Abstract
Psoriasis vulgaris is considered a chronic inflammatory disease, but its immunopathogenesis has not been well understood. The tumor necrosis factor alpha-induced protein 3 (TNFAIP3) gene functions in negative-feedback regulation of inflammation, and its single nucleotide polymorphism is associated with psoriasis. However, the relationship between the expression level of the TNFAIP3 gene in immune cells and psoriasis is not known so far. In the present study, TNFAIP3 mRNA expression levels in peripheral blood mononuclear cells from 44 patients with psoriasis vulgaris and 30 healthy controls were determined using real-time reverse transcription-PCR analysis. We found that expression of TNFAIP3 mRNA in all patients negatively correlated with the psoriatic area and severity index (PASI) (r = −0.5126; P = 0.0004) as well as with the percentage of body surface area affected by psoriasis (r = −0.5013; P = 0.0005). Patients were divided into mild and severe groups based on the mean PASI score. Expression of TNFAIP3 mRNA in the mild group was higher than that in the severe group (P = 0.0064). Moreover, compared with that in healthy controls, the expression of TNFAIP3 mRNA in the mild group was significantly upregulated (P = 0.0004), but the expression of TNFAIP3 mRNA in the severe group was not. These results suggest that the expression level of TNFAIP3 plays an important role in the pathology of psoriasis vulgaris and that the loss of upregulation of TNFAIP3 expression may contribute to the severity of psoriasis vulgaris.
INTRODUCTION
Psoriasis vulgaris is a systemic, immune-mediated disorder characterized by inflammatory skin manifestations and sustained activation of multiple immune cells, especially T cells (9, 18, 19). The activity of inflammation is paralleled with disease severity (12). However, the exact molecular mechanism that sustains inflammation in psoriasis has not been well understood.
Currently, accumulating evidence suggests that an abnormality of immunoregulatory genes be included in the molecular pathogenesis of psoriasis vulgaris. The immunoregulatory genes function in limiting the strength of T cell activation and expansion and include genes encoding inhibitory cytokines (such as transforming growth factor beta [TGF-β] and interleukin-10 [IL-10]), negative regulators of antigen receptor signaling (such as cytotoxic T lymphocyte antigen 4 [CTLA-4]), and repressive transcription factors (such as Foxp3) (22). Many of these genes have been discovered to be involved in the pathogenesis of psoriasis. For instance, serum levels of IL-10 were decreased in psoriasis and negatively correlated with the psoriatic area and severity index (PASI) (23). CTLA-4 is an inhibitory receptor expressed on activated T cells, downregulating T cell activation. Blockade of T lymphocyte costimulation with antibody against CTLA-4 reverses the cellular pathology of psoriatic plaques (1). Foxp3 is a master transcription factor for development and function of Treg cells and is defective in psoriasis (20).
The ubiquitin-editing enzyme tumor necrosis factor alpha-induced protein 3 (TNFAIP3), also known as A20, is a negative immunoregulatory protein. Its function is clearly demonstrated by the phenotype of TNFAIP3-deficient mice, which display cachexia and develop severe multiorgan inflammation causing premature lethality (26). Constitutive expression of TNFAIP3 is restricted in lymphoid tissues such as the thymus and spleen. In immune cells, the TNFAIP3 protein can be induced strongly under inflammatory conditions and acts as a negative-feedback regulator of NF-κB activation. Overexpression of TNFAIP3 can terminate NF-κB signaling transduced from tumor necrosis factor (TNF) receptors, Toll-like receptors, nucleotide-binding oligomerization domain-containing 2 (NOD2) receptors, and T cell receptors (8, 13). The TNFAIP3 gene can restrict activation of T cells by directly inhibiting NF-κB activation in T cells (8) or downregulating the T cell stimulatory capacity of dendritic cells (21).
TNFAIP3's NF-κB-inhibiting activities depend on its ubiquitin-editing function. The N terminus has deubiquitinating activity and can inhibit NF-κB signaling by removing K63 polyubiquitination chains from specific NF-κB signaling molecules. The C-terminal zinc finger-containing domain possesses E3 ubiquitin ligase activity and can turn off NF-κB activation by promoting K48 polyubiquitination of specific NF-κB signaling molecules, followed by proteasome-mediated degradation (7, 25, 27).
