis of additional white cells with germ tubes (c and d). The neutrophil then repeatedly contacts a group of surrounding opaque cells without phagocytosing these cells (e). Instead, two germ tube-forming white cells, which can be seen above the neutrophil in panel e, are ingested (f), while another white cell without a recognizable germ tube (below the neutrophil in panel f) is frequently contacted but not phagocytosed. The neutrophil then pushes an opaque cell aside to attack and ingest another germ tube-forming white cell (g) and migrates on and phagocytoses an additional germinating white cell (h). When the neutrophil finally disintegrates, most of the phagocytosed C. albicans cells apparently have been destroyed (i). Strain WOP4R22WH11G22B was used in this experiment. Bars, 20 μm. (B) Frequency of phagocytosis of opaque cells and white cells with and without a germ tube. A total of 495 phagocytosis events by 118 neutrophils were analyzed. Data are from four independent experiments including all four differentially labeled C. albicans strains and two biological replicates for each strain.
Interaction of dendritic cells with white and opaque C. albicans cells.
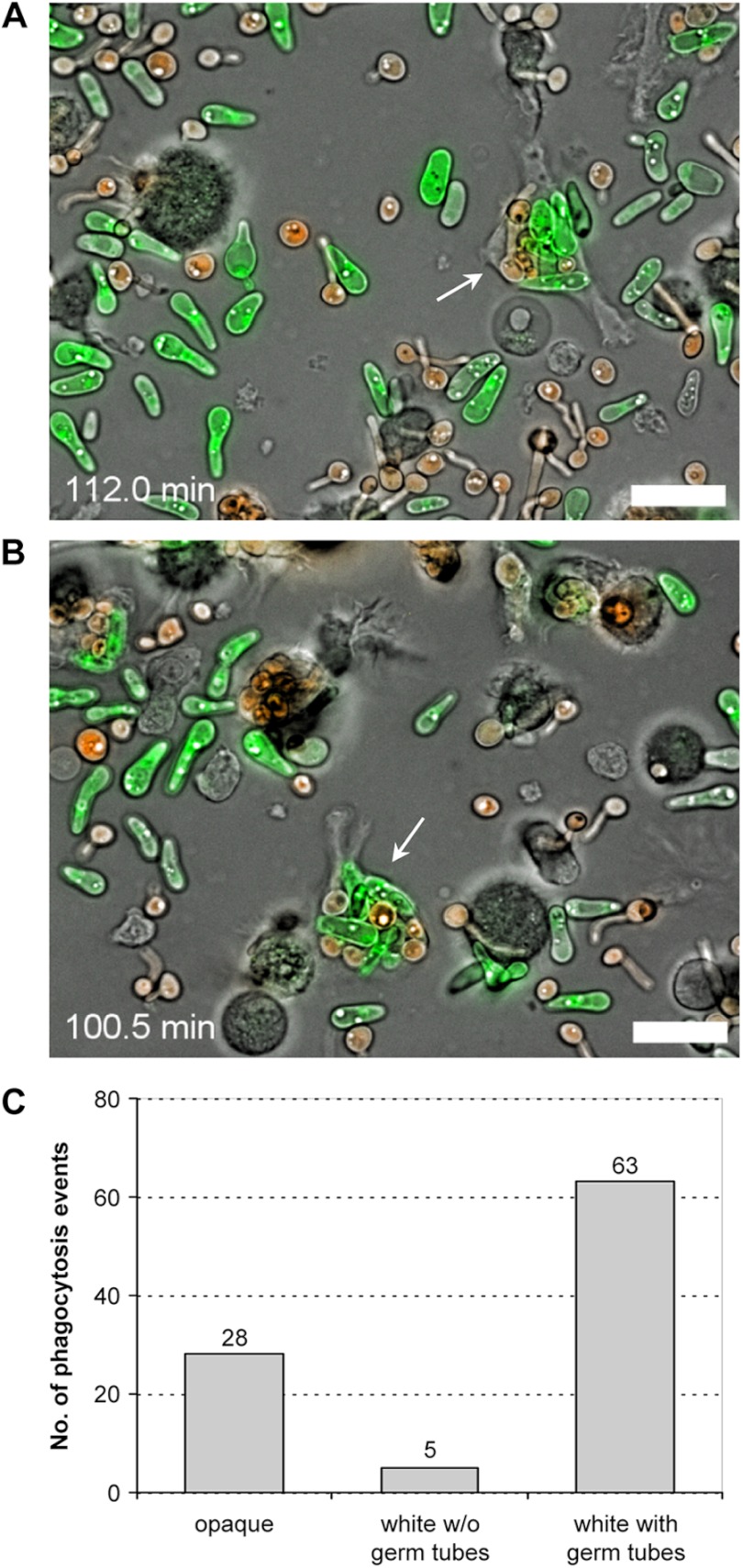
Dendritic cells are another important type of host defense cells that provide a linkage between the innate and adaptive immune system by phagocytosing microbial pathogens and presenting their antigens to other immune cells. They recognize and phagocytose both yeast and hyphal forms of C. albicans (32), but their behavior toward opaque cells has not been studied so far. We therefore examined the interaction of DCs with mixed populations of differentially labeled white and opaque cells under the same conditions as in the experiments with the neutrophils. Interestingly, the