Abstract
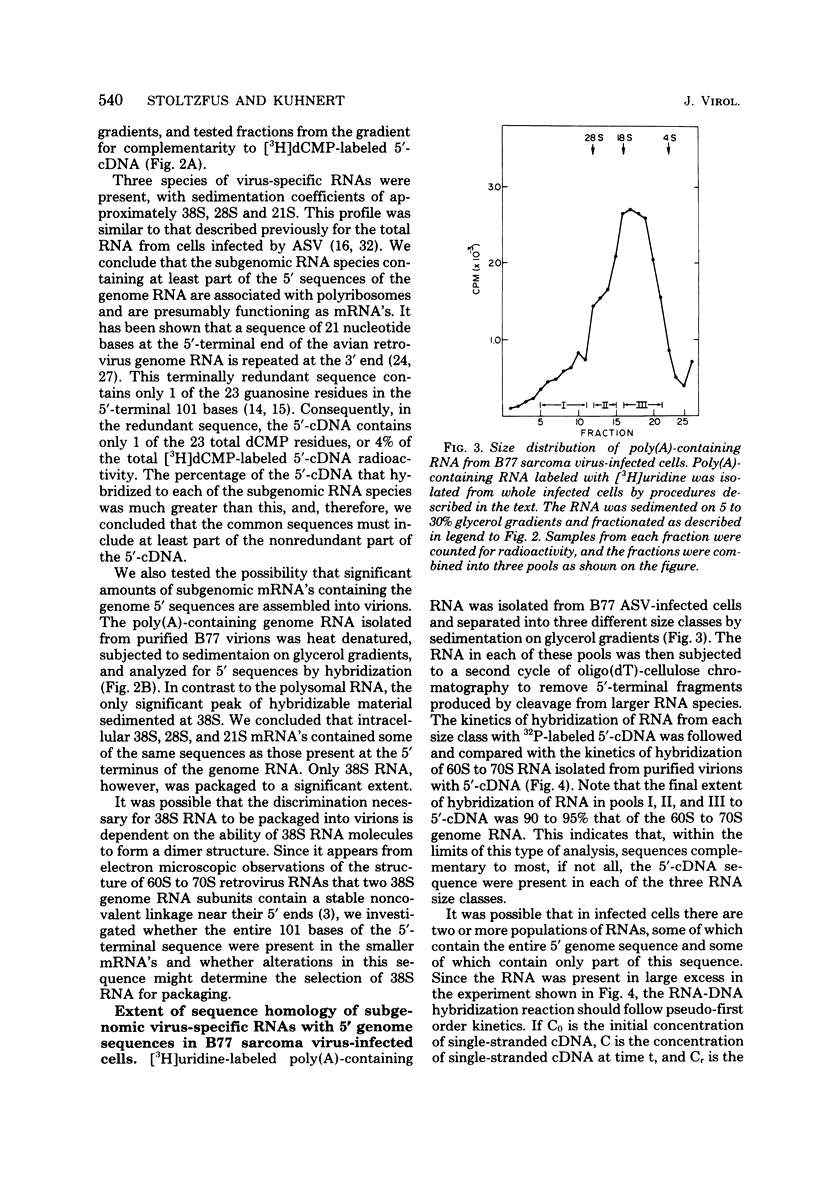
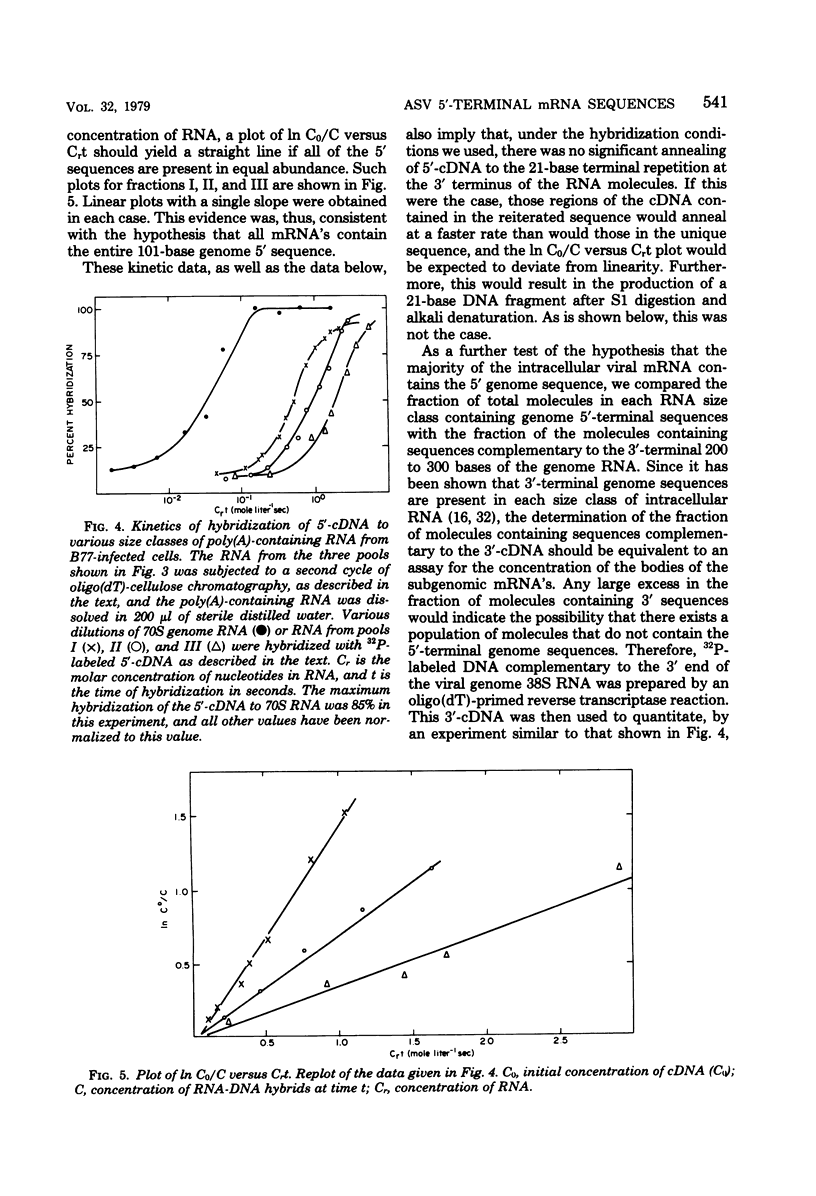
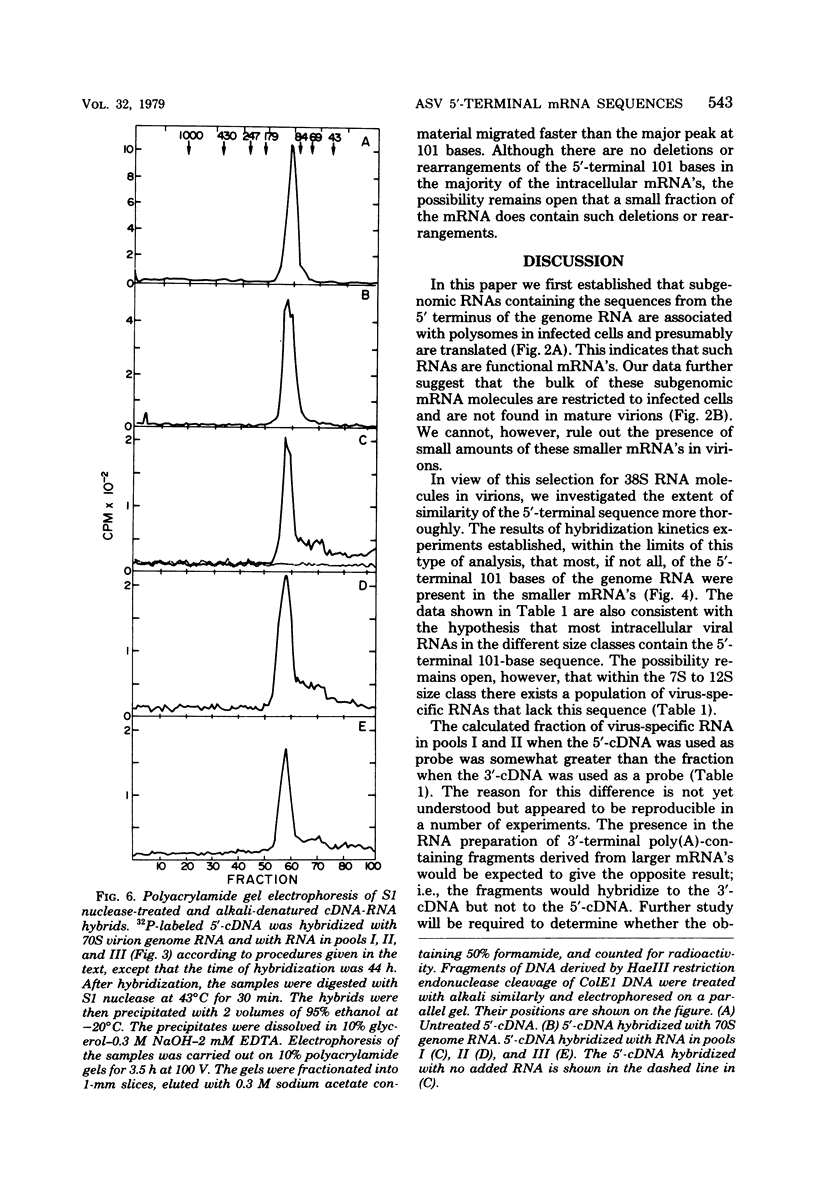
The polyribosomal fraction from chicken embryo fibroblasts infected with B77 avian sarcoma virus contained 38S, 28S, and 21S virus-specific RNAs in which sequences identical to the 5′-terminal 101 bases of the 38S genome RNA were present. The only polyadenylic acid-containing RNA species with 5′ sequences which was detectable in purified virions had a sedimentation coefficient of 38S. This evidence is consistent with the hypothesis that a leader sequence derived from the 5′ terminus of the RNA is spliced to the bodies of the 28S and 21S mRNA's, both of which have been shown previously to be derived from the 3′ terminal half of the 38S RNA. The entire 101-base 5′ terminal sequence of the genome RNA appeared to be present in the majority of the subgenomic intracellular virus-specific mRNA's, as established by several different methods. First, the extent of hybridization of DNA complementary to the 5′-terminal 101 bases of the genome to polyadenylic acid-containing subgenomic RNA was similar to the extent of its hybridization to 38S RNA from infected cells and from purified virions. Second, the fraction of the total cellular polyadenylic acid-containing RNA with 5′ sequences was similar to the fraction of RNA containing sequences identical to the extreme 3′ terminus of the genome RNA when calculated by the rate of hybridization of the appropriate complementary DNA probes. This suggests that most intracellular virus-specific RNA molecules contain sequences identical to those present in the 5′-terminal 101 bases of the genome. Third, the size of most of the radioactively labeled DNA complementary to the 5′-terminal 101 bases of the genome remained unchanged after the probe was annealed to either intracellular 38S RNA or to various size classes of subgenomic RNA and the hybrids were digested with S1 nuclease and denatured with