Abstract
In 2010, a large outbreak of dengue occurred in Santos, Brazil. The detection of the NS1 antigen was used for diagnosis in addition to the detection of IgG, IgM, and RNA. A large number of NS1 false-negative results were obtained. A total of 379 RNA-positive samples were selected for thorough evaluation. NS1 was reactive in 37.7% of cases. Most of the cases were characterized as a secondary infection by dengue 2 virus. Sequencing of NS1 positive and negative isolates did not reveal any mutation that could justify the diagnostic failure. Use of existing NS1 tests in the Brazilian population may present a low negative predictive value, and they should be used with caution, preferentially after performing a validation with samples freshly obtained during the ongoing epidemic.
INTRODUCTION
Dengue is a mosquito-borne viral infection found in tropical and subtropical regions around the world, following the geographical distribution of its vector, Aedes aegypti. There are four distinct serotypes, named dengue virus serotypes 1 to 4 (DENV-1 to DENV-4), all able to cause the same disease. Their genomic RNA is a single-stranded molecule of 11 kb in length, coding for three structural proteins, the nucleocapsid or core protein (C), a membrane-associated protein (M), and the envelope protein (E), and seven nonstructural (NS) proteins, NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5 (4).
WHO currently estimates a yearly incidence of 50 million to 100 million infections worldwide (25). The disease is now endemic in more than 100 countries in Africa, the Americas, the eastern Mediterranean, Southeast Asia, and the western Pacific, the last two regions being most seriously affected (19). Noticeably, dengue incidence is growing in the Americas with 1.1 million cases reported in 2011, of which 20,000 cases were classified as severe dengue. Markedly, Brazil is responsible for the heaviest burden, reporting 764,032 cases in this period (23). In fact, dengue is a priority health issue in Brazil, draining an enormous amount of resources that may allow temporary and limited control of outbreaks (23). Given that vector eradication seems an unreachable target (2), a great expectation is being deposited on the development of a vaccine (5).
Since, so far, there are no licensed vaccines or specific therapies for dengue, patient management relies on early diagnosis and good supportive care. Traditionally, a dengue diagnosis was made by cell culture isolation methods, which are laborious, time-consuming, and expensive and inappropriate for the management of individuals who are suspected of having dengue infection during outbreaks. More recently, high-throughput enzymatic immunoassays (EIAs) toward the detection of antibodies or antigens using a microplate format were adopted worldwide (12). In Brazil, the confirmatory algorithm relies on a 4-fold increase of IgG or IgM titers in paired serum samples collected at the onset of symptoms and 2 weeks after the initial symptoms or IgM detection on a sample taken at 7 or more days after the onset of the disease. This algorithm is an important surveillance tool but does not allow diagnosis to be made during the symptomatic phase, when most patients seek medical assistance (24).
In the last decade, dengue nonstructural protein 1 (NS1), produced in both membrane-associated and secreted forms, has been utilized as an early marker, since it is present in serum in between 1 and 9 days after the onset of clinical signs, with a peak from 3 to 5 days (1, 23, 26). The amount of secreted NS1 (sNS1) in the serum of individuals infected with DENV has been shown to directly correlate with viremia and the pathogenesis of dengue infection (10, 15).
The study of Guzman et al. evaluated well-characterized samples from many different countries by two NS1 assays. They observed an excellent specificity (100%) for both, whereas sensitivity was quite low (52 to 66%), according to the day the sample was drawn and the geographical region of the patient, but not disease severity (9). Studies performed in Brazil have also observed an excellent specificity but variable sensitivity of NS1 tests. While the work of Castro-Jorge and coworkers (3) reported a performance of Platelia (Bio-Rad) superior to real-time PCR (RT-PCR), Lima et al. verified a lower sensitivity by analyzing a large number of stored serum samples from confirmed cases (16). Of concern, the decreased sensitivity was particularly evident for samples containing DENV-3 (65 to 86%). The diminished sensitivity of NS1 kits was observed also for samples harboring DENV-2, from both Latin American (Nicaragua and Venezuela) and Asiatic (Vietnam and Thailand) countries (9). Corroborating these data, a kinetic study of NS1 secretion in the serum of Vietnamese children detected a significant reduction on the amount of NS1 according to dengue serotype and immune status (primary versus secondary dengue) (7). In general, the NS1 detection rate is higher in patients with primary dengue than those with secondary dengue. A possible basis for reduced sensitivity in secondary dengue is that NS1, along with other viral antigens, is sequestered in immunocomplexes when a substantial level of DENV-reactive IgG is present (13).