DCs did not distinguish between white and opaque cells and phagocytosed both cell types, albeit also with some preference for white cells. The DCs quickly started to take up C. albicans cells from the beginning of the coincubation, so that DCs with internalized fungi were already seen when video microscopy was initiated (see Videos S5 and S6 in the supplemental material). Often, individual DCs contained mixtures of white and opaque cells (Fig. 5A), and from a total of 96 phagocytosed cells counted in 21 DCs, 28 were opaque cells, 5 were white cells without a recognizable germ tube, and 63 were white cells with germ tubes (Fig. 5B).
Fig 5.
Dendritic cells phagocytose both white and opaque cells. (A, B) The pictures are from two biological replicates of one experiment (see Videos S5 and S6 in the supplemental material; time points are indicated) in which DCs were coincubated on glass slides with a mixture of RFP-expressing white cells and GFP-expressing opaque cells of strain WOP4G42WH11R22A. The arrows indicate DCs that have taken up several white and opaque cells. Bars, 20 μm. (C) Frequency of phagocytosis of opaque cells and white cells with and without a germ tube. A total of 96 phagocytosis events by 21 DCs were analyzed in the experiment described above.
Neutrophils phagocytose both white and opaque cells when embedded in a collagen matrix.
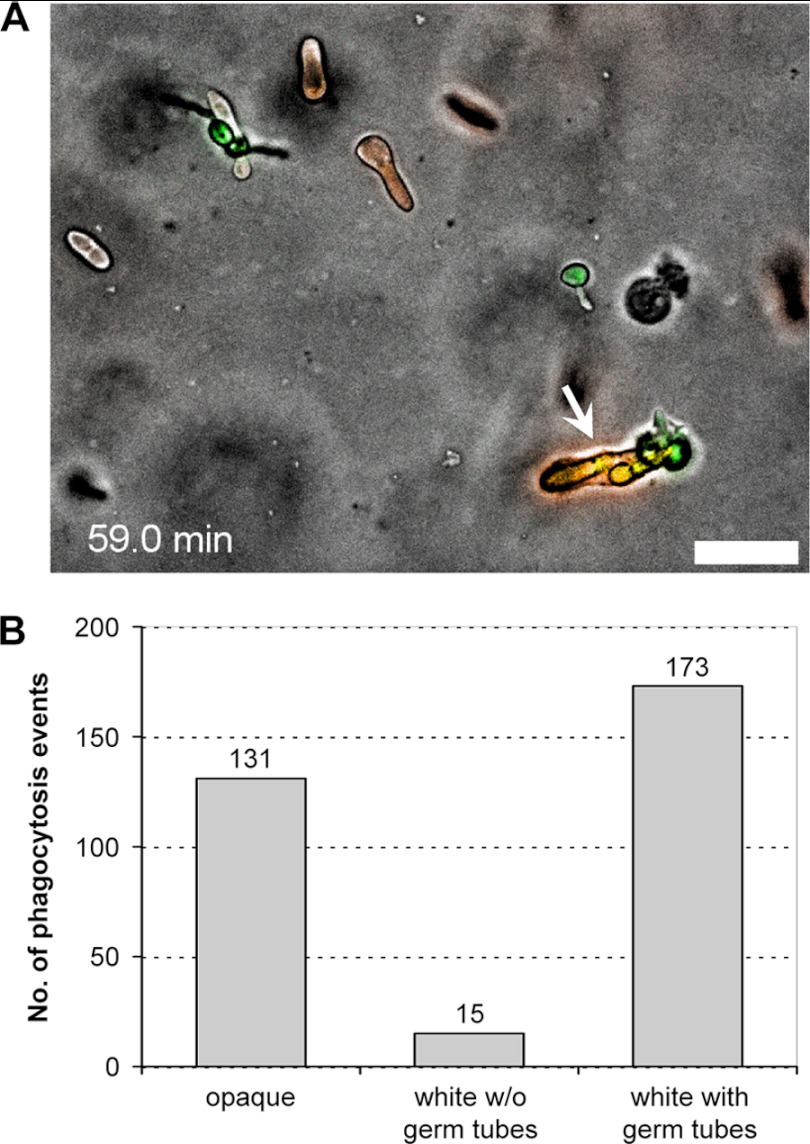
In a previous study, it was observed that neutrophils selectively phagocytosed Aspergillus fumigatus conidia but not C. albicans yeast cells (which did not germinate under the experimental conditions of that study) when coincubated with both fungi on a glass surface (31). However, when coincubated with the two types of fungal cells within a collagen matrix (a three-dimensional [3D] environment, as