alkali. However, after this procedure some DNA fragments of lower molecular weight were present. This was not the case when the DNA complementary to the 5′-terminal 101 bases of the genome was annealed to 38S genome RNA. These results suggest that, although the majority of the intracellular RNA contains the entire 101-base 5′-terminal leader sequence, a small population of virus-specific RNAs exist that contain either a shortened 5′ leader sequence or additional splicing in the terminal 101 bases.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aloni Y., Dhar R., Laub O., Horowitz M., Khoury G. Novel mechanism for RNA maturation: the leader sequences of simian virus 40 mRNA are not transcribed adjacent to the coding sequences. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3686–3690. doi: 10.1073/pnas.74.9.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemon K., Duesberg P., Vogt P. Evidence for crossing-over between avian tumor viruses based on analysis of viral RNAs. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4254–4258. doi: 10.1073/pnas.71.10.4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W., Davidson N. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell. 1976 Apr;7(4):595–607. doi: 10.1016/0092-8674(76)90210-5. [DOI] [PubMed] [Google Scholar]
- Berget S. M., Moore C., Sharp P. A. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billeter M. A., Parsons J. T., Coffin J. M. The nucleotide sequence complexity of avian tumor virus RNA. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3560–3564. doi: 10.1073/pnas.71.9.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Mandel J. L., Chambon P. Ovalbumin gene is split in chicken DNA. Nature. 1977 Nov 24;270(5635):314–319. doi: 10.1038/270314a0. [DOI] [PubMed] [Google Scholar]
- Bronson D. L., Elliott A. Y., Ritzi D. A comparison of four methods used to concentrate Rous sarcoma virus from tissue culture fluids. J Gen Virol. 1976 Dec;33(3):403–410. doi: 10.1099/0022-1317-33-3-403. [DOI] [PubMed] [Google Scholar]