Santos is the largest city of the São Paulo state coastal area, with approximately 500,000 inhabitants. Along with other neighboring municipalities, it receives thousands of visitors from the city of São Paulo and inland every weekend. In 2002, the first large dengue outbreak hit the region, with approximately 30,000 confirmed cases, mainly caused by DENV-3, but with cocirculation of DENV-1 and -2 as well. In 2010, another outbreak occurred in the area, this time the vast majority of the confirmed 35,000 cases were attributed to DENV-2 (17), although DENV-3 and -1 were rarely detected (18). This last epidemic was characterized by many cases of severe disease, with associated dengue hemorrhagic fever (DHF) and several deaths. Our laboratory provided support to one large hospital in Santos (Hospital Ana Costa). During routine analysis of the samples, we observed discrepancies between molecular tests (IgM and NS1 assays).
In this work, we report the clinical performance of NS1 kits in comparison to other markers such as IgM antibodies and viral RNA detection in serum samples of patients with confirmed dengue from Santos, São Paulo, Brazil.
MATERIALS AND METHODS
Patients and samples.
Samples were drawn between February and July of 2010, when an outbreak of DENV-2 occurred in the city of Santos. Patients with symptoms came to the Hospital Ana Costa emergency unit for attendance. For the purpose of this validation study, 379 samples harboring dengue RNA, evidenced by real-time PCR as described below, were selected. The day of fever and other clinic epidemiological data were collected from the patient chart.
In order to determine whether different epidemiological conditions like dengue serotype or dengue immunological status could interfere with the NS1 test performance during this epidemic, two other series of samples with laboratory-confirmed acute dengue were submitted to the same NS1 testing, both consisting of stored sera from previous outbreaks in Santos in 2002 (n = 82) and Goiânia in 2005 (n = 164) (8).
IgM detection.
We used two commercial tests for the detection of IgM antibodies. In 232 samples, the Duo test kit (Bio Diagnostics, Inc.) was used, which is a qualitative immunoassay for the simultaneous detection of both the antigen NS1 and IgG/IgM in serum, plasma, or whole blood. In the remaining 147 samples, the dengue IgM capture enzyme-linked immunosorbent assay (ELISA) kit (Panbio) was used.
IgG avidity.
The IgG avidity test was performed on acute- and convalescent-phase samples that were IgG positive by use of an in-house ELISA. In brief, antigens were prepared from Aedes albopictus C6/36 cells infected with DENV-1 to -4 and disrupted by sonication. The avidity index, expressed as a percentage, was calculated by determining the ratio of optical density with a 6 M urea treatment to the optical density without urea and then multiplying that value by 100 (6). A receiver-operating characteristic curve analysis performed using Analyse-it software (version 1.73) was used to evaluate the ability of the avidity test to distinguish between primary and secondary dengue infections. The cutoff point was defined as the highest sum of the estimates of sensitivity and specificity. Secondary infection was defined by an IgG avidity index cutoff point of 30%.
NS1 detection.
The Platelia dengue NS1 Ag kit (Bio-Rad) was employed according to manufacturer's instructions. This test is licensed for the qualitative or semiquantitative detection of the dengue NS1 antigen in human serum or plasma, in a sandwich format, microplate enzyme immunoassay.
Real-time PCR.