opposed to the two-dimensional [2D] environment on a glass slide), the neutrophils phagocytosed the C. albicans yeast cells instead of the A. fumigatus conidia, indicating that the environment influenced the recognition of different pathogens by the neutrophils (31). We therefore investigated whether neutrophils would also interact in a different manner with white and opaque cells of C. albicans when embedded in a collagen matrix. Interestingly, we found that under these conditions the neutrophils also recognized and phagocytosed the opaque cells, in addition to white cells with and without germ tubes. An example is shown in Fig. 6A and the corresponding video (see Video S7 in the supplemental material). Among 319 phagocytosis events counted in four independent experiments, opaque cells were phagocytosed 131 times (41%), germinating white cells were phagocytosed 173 times (54%), and white yeast cells without a recognizable germ tube (whose proportion in the population declined over time due to the induction of germ tube formation) were phagocytosed 15 times (5%) (Fig. 6B). Therefore, under appropriate conditions, neutrophils can also efficiently recognize and phagocytose opaque cells.
Fig 6.
Neutrophils phagocytose both white and opaque cells upon coincubation in a collagen matrix. (A) The picture was taken from Video S7 in the supplemental material at the indicated time point. The arrow shows a neutrophil that has taken up two GFP-expressing white cells and three RFP-expressing opaque cells of strain WOP4R22WH11G22B. Bar, 20 μm. (B) Frequency of phagocytosis of opaque cells and white cells with and without a germ tube. A total of 319 phagocytosis events by 274 neutrophils were analyzed. Data are from four independent experiments with two biological replicates.