- Bronson D. L., Elliott A. Y., Ritzi D. Concentration of Rous sarcoma virus from tissue culture fluids with polyethylene glycol. Appl Microbiol. 1975 Sep;30(3):464–471. doi: 10.1128/am.30.3.464-471.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L. T., Gelinas R. E., Broker T. R., Roberts R. J. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell. 1977 Sep;12(1):1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- Cordell B., Weiss S. R., Varmus H. E., Bishop J. M. At least 104 nucleotides are transposed from the 5' terminus of the avian sarcoma virus genome to the 5' termini of smaller viral mRNAs. Cell. 1978 Sep;15(1):79–91. doi: 10.1016/0092-8674(78)90084-3. [DOI] [PubMed] [Google Scholar]
- Darlix J. L., Spahr P. F., Bromley P. A., Jaton J. C. In vitro, the major ribosome binding site on Rous sarcoma virus RNA does not contain the nucleotide sequence coding for the N-terminal amino acids of the gag gene product. J Virol. 1979 Feb;29(2):597–611. doi: 10.1128/jvi.29.2.597-611.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich R., Kung H. J., Baker B., Varmus H. E., Goodman H. M., Bishop J. M. Characterization of DNA complementary to nucleotide sequences at the 5'-terminus of the avian sarcoma virus genome. Virology. 1977 Jun 1;79(1):198–215. doi: 10.1016/0042-6822(77)90345-2. [DOI] [PubMed] [Google Scholar]
- Haseltine W. A., Maxam A. M., Gilbert W. Rous sarcoma virus genome is terminally redundant: the 5' sequence. Proc Natl Acad Sci U S A. 1977 Mar;74(3):989–993. doi: 10.1073/pnas.74.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward W. S. Size and genetic content of viral RNAs in avian oncovirus-infected cells. J Virol. 1977 Oct;24(1):47–63. doi: 10.1128/jvi.24.1.47-63.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Flavell R. A. The rabbit beta-globin gene contains a large large insert in the coding sequence. Cell. 1977 Dec;12(4):1097–1108. doi: 10.1016/0092-8674(77)90172-6. [DOI] [PubMed] [Google Scholar]