Genetic studies have revealed a crucial link between polymorphisms in the TNFAIP3 gene locus and psoriasis (24). However, the relationship between the expression level of TNFAIP3 and psoriasis is still unclear. Hence, we examined expression of TNFAIP3 mRNA in peripheral blood mononuclear cells (PBMC) from patients with psoriasis vulgaris and analyzed the correlation between its expression level and disease severity. We also examined the expression of TNFAIP3 mRNA in healthy individuals and compared it with the TNFAIP3 expression level in patients with mild or severe disease.
MATERIALS AND METHODS
Human subjects.
Forty-four patients with psoriasis vulgaris were enrolled in this study. Twenty patients were in the progressive stage, with new psoriatic lesions presenting themselves. Twenty-four patients were in the stationary stage, with lesions stopping growth. Psoriasis severity can be measured by the amount of skin affected by psoriasis (11). The percentage of body surface area (%BSA) affected by psoriasis was calculated for all patients by the same physician, who was blinded to the A20 mRNA results. Another index used to measure the disease severity was the PASI. In the PASI scoring system, the areas of psoriatic involvement of the head (H), trunk (T), upper limbs (U), and lower limbs (L) are given numerical values from 0 to 6, and for each area, erythema (E), infiltration (I), and desquamation (D) are assessed on a scale of 0 to 4. The PASI is calculated by use of the following formula: PASI = 0.1(EH + IH + DH)AH + 0.3(ET + IT + DT)AT + 0.2(EU + IU + DU)AU + 0.4(EL + IL + DL)AL (11). The PASI was evaluated for all patients by the same physician, who was blinded to the A20 mRNA results. The patients were divided into a mild group and a severe group based on the mean PASI score. A patient who had a PASI score lower than the mean value was classified into the mild group. A patient who had a PASI score equal to or higher than the mean was classified into the severe group. Thirty sex- and age-matched healthy controls were also recruited. In order to exclude the influence of immunosuppressive drugs on TNFAIP3 expression, peripheral blood was sampled from all patients before they took any immunosuppressive drug. All of the blood samples from the patients and healthy controls were used with informed consent and approval from the Ethics Committee of Shandong University, China. The characteristics of the patients and healthy subjects are summarized in Table 1.
Table 1.
Characteristics of the studied subjects
| Characteristic | Valuea |
|
|---|---|---|
| Psoriasis patients (n = 44) | Healthy controls (n = 30) | |
| No. (%) of females | 17 (38.6) | 11 (36.7) |
| No. (%) of males | 27 (61.4) | 19 (63.3) |
| Mean age (range) (yr) | 31 (8–67) | 28 (15–58) |
| No. (%) of individuals with psoriasis type | ||
| Plaque type | 26 (59.1) | 0 |
| Guttate type | 18 (40.9) | 0 |
| No. (%) of individuals with psoriasis stage | ||
| Stationary | 24 (54.5) | 0 |
| Progressive | 20 (45.5) | 0 |
| Mean %BSA (range) | 6.34 (0.5–38) | |
| Mean PASI (range) | 4.84 (0.2–18) | |
| Mean total lesion area (range) (cm2) | 8.5 (1–34) | |
| Cell count (109/liter) | ||
| White blood cells | 7.17 ± 2.17 | |
| Red blood cells | 4.59 ± 0.58 | |
| Platelets | 231 ± 54.8 | |
| Cell % | ||
| Neutrophils | 60.95 ± 7.82 | |
| Lymphocytes | 30.05 ± 6.49 | |
| Monocytes | 6.13 ± 2.58 | |
| Eosinophils | 1.99 ± 2.21 | |
| Basophils | 0.061 ± 0.075 | |
| ESR (mm/h) | 6.27 ± 6.07 | |
| No. (%) of individuals | ||
| RF negative | 44 (100.0) | |
| RF positive | 0 (0) | |
| ASO negative | 27 (61.4) | |
| ASO positive | 17 (38.6) | |
Unless stated otherwise, values are means ± standard deviations.
Laboratory assessments.