Dengue RNA from all 4 serotypes was detected and quantified by an in-house real-time PCR method. RNA was extracted from 140 μl of plasma using the Qiagen viral RNA kit. All RT-PCRs were performed in duplicate, with an input of 7.5 μl of an RNA template in a final reaction volume of 10 μl. Amplification was carried out by employing SuperScript III Platinum SYBR green one-step quantitative reverse transcriptase PCR (qRT-PCR) with the ROX kit (Invitrogen, Inc., EUA) and pan-dengue primers (11), covering all 4 serotypes, at 0.4 μM. Cycling conditions were as follows: a 10-min reverse transcription step at 60°C and then 1 min for Taq polymerase activation at 95°C, followed by 45 cycles of PCR at 95°C without holding time (denaturation), 60°C for 3 s (annealing), and 72°C for 10 s (extension), run on an ABI 7300 real-time PCR system (Applied Biosystems, Brazil). Bovine diarrhea virus (BVDV), a flavivirus, was grown in the bovine kidney cell line MDBK, and the supernatant was used as an internal control. It was added to the samples before extraction and also submitted to a parallel real-time PCR assay in order to control RNA extraction and reverse transcription. The supernatant from DENV-3 cell cultures was included as an external control in every RT-PCR run. The positive controls (DENV-3) were isolated from the Aedes albopictus mosquito cell line C6/36. The DENV-3 supernatant was previously quantified by a commercial dengue real-time PCR kit (RealArt; Artus/Qiagen, Germany) (14) and used to generate a standard curve. The detection limit of this assay was determined by probit analysis on the quantified DENV-3 standard and estimated to be 100 copies/ml (95% limit of detection [LOD]). Dengue serotypes were ascribed by using a multiplex PCR generating different-molecular-weight fragments according to the dengue serotype, as described previously (11).
Multiplex PCR for dengue genotyping. (i) RNA extraction.
RNA was extracted in duplicate from 140 μl of plasma by employing a viral RNA kit (Qiagen, Germany). Elution was performed in 60 μl according to the manufacturer's instructions.
(ii) cDNA synthesis and multiplex RT-PCR.
cDNA was synthesized from 22 μl of RNA, extracted as described above, plus 2.5 μM random hexamers (6-mer; Amersham, Brazil), 1 mM dithiothreitol, 1 U/μl RNase inhibitor (Invitrogen, Brazil), and 2.5 U of Moloney murine leukemia virus RT (Invitrogen, Brazil). This mixture was incubated for 5 min at 65°C and then for 30 min at 37°C, and RT was inactivated by a final incubation of 5 min at 95°C. PCR was performed as described elsewhere (8). Briefly, 5 μl of cDNA was added to 20 μl of a PCR mixture consisting of the primers D1, TS1, and TS2 at 0.5 μM and the primers TS3 and TS4 at 0.125 μM, 3 mM MgCl2, 1× PCR buffer, 200 μM deoxynucleoside triphosphates, and 1.25 U of Platinum Taq polymerase (Invitrogen, São Paulo, Brazil). The thermocycler (Mastercycler gradient; Eppendorf, Hamburg, Germany) profile was as follows: 94°C for 2 min, 40 cycles of 94°C for 45 s, 55°C for 1 min, and 72°C for 1 min, followed by a final extension at 72°C for 7 min. Fragments of different lengths were obtained from dengue virus serotypes as follows: 482 bp for DENV-1, 119 bp for DENV-2, 290 bp for DENV-3, and 389 bp for DENV-4. Total RNAs extracted from C6/36 cell cultures infected with the respective dengue virus serotypes were used as controls. PCR products were run on 2% agarose gels, stained with ethidium bromide, and documented on a UV apparatus.
(iii) PCR amplification and sequencing.
A fragment of 1,030 bp from the envelope region, corresponding to nucleotides 2400 to 3430 of the DENV-2 complete genome (GenBank reference sequence GQ199890), was amplified by PCR using the following specific primers: D2 NS1_2420_F, 5′GATAGTGGTTGCGTTGTGAGCTG3′, and D2_NS1_3450_R, 5′GGCTGTGACCARRGARYTGACCARAT3′. The thermal profile for amplification was 38 cycles of 45 s of denaturation at 94°C, 1 min of annealing at 56°C, and 1 min of extension at 72°C. PCR products were purified for elimination of primer dimers and excess oligonucleotides with the PureLink quick gel extraction kit (Invitrogen), with few adaptations for direct purification of PCR products in solution. Sequencing reactions were performed with the BigDye terminator kit version 3.0 (Applied Biosystems, Bedford, MA). The electropherograms were analyzed, and consensus sequences were generated with CodonCode Aligner version 3.0.
RESULTS
Of the 379 patients, 266 (70%) were classified as dengue without warning signs, 94 (25%) as dengue with warning signs, and 19 (5%) as severe dengue. NS1 was positive in 37.7% (143/379) of patients and IgM in 53.5% (203/379) of patients as depicted in Table 1. Of the 143 NS1-positive patients, 142/379 (37.4%) were positive for Platelia and 50/232 (21.5%) for Duo NS1. In these samples, the concordance between Platelia and Duo NS1 was 79.3% (184/232), with 48/232 positive and 136/232 negative for both.