DISCUSSION
It is currently not understood why C. albicans has integrated a morphological switch into its life cycle to undergo sexual development. In contrast to the model yeast Saccharomyces cerevisiae, where haploid a or α cells derived from diploid a/α cells can immediately mate with cells of the opposite mating type, C. albicans cells that have become homozygous for the mating type locus (MTLa/a or MTLα/α) first have to switch from the normal yeast morphology (termed white in this context) to the opaque cell form in order to become mating competent (33, 34). It is commonly assumed that white-opaque switching has evolved in C. albicans as an adaptation to the specific requirements of life in human and warm-blooded animal hosts. The phase-specific expression of adhesins and metabolic genes may allow opaque cells an efficient colonization of host niches to which white cells are less well adapted and where mating may occur. Previous studies have also indicated that white-opaque switching affects the interaction of C. albicans with the host immune system. It has been shown that opaque cells are less efficiently phagocytosed than white cells by Drosophila S2 cells and mouse macrophages (20), suggesting that switching to the opaque phase may be an immune evasion mechanism. In humans, neutrophils play a major role in the prevention of Candida infections. The finding that opaque cells do not produce a chemoattractant for neutrophils that is secreted by white cells pointed to the possibility that opaque cells can also avoid detection by these important first-line host defense cells (19). However, earlier work had shown that opaque cells are in fact more susceptible than white cells to killing by neutrophils and also stimulate the production of reactive oxygen species, which is one mechanism by which these phagocytes destroy invading pathogens, more strongly than do white cells (17). The findings of our present study may explain these conflicting results, and they also provide more detailed insight into the differential interaction of the various morphological forms of C. albicans with cells of the host immune system. Similar to the observations made with Drosophila S2 cells and mouse macrophages (20), we found that neutrophils selectively phagocytosed white cells and largely ignored opaque cells when coincubated with both cell types on a glass slide. In contrast, Kolotila and Diamond reported that white and opaque cells were phagocytosed with equal efficiency by neutrophils, and opaque cells were even more efficiently killed than white cells because of their increased susceptibility to reactive oxygen species (17). Of note, these authors used opsonized white and opaque cells that were incubated with PMNs for 1 h at 37°C in tubes under shaking. Opsonization may facilitate recognition of opaque cells by neutrophils, and we indeed also found increased phagocytosis of opaque cells when the experiments were performed in culture medium containing active FCS instead of heat-inactivated FCS, although germinating white cells were still much more efficiently phagocytosed (91%) than opaque cells (7%) and nongerminating white yeast cells (2%) (unpublished data). In addition, coincubation under shaking, as was done in the experiments by Kolotila and Diamond (17), may have altered the behavior of the neutrophils compared to their behavior in our experiments on glass slides, similar to the different results that we obtained when neutrophils and C. albicans cells were coincubated within a collagen matrix. Therefore, the environment plays a major role in the recognition of the different morphologies of C. albicans. Although both the surface of a glass slide and a collagen matrix are artificial environments, similar environmental differences (e.g., between mucosal surfaces and deep tissue) are likely to exist in vivo, and switching to the opaque phase may allow immune evasion in some, but not all, host niches. Our results also demonstrate that opaque cells cannot avoid recognition by all types of phagocytic cells. In contrast to human neutrophils, Drosophila S2 cells, and mouse macrophages, DCs efficiently phagocytosed both white and opaque cells.
An important finding of our study was that during the coincubation on glass slides, white cells were attacked by the neutrophils only after they had started to germinate, indicating that under these conditions the neutrophils recognized a structure that is selectively expressed on the hyphal form of C. albicans. Phagocytes engage multiple receptors that bind different molecules on the fungal surface (35). Apparently, the involvement of specific receptors depends on the environment, because opaque cells were not recognized on a glass surface but efficiently phagocytosed when encountered within a collagen matrix. In line with these observations, it was previously found that carboxyfluorescein succinimidyl ester-labeled, nongerminating C. albicans yeast cells were inefficiently phagocytosed by PMNs and a peritoneal macrophage-derived cell line on a glass surface but efficiently phagocytosed within a collagen matrix, while the reverse was true for A. fumigatus conidia (31). Our results are also in agreement with those of a previous report in which the interactions of yeast and filamentous forms of C. albicans with neutrophils were compared (36). In that study, C. albicans filaments, but not yeast cells, induced the targeted motility of human neutrophils, and the neutrophils engulfed only filaments and germ tubes but did not phagocytose yeast cells. However, neutrophils can also kill C. albicans by extracellular (neutrophil extracellular trap formation) mechanisms, and another study found that hyphae and yeast cells were killed with equal efficiency (37).