For the psoriasis patients, laboratory parameters included full blood counts performed on a Coulter LH 750 hematology analyzer (Beckman Coulter, Miami, FL), determination of the erythrocyte sedimentation rate (ESR) using the Westergren test, and detection of rheumatoid factor (RF) and antistreptolysin O (ASO) by the latex fixation test.
RNA and cDNA preparation from PBMC.
PBMC were separated by density gradient centrifugation from peripheral blood anticoagulated with sodium citrate. Total RNA was extracted from PBMC (5 × 105) by use of TRIzol (Invitrogen, CA) and quantified by photometric measurement. One microgram of RNA was reverse transcribed to cDNA by use of a reverse transcription system kit (Promega, WI).
Quantitative real-time PCR and standard PCR.
The expression of TNFAIP3 mRNA was evaluated by quantitative real-time PCR. The level of β-actin mRNA was also detected as an internal control for each sample. Primers used in real-time PCR were as follows: TNFAIP3 forward, 5′-CGTCCAGGTTCCAGAACACCATTC-3′; TNFAIP3 reverse, 5′-TGCGCTGGCTCGATCTCAGTTG-3′; β-actin forward, 5′-GACTACCTCATGAAGATCCTCACC-3′; and β-actin reverse, 5′-TCTCCTTAATGTCACGCACGATT-3′. The primers were designed to be exon-exon junction spanning to increase the specificity for mRNA rather than genomic DNA and were confirmed via nucleotide BLAST searches (NCBI).
Real-time PCR was performed in an ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA), using a SYBR green I real-time PCR kit in accordance with the instructions of the manufacturer (TaKaRa, Dalian, China). The kit consisted of ROX reference dye (50×) and a master mix (2×) including hot-starting Taq enzyme, a deoxynucleoside triphosphate (dNTP) mixture, Mg2+, and SYBR green I dye. Each real-time PCR mixture contained 10 μl of 2× master mix, 2.0 μl of forward and reverse primers (10 μM concentration of each), 0.4 μl of 50× ROX reference dye, 2.0 μl of sample cDNA, and 5.6 μl of nuclease-free water, to make a total volume of 20 μl. Each sample was run in triplicate with a two-step PCR protocol (95°C for 10 s followed by 40 cycles of 95°C for 5 s and 60°C for 40 s). All of the PCR products were run in an agarose gel and were confined to a single band of the expected size. A melting curve analysis was also performed, and a single peak was present, confirming the specificity of the products. The expression of the TNFAIP3 gene was normalized to that of β-actin and determined using the comparative 2−ΔΔCT method, which is a relative quantification method.
To confirm the results of the quantitative PCR assay, standard PCRs to amplify the TNFAIP3 gene from PBMC from some subjects were performed with primers different from those used in real-time PCR. The following sequence-specific primers were used for standard PCR amplification: (i) for the internal control β-actin gene, 5′-AGGCCAACCGCGAGAAGATG-3′ (forward) and 5′-CACACGGAGTACTTGCGCTCAG-3′; and (ii) for the TNFAIP3 gene, 5′-TCAAGCTGCACGGACTCCTG-3′ and 5′-GACCCACCTGTTTCCGGTTAG-3′. Reverse transcription-PCR (RT-PCR) analysis of β-actin was performed with 26 cycles of 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C, followed by 5 min at 72°C. The product was 682 bp. RT-PCR analysis of TNFAIP3 was performed with 30 cycles of 30 s at 94°C, 30 s at 56°C, and 30 s at 72°C, followed by 5 min at 72°C. The product was 615 bp. The PCR products were separated electrophoretically in a 1.5% agarose gel and visualized by ethidium bromide staining.
Statistical analysis.
Data are presented as means ± standard deviations. The Mann-Whitney test was used to analyze the difference in TNFAIP3 mRNA levels between subject groups. Correlation analysis was carried out using Spearman's rank test. A probability level of 0.05 was considered to indicate a significant difference. All analyses were performed with GraphPad Prism software, version 5.0.
RESULTS
Laboratory measurements of patients with psoriasis vulgaris.