Table 1.
Results of NS1 and IgM tests according to the days since the onset of symptoms in 379 dengue RNA-positive samples from the Santos 2010 outbreak
| Days since the onset of symptoms | No. of NS1-positive samples/total no. of total samples (%) | No. of IgM-positive samples/total no. of samples (%) |
|---|---|---|
| 1 | 10/24 (42) | 6/24 (25) |
| 2 | 34/63 (54) | 17/63 (27) |
| 3 | 47/91 (52) | 29/91 (32) |
| 4 | 23/62 (37) | 38/62 (61) |
| 5 | 9/44 (20) | 32/44 (73) |
| 6 | 7/38 (18) | 35/38 (92) |
| 7 | 9/24 (38) | 18/24 (75) |
| >8 | 4/33 (12) | 28/33 (85) |
Analyzing NS1 positivity according to the days since the onset of symptoms, positive results were obtained since the first day, with the best performance peaking in 3 days of symptoms. From day 4 on, a decrease in the detection of NS1 was observed. IgM antibodies were detected from the first day of symptoms, having the best performance on the sixth day of symptoms. These data are shown in Table 1.
The IgG avidity index allowed the classification of these samples into primary (13/379) or secondary dengue (260/379), while in 106 samples, the absence of dengue-specific IgG in the acute-phase serum sample precluded the performance of the IgG avidity test. However, 35 out of those 106 were classified as primary infection as well, on the basis of absence of IgG after 4 days of fever.
In order to verify potential diagnostic variability due to different epidemiological settings, Platelia NS1 was also applied to stored samples collected previously during another outbreak in Santos in 2002 and in Goiânia, Brazil, in 2005. These samples were tested in concomitance to samples of the Santos 2010 outbreak (Table 2).
Table 2.
NS1 results in primary and secondary acute dengue casesa
| Days since the onset of symptoms | No. of NS1-positive samples/total no. of samples (%) |
||
|---|---|---|---|
| Santos 2002 | Goiânia 2005 | Santos 2010 | |
| Primary infection | |||
| 1–3 | 2/2 (100) | 15/16 (93.7) | 2/4 (50) |
| 4–5 | 6/7 (85) | 18/19 (94.7) | 19/25 (76) |
| 6–7 | 6/7 (85) | 19/19 (100) | 8/14 (57) |
| 8 | 1/1 (100) | 5/7 (71.4) | 3/5 (60) |
| Total | 15/17 (88) | 57/61 (93.4) | 32/48 (67) |
| Secondary infection | |||
| 1–3 | 6/9 (66) | 12/19 (63.1) | 39/108 (36) |
| 4–5 | 10/16 (62.5) | 24/33 (72.7) | 8/76 (11) |
| 6–7 | 19/331 (61) | 31/37 (83.7) | 7/48 (15) |
| 8 | 5/10 (50) | 12/14 (85.7) | 0/28 (0) |
| Total | 40/66 (60) | 79/103 (76.6) | 54/260 (38) |
Results are according to days of disease in a cohort of 82 samples from the Santos 2002 outbreak, 164 samples from Goiânia 2005 and 308 samples from Santos 2010 (71 samples from the current outbreak could not be classified into primary or secondary dengue due to the absence of IgG before 4 days of symptoms). All cases included in this analysis had the dengue diagnosis confirmed by IgM and/or were RNA positive.
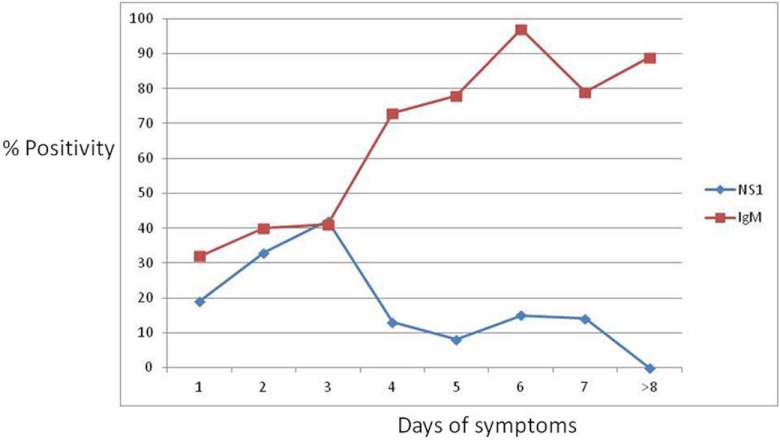
Among the 260 secondary infections, 167 (64.2%) were IgM positive and 54 (20.7%) were NS1 positive. If we restrict the analysis of the sensitivity of NS1 antigen and IgM antibodies testing for those patients with high IgG avidity, our criteria for assigning them as secondary dengue, NS1 positivity is generally low, decreasing with the days of disease (Fig. 1).