An unresolved question is which hypha-specific structure is recognized by the neutrophils under conditions in which they do not phagocytose opaque cells and nongerminating yeast cells. An attractive candidate is the pH-regulated antigen Pra1, which is predominantly expressed in a highly glycosylated form on the surface of C. albicans hyphae, but not yeast cells, and bound by the αMβ2 receptor on PMNs. Pra1 also acts as a chemoattractant for PMNs, and deletion of PRA1 decreased PMN migration and PMN adhesion to and killing of C. albicans (38, 39). It has also been reported that dendritic cells can recognize C. albicans by an αMβ2-independent mechanism (39), which would explain our finding that dendritic cells phagocytosed opaque cells under conditions in which they were not attacked by neutrophils. However, forced expression of PRA1 in opaque cells did not result in enhanced phagocytosis by neutrophils (our unpublished data), although we cannot exclude the possibility that Pra1 was not properly expressed on the surface of the transformed opaque cells. Hyphae express many other cell surface proteins that are not found on yeast cells. One of these, Hyr1, confers even increased resistance to killing by neutrophils and other phagocytes (40, 41). Therefore, it remains to be established which hypha-specific antigen(s) and corresponding receptor(s) on neutrophils are responsible for the selective phagocytosis of germinating C. albicans.
Another open question is why the neutrophils phagocytosed opaque cells within the three-dimensional environment of the collagen matrix but not on the two-dimensional glass surface. Previous experiments have shown that the change in behavior of the neutrophils is immediate; i.e., after leaving the 3D environment, neutrophils immediately change into 2D mode (31). This would be too fast to explain it by any significant change in surface composition. We speculate that the altered behavior of the neutrophils is dependent on the sensing of the 3D environment, which has been shown to influence cell physiology (42, 43). On the other hand, it is also possible that a surface structure that is recognized by the neutrophils may be specifically presented on opaque cells within the 3D environment.
The switch to hyphal growth is a major virulence determinant of C. albicans that facilitates tissue invasion. As opaque cells do not undergo filamentation under most conditions that induce hypha formation in white cells, it is tempting to speculate that switching to the opaque form may render C. albicans less aggressive toward its host but at the same time allow escape from the host immune system in certain host niches, where mating and genetic exchange may then occur undisturbed.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sabine Keppler-Ross for the gift of plasmid pCaADH1-yEmRFP.
This study was supported by the Deutsche Forschungsgemeinschaft (SFB 479).
Footnotes
Published ahead of print 2 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00266-12.