The demographic characteristics, clinical manifestations, and laboratory measurements of patients are presented in Table 1. The mean %BSA affected was 6.34%, with a range of 0.5% to 38%, while the mean PASI value was 4.84, with a range of 0.2 to 18. Blood cell counts showed that the numbers of red blood cells, white blood cells, neutrophils, basophils, and eosinophils were within their normal ranges for all patients. Two patients (4.5%) had more lymphocytes and monocytes than would fall within the normal ranges, and three patients (6.8%) had platelets beyond the normal level. The range of ESRs was from 0 to 20 mm/h, and the mean was 6.27 mm/h. No patient tested positive for RF. A total of 38.6% of patients tested positive for ASO.
Analysis of relationships between TNFAIP3 mRNA expression and characteristics or laboratory parameters for patients with psoriasis vulgaris.
We examined the expression of TNFAIP3 mRNA in PBMC from 44 patients with psoriasis by using real-time RT-PCR and analyzed the association of TNFAIP3 expression with demographic characteristics, clinical manifestations, and laboratory parameters. The results revealed that TNFAIP3 mRNA expression levels negatively correlated with the %BSA (r = −0.5013; P = 0.0005) and PASI (r = −0.5126; P = 0.0004) (Fig. 1 and 2). No statistically significant relationships were found between TNFAIP3 mRNA expression levels and other characteristics, clinical manifestations, or laboratory parameters for the patients with psoriasis vulgaris.
Fig 1.
Negative correlation between TNFAIP3 mRNA expression levels and %BSA for all patients with psoriasis vulgaris (n = 44).
Fig 2.
Negative correlation between TNFAIP3 mRNA expression levels and PASI for all patients with psoriasis vulgaris (n = 44).
Comparison of TNFAIP3 mRNA expression in PBMC from psoriasis patients in the mild or severe group and those from healthy controls.
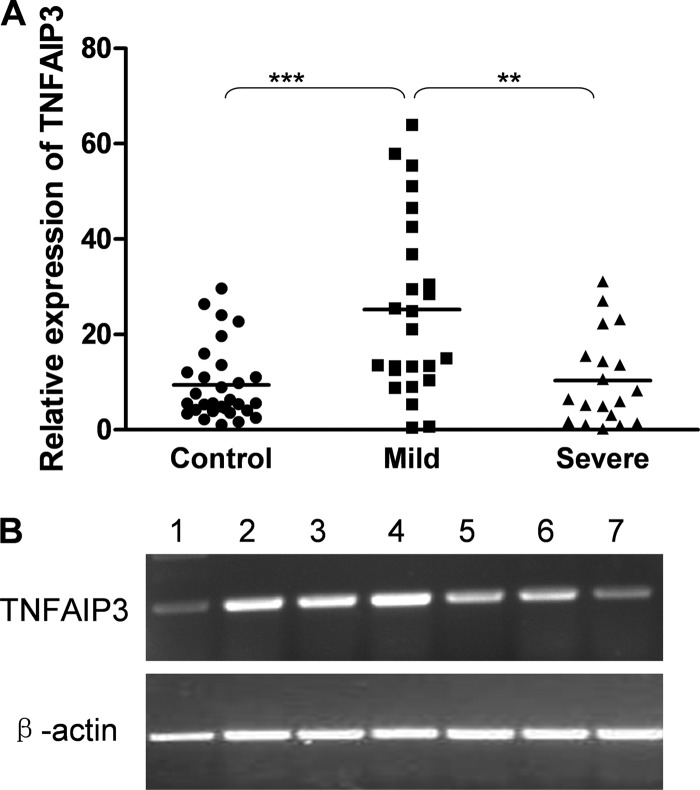
We examined the expression of TNFAIP3 mRNA in PBMC from 30 healthy control individuals by using real-time RT-PCR. Psoriasis patients were divided into a mild group (n = 25) and a severe group (n = 19) based on the mean PASI score (4.84). The mean TNFAIP3 mRNA expression level in the mild group (25.22) was significantly upregulated compared with those for healthy controls (9.45) (P = 0.0004) and the severe group (10.36) (P = 0.0064). There was no significant difference in TNFAIP3 expression between the severe group and healthy controls (P = 0.9591). The TNFAIP3 expression of some subjects was analyzed by standard PCR. The direction of expression was consistent in the standard PCR experiment and the real-time PCR assay (Fig. 3).