Fig 1.
Rate (%) of reactivity for NS1 and IgM according to the days since the onset of symptoms for secondary infection (n = 260).
In 367 samples, we did a multiplex PCR toward dengue RNA genotyping: 88/367 (24%) were positive, 84/88 (96%) contained dengue type 2 RNA, 3/88 (3%) type 1, and 1/88 (1%) type 3. In samples harboring dengue type 2 RNA, 52/84 (62%) samples were positive for NS1.
The criterion for sample inclusion in this study was a positive real-time PCR. The viral load had a mean of 113,385 copies/ml and a median of 307 copies/ml. The relationship between IgM, NS1, and the viral load is shown in Table 3.
Table 3.
IgM and NS1 positivity rates stratified by viral load (copies/ml)a
| Real-time copies/ml | No. of IgM-positive samples/total no. of samples (%) | No. NS1-positive samples/total no. of samples (%) |
|---|---|---|
| <305 | 125/172 (72) | 26/172 (15) |
| 305–10,000 | 51/108 (47) | 51/108 (47) |
| 10,000–100,000 | 12/36 (33) | 22/36 (61) |
| >100,000 | 14/63 (22) | 44/63 (70) |
The differences on the positivity rate across viral load categories were statistically significant for both markers (P < 0,0001).
On an attempt to identify putative NS1 mutations that could justify a lack of detection, 14 samples (8/14 NS1 positive, 5/14 IgM positive, and one negative for both markers) (Table 4) had the NS1 gene partially sequenced. Seven samples were sequenced after viral culture on the C63/6 mosquito cell line, used to increase the viral load. Supernatants from the first passage were harvested for RNA extraction and NS1 gene sequencing. The remaining seven viruses were amplified directly from RNA extracted from serum samples. All samples belonged to serotype 2, although only 8/14 (57%) were genotyped as DENV-2 by multiplex PCR (Table 4). Sequencing revealed a high level of similarity among sequences, with no observed distinction between viruses that were positive and those tested as negative in NS1 and IgM tests. Also, no amino acid substitution that could explain the decrease in sensitivity of the serological tests was found by comparison to viruses that circulated in Brazil in previous epidemics (2004 to 2008) (data not shown).
Table 4.
Profile of sequenced samples
| GenBank accession no. | NS1 | IgM | Copies/ml | Serotype |
|---|---|---|---|---|
| JX417131 | Positive | Positive | 69,366 | 2 |
| JX417132 | Positive | Positive | 11,994 | 2 |
| JX417133 | Negative | Positive | 492 | 2 |
| JX417134 | Negative | Negative | 666 | 2 |
| JX417135 | Negative | Negative | 2,840,000 | 2 |
| JX417136 | Positive | Negative | 45,839 | 2 |
| JX417137 | Positive | Negative | 14,200,000 | 2 |
| JX417138 | Positive | Negative | 896,004 | 2 |
| JX417139 | Negative | Negative | 2,040,000 | 2 |
| JX417140 | Positive | Negative | 2,070,000 | 2 |
| JX417141 | Negative | Negative | 1,210,000 | 2 |
| JX417142 | Positive | Positive | 4,550,000 | 2 |
| JX417143 | Positive | Negative | 11,800,000 | 2 |
| JX417144 | Negative | Positive | 400,455 | 2 |
DISCUSSION
NS1 antigen is strongly immunogenic and bears a relationship to viremia of dengue. Several assays, both enzymatic immunoassays and immunochromatographic rapid tests, are able to detect it in plasma, serum, and whole blood. In general, their performance was found to be excellent, and the routine diagnosis of dengue is made worldwide by such methods. However, many groups in different countries reported a low sensitivity of NS1 assays compared to molecular methods, mainly on populations that had experienced several sequential outbreaks of this disease (9, 10). As this finding is definitely more pronounced in secondary infections, it has been suggested that NS1 could be sequestered into immunocomplexes with IgG (9, 13). In the present series, at least 68% of patients had secondary dengue infection, which could explain the diminished performance (less than 40% of positivity) of the NS1 test. In contrast, in the 2002 Santos epidemic, NS1 was found to perform reasonably well, as 60% of the confirmed secondary infections were reactive by the Platelia assay. Similar samples from Goiânia displayed an even higher NS1 reactivity rate (76.6%) on secondary dengue cases (Table 2).