REFERENCES
- 1. Kumamoto CA, Vinces MD. 2005. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell. Microbiol. 7:1546–1554 [DOI] [PubMed] [Google Scholar]
- 2. Lockhart SR, Pujol C, Daniels KJ, Miller MG, Johnson AD, Pfaller MA, Soll DR. 2002. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 162:737–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miller MG, Johnson AD. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293–302 [DOI] [PubMed] [Google Scholar]
- 4. Forche A, Alby K, Schaefer D, Johnson AD, Berman J, Bennett RJ. 2008. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 6:e110 doi:10.1371/journal.pbio.0060110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alby K, Bennett RJ. 2009. Stress-induced phenotypic switching in Candida albicans. Mol. Biol. Cell 20:3178–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang G, Srikantha T, Sahni N, Yi S, Soll DR. 2009. CO2 regulates white-to-opaque switching in Candida albicans. Curr. Biol. 19:330–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang G, Yi S, Sahni N, Daniels KJ, Srikantha T, Soll DR. 2010. N-Acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 6:e1000806 doi:10.1371/journal.ppat.1000806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morschhäuser J. 2010. Regulation of white-opaque switching in Candida albicans. Med. Microbiol. Immunol. 199:165–172 [DOI] [PubMed] [Google Scholar]
- 9. Ramírez-Zavala B, Reuß O, Park Y-N, Ohlsen K, Morschhäuser J. 2008. Environmental induction of white-opaque switching in Candida albicans. PLoS Pathog. 4:e1000089 doi:10.1371/journal.ppat.1000089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soll DR. 1992. High-frequency switching in Candida albicans. Clin. Microbiol. Rev. 5:183–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lan CY, Newport G, Murillo LA, Jones T, Scherer S, Davis RW, Agabian N. 2002. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 99:14907–14912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsong AE, Miller MG, Raisner RM, Johnson AD. 2003. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell 115:389–399 [DOI] [PubMed] [Google Scholar]
- 13. Tuch BB, Mitrovich QM, Homann OR, Hernday AD, Monighetti CK, De La Vega FM, Johnson AD. 2010. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet. 6:e1001070 doi:10.1371/journal.pgen.1001070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kvaal C, Lachke SA, Srikantha T, Daniels K, McCoy J, Soll DR. 1999. Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect. Immun. 67:6652–6662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kvaal CA, Srikantha T, Soll DR. 1997. Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infect. Immun. 65:4468–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anderson J, Cundiff L, Schnars B, Gao MX, Mackenzie I, Soll DR. 1989. Hypha formation in the white-opaque transition of Candida albicans. Infect. Immun. 57:458–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kolotila MP, Diamond RD. 1990. Effects of neutrophils and in vitro oxidants on survival and phenotypic switching of Candida albicans WO-1. Infect. Immun. 58:1174–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lilic D. 2012. Unravelling fungal immunity through primary immune deficiencies. Curr. Opin. Microbiol. 15:420–426 [DOI] [PubMed] [Google Scholar]
- 19. Geiger J, Wessels D, Lockhart SR, Soll DR. 2004. Release of a potent polymorphonuclear leukocyte chemoattractant is regulated by white-opaque switching in Candida albicans. Infect. Immun. 72:667–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lohse MB, Johnson AD. 2008. Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes. PLoS One 3:e1473 doi:10.1371/journal.pone.0001473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169:189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bedell GW, Soll DR. 1979. Effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: evidence for zinc-resistant and -sensitive pathways for mycelium formation. Infect. Immun. 26:348–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Soll DR, Morrow B, Srikantha T. 1993. High-frequency phenotypic switching in Candida albicans. Trends Genet. 9:61–65 [DOI] [PubMed] [Google Scholar]
- 24. Reuß O, Vik Å, Kolter R, Morschhäuser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127 [DOI] [PubMed] [Google Scholar]