Fig 3.
TNFAIP3 mRNA expression levels in PBMC from patients with psoriasis vulgaris and in those from healthy controls. (A) Real-time RT-PCR was performed to quantify expression of the TNFAIP3 gene in PBMC from patients with mild psoriasis (n = 25) and severe psoriasis (n = 19), as well as in those from healthy controls (n = 30). Horizontal lines indicate means (9.45 for the control group, 25.22 for the mild group, and 10.36 for the severe group). TNFAIP3 mRNA expression levels in the mild group were significantly upregulated compared with those for healthy controls and the severe group. There was no significant difference in TNFAIP3 expression between the severe group and healthy controls. (B) Standard RT-PCR was performed to quantify expression of the TNFAIP3 gene in PBMC from healthy controls (lanes 1 and 2) and patients with mild psoriasis (lanes 3 and 4) or severe psoriasis (lanes 5, 6, and 7). The expression level of TNFAIP3 was consistent with that resulting from real-time PCR. **, P < 0.01; ***, P < 0.001.
DISCUSSION
Psoriasis is an inflammatory disease associated with dysfunction of multiple immunoregulatory genes. The TNFAIP3 gene plays a crucial role in limiting inflammation mediated by immune cells (8, 21). In this study, we examined the expression level of the TNFAIP3 gene in PBMC from patients with psoriasis vulgaris and found that it was negatively correlated with disease severity. Compared to that in healthy controls, the expression of TNFAIP3 was upregulated in patients with mild disease but not in patients with severe disease. These results suggest that the expression level of the TNFAIP3 gene plays a vital role in the pathology of psoriasis vulgaris and that a loss of upregulation may lead to severe psoriasis vulgaris.
The expression level of the TNFAIP3 gene may take part directly in the pathological mechanism of psoriasis vulgaris. The severity of psoriasis can be determined by the %BSA and the PASI. The %BSA describes the lesion area, while the PASI takes into account the area coverage and lesion appearance (11). The expression of TNFAIP3 mRNA reversely correlated with the PASI as well as with the %BSA. This means that the expression level of the TNFAIP3 gene may be involved in the severity of disease. The expression level of TNFAIP3 was significantly lower in severe disease than in mild disease. This suggests that low expression of TNFAIP3 in psoriasis may affect the protein's function such that the inflammation cannot be restricted and results in severe psoriasis vulgaris.
The low TNFAIP3 expression level in severe psoriasis may be caused by a loss or inefficiency of expression upregulation. An upregulation of TNFAIP3 expression was induced in patients with mild psoriasis vulgaris compared with that in healthy controls. This upregulation may have been due to the chronic inflammation existing in psoriasis patients. Persistent inflammation in psoriasis is characterized by elevated serum levels of multiple proinflammatory cytokines, including TNF-α, gamma interferon (IFN-γ), IL-1, IL-2, IL-6, IL-7, IL-8, IL-12, IL-17, and IL-18 (3, 4). Some proinflammatory cytokines, such as TNF-α, IL-1, and IL-17, can stimulate PBMC, leading to activation of NF-κB (8, 28). The activated NF-κB is translocated into the nucleus to bind to kappa B elements on the TNFAIP3 promoter, enhancing transcription of the TNFAIP30 gene (14). Therefore, sustained inflammation accounts for the increased TNFAIP3 expression in PBMC of patients in the mild group. But TNFAIP3 expression was not upregulated in the severe group compared to healthy controls, although the proinflammatory cytokines are also elevated in patients with severe psoriasis (4, 16). The loss of upregulation may contribute to the inefficiency of TNFAIP3 expression and affect its function in limiting inflammation, resulting in severe disease.