- Joho R. H., Billeter M. A., Weissmann C. Mapping of biological functions on RNA of avian tumor viruses: location of regions required for transformation and determination of host range. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4772–4776. doi: 10.1073/pnas.72.12.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon P., Duesberg P. H. Subgenomic, cellular Rous sarcoma virus RNAs contain oligonucleotides from the 3' half and the 5' terminus of virion RNA. Nature. 1977 Dec 15;270(5638):631–634. doi: 10.1038/270631a0. [DOI] [PubMed] [Google Scholar]
- Oppermann H., Bishop J. M., Varmus H. E., Levintow L. A joint produce of the genes gag and pol of avian sarcoma virus: a possible precursor of reverse transcriptase. Cell. 1977 Dec;12(4):993–1005. doi: 10.1016/0092-8674(77)90164-7. [DOI] [PubMed] [Google Scholar]
- Pawson T., Harvey R., Smith A. E. The size of Rous sarcoma virus mRNAs active in cell-free translation. Nature. 1977 Aug 4;268(5619):416–420. doi: 10.1038/268416a0. [DOI] [PubMed] [Google Scholar]
- Pawson T., Martin G. S., Smith A. E. Cell-free translation of virion RNA from nondefective and transformation-defective Rous sarcoma viruses. J Virol. 1976 Sep;19(3):950–967. doi: 10.1128/jvi.19.3.950-967.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. E., Zamecnik P. C., Weith H. L. Rous sarcoma virus genome is terminally redundant: the 3' sequence. Proc Natl Acad Sci U S A. 1977 Mar;74(3):994–998. doi: 10.1073/pnas.74.3.994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T. E., Carbon J., Berg P. Construction and analysis of viable deletion mutants of simian virus 40. J Virol. 1976 May;18(2):664–671. doi: 10.1128/jvi.18.2.664-671.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T. E., Rhodes C., Rigby P. W., Berg P. Biochemical method for mapping mutational alterations in DNA with S1 nuclease: the location of deletions and temperature-sensitive mutations in simian virus 40. Proc Natl Acad Sci U S A. 1975 Mar;72(3):989–993. doi: 10.1073/pnas.72.