The poor performance of the NS1 test in the 2010 epidemic does not appear to be related to a specific NS1 assay, as the two different assays evaluated here presented similar performances. It has been hypothesized that the NS1 test can have different sensitivity according to the dengue virus serotype, and a reduced sensitivity of NS1 assays for DENV-2 has already been demonstrated (9). Indeed, a better performance of the NS1 test was found for samples from the previous epidemics of DENV-1/-3 in Santos and DENV-3 in Goiânia compared to the 2010 DENV-2 epidemic. However, differences in the epidemiological background among populations, in the succession and penetrance of dengue serotypes over time, make it difficult to evaluate the real impact of serotypes in NS1 performance.
Finally, to rule out the hypothesis that amino acid change(s) in NS1 protein could negatively affect the performance of the NS1 test in the 2010 DENV-2 outbreak by abrogating the binding of the monoclonal antibodies in the commercial kits, we sequenced the NS1 gene from 14 cases. Sequence comparison of NS1-positive and -negative samples in this study and also DENV-2 sequences from two previous epidemics in Brazil (Rio de Janeiro) did not reveal any amino acid difference.
NS1 tests are fast and easy to perform, becoming a valuable tool for the diagnosis of dengue acute infection. However, our data indicate that current NS1 assays should not be used in the Brazilian population before validation with samples from the occurring outbreak, as numerous false-negative results can occur. Employing an assay coupling NS1 and IgM may mitigate this reduced sensitivity. Another alternative would be to adopt RNA testing as the gold standard and do IgM testing on follow-up samples from RNA-negative cases. As more automated and high-throughput nucleic acid test (NAT) platforms are increasingly available, this seems to be a realistic approach in the short term.
In recent years, a few cases of transmission of dengue by blood transfusion were reported, two of them causing severe dengue in the recipients (21, 22), even though dengue was named as one of the major emerging threats to the blood supply (20). However, the intention to screen blood donations for dengue was previously hampered by the lack of validated assays. Currently, NS1 EIAs are conceived as high-throughput alternatives. Nevertheless, the sensitivity presented here, of approximately 40%, is absolutely incompatible with the standards of blood screening, where adopted assays usually require sensitivity above 99%.
In conclusion, we showed that NS1 test sensitivity can vary in different epidemiological settings. Secondary infections and/or DENV-2 may have contributed to the high number of false-negative results during the epidemic of 2010, but the biological causes for the observed failure of NS1 testing remain to be clarified.
ACKNOWLEDGMENTS
This study was supported by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP grant 2010/12313-5).
None of us has a commercial association or other associations that might pose a conflict of interest.