The loss of upregulation of TNFAIP3 expression in severe psoriasis vulgaris can be explained by a polymorphism or promoter methylation of the TNFAIP3 gene. Some single nucleotide polymorphisms (SNPs) can influence expression of the TNFAIP3 gene, such as two intronic SNPs (rs610604 and rs5029930) associated with lower levels of gene expression (5, 15, 17). Additionally, a haplotype (TT > A) resulting in reduced TNFAIP3 expression was recently identified in a conserved regulatory region downstream of TNFAIP3 (2). Therefore, the loss of upregulation of TNFAIP3 expression may be attributed partially to polymorphism of the TNFAIP3 gene. The other factor affecting expression of the TNFAIP3 gene is promoter methylation. TNFAIP3 is targeted by promoter methylation in some hematological malignancies (6, 10). Thus, the inefficient expression of the TNFAIP3 gene in severe psoriasis vulgaris may be due in part to its aberrant methylation. This remains to be investigated further.
This study is limited by the sole use of a real-time RT-PCR method for measurement of TNFAIP3 expression and lacks protein data and data from local psoriatic tissues. Assessment of TNFAIP3 expression in subsets of peripheral blood mononuclear cells, such as CD4+ T cells, may improve the negative correlation between TNFAIP3 expression and the disease severity of psoriasis vulgaris, since psoriasis is considered to be a chronic T cell-mediated inflammatory disease (9, 18) and because the TNFAIP3 gene functions in limitation of T cell activation (8). Therefore, quantification of the TNFAIP3 protein by flow cytometry will be an emphasis in our future research work.
In summary, we report that TNFAIP3 expression in PBMC from patients with psoriasis vulgaris is reversely correlated with disease activity. Compared to healthy controls, patients with mild disease had an upregulation of TNFAIP3 expression, while those with severe disease did not. Our study suggests that the expression level of the TNFAIP3 gene is directly involved in the pathological mechanisms of psoriasis and that a loss of upregulation of TNFAIP3 expression in immune cells may induce severe disease. These data support the hypothesis that the TNFAIP3 gene may be a target for therapy of psoriasis disease.
ACKNOWLEDGMENTS
This work was supported by grants from the National Nature Science Foundation of China (grants 81071705 and 30500591), the Award Funds for Excellent Young and Middle-Aged Scientists of Shandong Province (grant BS2009YY002), and the National 973 Program of China (grant 2011CB503906).
Footnotes
Published ahead of print 10 October 2012
REFERENCES
- 1. Abrams JR, et al. 2000. Blockade of T lymphocyte costimulation with cytotoxic T lymphocyte-associated antigen 4-immunoglobulin (CTLA4Ig) reverses the cellular pathology of psoriatic plaques, including the activation of keratinocytes, dendritic cells, and endothelial cells. J. Exp. Med. 192: 681– 694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adrianto I, et al. 2011. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat. Genet. 43: 253– 258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson KS, et al. 2010. Elevation of serum epidermal growth factor and interleukin 1 receptor antagonist in active psoriasis vulgaris. Br. J. Dermatol. 163: 1085– 1089 [DOI] [PubMed] [Google Scholar]
- 4. Arican O, Aral M, Sasmaz S, Ciragil P. 2005. Serum levels of TNF-alpha, IFN-gamma, IL-6, IL-8, IL-12, IL-17, and IL-18 in patients with active psoriasis and correlation with disease severity. Mediators Inflamm. 2005: 273– 279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boonyasrisawat W, et al. 2007. Tag polymorphisms at the A20 (TNFAIP3) locus are associated with lower gene expression and increased risk of coronary artery disease in type 2 diabetes. Diabetes 56: 499– 505 [DOI] [PubMed] [Google Scholar]
- 6. Chanudet E, et al. 2010. A20 is targeted by promoter methylation, deletion and inactivating mutation in MALT lymphoma. Leukemia 24: 483– 487 [DOI] [PubMed] [Google Scholar]
- 7. Coornaert B, Carpentier I, Beyaert R. 2009. A20: central gatekeeper in inflammation and immunity. J. Biol. Chem. 284: 8217– 8221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Düwel M, et al. 2009. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. J. Immunol. 182: 7718– 7728 [DOI] [PubMed] [Google Scholar]