- 25. Park YN, Strauß A, Morschhäuser J. 2004. The white-phase-specific gene WH11 is not required for white-opaque switching in Candida albicans. Mol. Genet. Genomics 272:88–97 [DOI] [PubMed] [Google Scholar]
- 26. Morschhäuser J, Michel S, Hacker J. 1998. Expression of a chromosomally integrated, single-copy GFP gene in Candida albicans, and its use as a reporter of gene regulation. Mol. Gen. Genet. 257:412–420 [DOI] [PubMed] [Google Scholar]
- 27. Reuß O, Morschhäuser J. 2006. A family of oligopeptide transporters is required for growth of Candida albicans on proteins. Mol. Microbiol. 60:795–812 [DOI] [PubMed] [Google Scholar]
- 28. Strauß A, Michel S, Morschhäuser J. 2001. Analysis of phase-specific gene expression at the single-cell level in the white-opaque switching system of Candida albicans. J. Bacteriol. 183:3761–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Keppler-Ross S, Noffz C, Dean N. 2008. A new purple fluorescent color marker for genetic studies in Saccharomyces cerevisiae and Candida albicans. Genetics 179:705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Labeur MS, Roters B, Pers B, Mehling A, Luger TA, Schwarz T, Grabbe S. 1999. Generation of tumor immunity by bone marrow-derived dendritic cells correlates with dendritic cell maturation stage. J. Immunol. 162:168–175 [PubMed] [Google Scholar]
- 31. Behnsen J, Narang P, Hasenberg M, Gunzer F, Bilitewski U, Klippel N, Rohde M, Brock M, Brakhage AA, Gunzer M. 2007. Environmental dimensionality controls the interaction of phagocytes with the pathogenic fungi Aspergillus fumigatus and Candida albicans. PLoS Pathog. 3:e13 doi:10.1371/journal.ppat.0030013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. d'Ostiani CF, Del Sero G, Bacci A, Montagnoli C, Spreca A, Mencacci A, Ricciardi-Castagnoli P, Romani L. 2000. Dendritic cells discriminate between yeasts and hyphae of the fungus Candida albicans. Implications for initiation of T helper cell immunity in vitro and in vivo. J. Exp. Med. 191:1661–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johnson A. 2003. The biology of mating in Candida albicans. Nat. Rev. Microbiol. 1:106–116 [DOI] [PubMed] [Google Scholar]
- 34. Soll DR, Lockhart SR, Zhao R. 2003. Relationship between switching and mating in Candida albicans. Eukaryot. Cell 2:390–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Netea MG, Brown GD, Kullberg BJ, Gow NA. 2008. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 6:67–78 [DOI] [PubMed] [Google Scholar]
- 36. Wozniok I, Hornbach A, Schmitt C, Frosch M, Einsele H, Hube B, Loffler J, Kurzai O. 2008. Induction of ERK-kinase signalling triggers morphotype-specific killing of Candida albicans filaments by human neutrophils. Cell. Microbiol. 10:807–820 [DOI] [PubMed] [Google Scholar]
- 37. Urban CF, Reichard U, Brinkmann V, Zychlinsky A. 2006. Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell. Microbiol. 8:668–676 [DOI] [PubMed] [Google Scholar]
- 38. Soloviev DA, Fonzi WA, Sentandreu R, Pluskota E, Forsyth CB, Yadav S, Plow EF. 2007. Identification of pH-regulated antigen 1 released from Candida albicans as the major ligand for leukocyte integrin αMβ2. J. Immunol. 178:2038–2046 [DOI] [PubMed] [Google Scholar]
- 39. Soloviev DA, Jawhara S, Fonzi WA. 2011. Regulation of innate immune response to Candida albicans infections by αMβ2-Pra1p interaction. Infect. Immun. 79:1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dwivedi P, Thompson A, Xie Z, Kashleva H, Ganguly S, Mitchell AP, Dongari-Bagtzoglou A. 2011. Role of Bcr1-activated genes Hwp1 and Hyr1 in Candida albicans oral mucosal biofilms and neutrophil evasion. PLoS One 6:e16218 doi:10.1371/journal.pone.0016218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luo G, Ibrahim AS, Spellberg B, Nobile CJ, Mitchell AP, Fu Y. 2010. Candida albicans Hyr1p confers resistance to neutrophil killing and is a potential vaccine target. J. Infect. Dis. 201:1718–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Klemke M, Kramer E, Konstandin MH, Wabnitz GH, Samstag Y. 2010. An MEK-cofilin signalling module controls migration of human T cells in 3D but not 2D environments. EMBO J. 29:2915–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lammermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Soldner R, Hirsch K, Keller M, Forster R, Critchley DR, Fassler R, Sixt M. 2008. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature 453:51–55 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.