Footnotes
Published ahead of print 24 October 2012
REFERENCES
- 1. Alcon S, et al. 2002. Enzyme-linked immunosorbent assay specific to dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J. Clin. Microbiol. 40:376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barreto ML, et al. 2011. Successes and failures in the control of infectious diseases in Brazil: social and environmental context, policies, interventions, and research needs. Lancet 377:1877–1889 [DOI] [PubMed] [Google Scholar]
- 3. Castro-Jorge LA, et al. 2010. Clinical evaluation of the NS1 antigen-capture ELISA for early diagnosis of dengue virus infection in Brazil. J. Med. Virol. 82:1400–1405 [DOI] [PubMed] [Google Scholar]
- 4. Chambers TJ, Hahn CS, Galler R, Rice CM. 1990. Flavivirus genome organization, expression and replication. Annu. Rev. Microbiol. 44:649–688 [DOI] [PubMed] [Google Scholar]
- 5. Coller BA, Clements DE. 2011. Dengue vaccines: progress and challenges. Curr. Opin. Immunol. 23:391–398 [DOI] [PubMed] [Google Scholar]
- 6. de Souza VA, et al. 2004. Use of an immunoglobulin G avidity test to discriminate between primary and secondary dengue virus infections. J. Clin. Microbiol. 42:1782–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duyen HT, et al. 2011. Kinetics of plasma viremia and soluble nonstructural protein 1 concentrations in dengue: differential effects according to serotype and immune status. J. Infect. Dis. 203:1292–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guilarde AO, et al. 2008. Dengue and dengue hemorrhagic fever among adults: clinical outcomes related to viremia, serotypes, and antibody response. J. Infect. Dis. 197:817–824 [DOI] [PubMed] [Google Scholar]
- 9. Guzman MG, et al. 2010. Multi-country evaluation of the sensitivity and specificity of two commercially-available NS1 ELISA assays for dengue diagnosis. PLoS Negl. Trop. Dis. 4:e811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hang VT, et al. 2009. Diagnostic accuracy of NS1 ELISA and lateral flow rapid tests for dengue sensitivity, specificity and relationship to viraemia and antibody responses. PLoS Negl. Trop. Dis. 3:e360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harris E, et al. 1998. Typing of dengue viruses in clinical specimens and mosquitoes by single-tube multiplex reverse transcriptase PCR. J. Clin. Microbiol. 36:2634–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kao CL, King CC, Chao DY, Wu HL, Chang GJ. 2005. Laboratory diagnosis of dengue virus infection: current and future perspectives in clinical diagnosis and public health. J. Microbiol. Immunol. Infect. 38:5–16 [PubMed] [Google Scholar]
- 13. Koraka P, et al. 2003. Detection of immune-complex-dissociated nonstructural-1 antigen in patients with acute dengue virus infections. J. Clin. Microbiol. 41:4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Levi JE, et al. 2007. Evaluation of a commercial real-time PCR kit for detection of dengue virus in samples collected during an outbreak in Goiania, Central Brazil, in 2005. J. Clin. Microbiol. 45:1893–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Libraty DH, et al. 2002. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J. Infect. Dis. 186:1165–1168 [DOI] [PubMed] [Google Scholar]
- 16. Lima MRQ, Nogueira RM, Schatzmayr HG, dos Santos FB. 2010. Comparison of three commercially available dengue NS1 antigen capture assays for acute diagnosis of dengue in Brazil. PLoS Negl. Trop. Dis. 4:e738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Romano CM, et al. 2010. Characterization of Dengue virus type 2: new insights on the 2010 Brazilian epidemic. PLoS One 5:e11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Secretaria da Saúde do Estado de São Paulo Official data from dengue epidemics in the state of São Paulo, Brazil. http://www.cve.saude.sp.gov.br/htm/zoo/dengue_dados.html
- 19. Simmons CP, Farrar JJ, Nguyen VV, Wills B. 2012. Dengue. N. Engl. J. Med. 366:1423–1432 [DOI] [PubMed] [Google Scholar]
- 20. Stramer SL, et al. 2009. Emerging infectious disease agents and their potential threat to transfusion safety. Transfusion 49(Suppl 2):1S–29S [DOI] [PubMed] [Google Scholar]
- 21. Stramer SL, et al. 2012. Dengue viremia in blood donors identified by RNA and detection of dengue transfusion transmission during the 2007 dengue outbreak in Puerto Rico. Transfusion 52:1657–1666 [DOI] [PubMed] [Google Scholar]
- 22. Tambyah PA, Koay ES, Poon ML, Lin RV, Ong BK. 2008. Dengue hemorrhagic fever transmitted by blood transfusion. N. Engl. J. Med. 359:1526–1527 [DOI] [PubMed] [Google Scholar]
- 23. Teixeira MG. 2012. Few characteristics of dengue′s fever epidemiology in Brazil. Rev. Inst. Med. Trop. Sao Paulo 54(Suppl 18):S1–S4 [DOI] [PubMed] [Google Scholar]
- 24. Teles FR, Prazeres DM, Lima-Filho JL. 2005. Trends in dengue diagnosis. Rev. Med. Virol. 15:287–302 [DOI] [PubMed] [Google Scholar]
- 25. WHO World statistics on dengue cases by region, country, and severity. http://www.who.int/topics/dengue/en/ Accessed July 2012
- 26. Young PR, Hilditch PA, Bletchly C, Halloran W. 2000. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J. Clin. Microbiol. 8:1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]