- 9. Elder JT. 2009. Genome-wide association scan yields new insights into the immunopathogenesis of psoriasis. Genes Immun. 10: 201– 209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frenzel LP, et al. 2011. Sustained NF-kappaB activity in chronic lymphocytic leukemia is independent of genetic and epigenetic alterations in the TNFAIP3 (A20) locus. Int. J. Cancer 128: 2495– 2500 [DOI] [PubMed] [Google Scholar]
- 11. Henseler T, Schmitt-Rau K. 2008. A comparison between BSA, PASI, PLASI and SAPASI as measures of disease severity and improvement by therapy in patients with psoriasis. Int. J. Dermatol. 47: 1019– 1023 [DOI] [PubMed] [Google Scholar]
- 12. Kanelleas A, et al. 2011. The role of inflammatory markers in assessing disease severity and response to treatment in patients with psoriasis treated with etanercept. Clin. Exp. Dermatol. 36: 845– 850 [DOI] [PubMed] [Google Scholar]
- 13. Kingeter LM, Paul S, Maynard SK, Cartwright NG, Schaefer BC. 2010. Cutting edge: TCR ligation triggers digital activation of NF-kappaB. J. Immunol. 185: 4520– 4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krikos A, Laherty CD, Dixit VM. 1992. Transcriptional activation of the tumor necrosis factor alpha-inducible zinc finger protein, A20, is mediated by kappa B elements. J. Biol. Chem. 267: 17971– 17976 [PubMed] [Google Scholar]
- 15. Lodolce JP, et al. 2010. African-derived genetic polymorphisms in TNFAIP3 mediate risk for autoimmunity. J. Immunol. 184: 7001– 7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mizutani H, Ohmoto Y, Mizutani T, Murata M, Shimizu M. 1997. Role of increased production of monocytes TNF-alpha, IL-1beta and IL-6 in psoriasis: relation to focal infection, disease activity and responses to treatments. J. Dermatol. Sci. 14: 145– 153 [DOI] [PubMed] [Google Scholar]
- 17. Nair RP, et al. 2009. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat. Genet. 41: 199– 204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nograles KE, Davidovici B, Krueger JG. 2010. New insights in the immunologic basis of psoriasis. Semin. Cutan. Med. Surg. 29: 3– 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reich K. 2012. The concept of psoriasis as a systemic inflammation: implications for disease management. J. Eur. Acad. Dermatol. Venereol. 26(Suppl 2):3–11 [DOI] [PubMed] [Google Scholar]
- 20. Song QH, et al. 2012. An association study of single nucleotide polymorphisms of the FOXP3 intron-1 and the risk of psoriasis vulgaris. Indian J. Biochem. Biophys. 49: 25– 35 [PubMed] [Google Scholar]
- 21. Song XT, et al. 2008. A20 is an antigen presentation attenuator, and its inhibition overcomes regulatory T cell-mediated suppression. Nat. Med. 14: 258– 265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun H, et al. 2008. TIPE2, a negative regulator of innate and adaptive immunity that maintains immune homeostasis. Cell 133: 415– 426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takahashi H, Tsuji H, Hashimoto Y, Ishida-Yamamoto A, Iizuka H. 2010. Serum cytokines and growth factor levels in Japanese patients with psoriasis. Clin. Exp. Dermatol. 35: 645– 649 [DOI] [PubMed] [Google Scholar]
- 24. Vereecke L, Beyaert R, van Loo G. 2011. Genetic relationships between A20/TNFAIP3, chronic inflammation and autoimmune disease. Biochem. Soc. Trans. 39: 1086– 1091 [DOI] [PubMed] [Google Scholar]
- 25. Vereecke L, Beyaert R, van Loo G. 2009. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol. 30: 383– 391 [DOI] [PubMed] [Google Scholar]
- 26. Vereecke L, et al. 2010. Enterocyte-specific A20 deficiency sensitizes to tumor necrosis factor-induced toxicity and experimental colitis. J. Exp. Med. 207: 1513– 1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Verstrepen L, et al. 2010. Expression, biological activities and mechanisms of action of A20 (TNFAIP3). Biochem. Pharmacol. 80: 2009– 2020 [DOI] [PubMed] [Google Scholar]
- 28. Xie S, et al. 2010. IL-17 activates the canonical NF-kappaB signaling pathway in autoimmune B cells of BXD2 mice to upregulate the expression of regulators of G-protein signaling 16. J. Immunol. 184: 2289– 2296 [DOI] [PMC free article] [PubMed] [Google Scholar]