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll E., Billeter M. A., Palmenberg A., Weissmann C. Avian myeloblastosis virus RNA is terminally redundant: implications for the mechanism of retrovirus replication. Cell. 1977 Sep;12(1):57–72. doi: 10.1016/0092-8674(77)90185-4. [DOI] [PubMed] [Google Scholar]
- Stoltzfus C. M., Snyder P. N. Structure of B77 sarcoma virus RNA: stabilization of RNA after packaging. J Virol. 1975 Nov;16(5):1161–1170. doi: 10.1128/jvi.16.5.1161-1170.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal J., Kung H. J., Varmus H. E., Bishop J. M. Characterization of DNA complementary to nucleotide sequences adjacent to poly(A) at the 3'-terminus of the avian sarcoma virus genome. Virology. 1977 Jun 1;79(1):183–197. doi: 10.1016/0042-6822(77)90344-0. [DOI] [PubMed] [Google Scholar]
- Taylor J. M., Illmensee R. Site on the RNA of an avian sarcoma virus at which primer is bound. J Virol. 1975 Sep;16(3):553–558. doi: 10.1128/jvi.16.3.553-558.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walseth T. F., Johnson R. A. The enzymatic preparation of [alpha-(32)P]nucleoside triphosphates, cyclic [32P] AMP, and cyclic [32P] GMP. Biochim Biophys Acta. 1979 Mar 28;562(1):11–31. doi: 10.1016/0005-2787(79)90122-9. [DOI] [PubMed] [Google Scholar]
- Wang L. H., Duesberg P., Beemon K., Vogt P. K. Mapping RNase T1-resistant oligonucleotides of avian tumor virus RNAs: sarcoma-specific oligonucleotides are near the poly(A) end and oligonucleotides common to sarcoma and transformation-defective viruses are at the poly(A) end. J Virol. 1975 Oct;16(4):1051–1070. doi: 10.1128/jvi.16.4.1051-1070.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S. R., Varmus H. E., Bishop J. M. The size and genetic composition of virus-specific RNAs in the cytoplasm of cells producing avian sarcoma-leukosis viruses. Cell. 1977 Dec;12(4):983–992. doi: 10.1016/0092-8674(77)90163-5. [DOI] [PubMed] [Google Scholar]



