Abstract
Pseudomonas aeruginosa NCGM1588 has a novel chromosomal class 1 integron, In151, which includes the aac(6′)-Iaj gene. The encoded protein, AAC(6′)-Iaj, was found to consist of 184 amino acids, with 70% identity to AAC(6′)-Ia. Escherichia coli transformed with a plasmid containing the aac(6′)-Iaj gene acquired resistance to all aminoglycosides tested except gentamicin. Of note, aac(6′)-Iaj contributed to the resistance to arbekacin. Thin-layer chromatography revealed that AAC(6′)-Iaj acetylated all aminoglycosides tested except gentamicin. These findings indicated that AAC(6′)-Iaj is a functional acetyltransferase that modifies the amino groups at the 6′ positions of aminoglycosides and contributes to aminoglycoside resistance of P. aeruginosa NCGM1588, including arbekacin.
INTRODUCTION
The major mechanism of resistance to aminoglycosides is the production of aminoglycoside-modifying enzymes (1). The aminoglycoside 6′-N-acetyltransferases [AAC(6′)s] are of particular interest because they can modify a number of clinically important aminoglycosides, including amikacin, gentamicin, netilmicin, and tobramycin. The AAC(6′)-I type confers resistance to amikacin through acetylation of the drug, whereas the AAC(6′)-II type acetylates gentamicin. To date, 43 genes, designated aac(6′)-Ia to aacA43, which encode AAC(6′)-I enzymes, have been cloned and characterized (1–3). Genes encoding aminoglycoside-modifying enzymes are often located on integrons (4), sequences that can integrate gene cassettes through site-specific recombination (5), in both plasmid and genomic DNA (4). Class 1 integrons participate in multidrug resistance in Pseudomonas aeruginosa (6–8).
Pseudomonas aeruginosa is a nosocomial pathogen that exhibits a remarkable ability to acquire resistance to several antibiotics. In Japan, the most serious problem has been the emergence of multidrug-resistant (MDR) P. aeruginosa strains, which are defined as having resistance to carbapenems, amikacin, and fluoroquinolones (9, 10).
Previously, we described a nosocomial outbreak caused by an MDR P. aeruginosa strain, IMCJ2.S1 (present name, NCGM2.S1) in a hospital in the eastern part of Japan (10). IMCJ2.S1 was found to harbor an aminoglycoside 6′-N-acetyltransferase gene, aac(6′)-Iae, in a chromosomal integron (9). A study in Japan in 2008 revealed two MDR P. aeruginosa clinical isolates harboring aac(6′)-Iaf (11). In 2011, a clinical isolate of MDR P. aeruginosa negative for aac(6′)-Iae and aac(6′)-Iaf was found. The isolate contained a novel aminoglycoside 6′-N-acetyltransferase gene, aac(6′)-Iaj. Here, we report the structure of this gene and the properties of its product.
MATERIALS AND METHODS
Bacterial strains and plasmids.
A clinical isolate of P. aeruginosa, NCGM1588, was obtained from the respiratory tract of a patient in 2011in a hospital in Osaka, Japan. P. aeruginosa IMCJ2.S1, which is a representative strain of a cluster endemic to Japan, was used as a control (10, 12). Escherichia coli DH5α (TaKaRa Bio, Shiga, Japan) and E. coli BL21-CodonPlus (DE3)-RIP (Agilent Technologies, Santa Clara, CA) were used as hosts for recombinant plasmids and for expression of aac(6′)-Iaj, respectively. Plasmids pSTV28 and pQE2 were used for cloning of aac(6′)-Iaj and purification of recombinant AAC(6′)-Iaj, respectively (11).
Antimicrobial agents.
Amikacin (AMK), ceftazidime (CAZ), ciprofloxacin (CIP), colistin (CST), lividomycin A (LIV), piperacillin (PIP), polymyxin B (PMB), sisomicin (SIS), and tobramycin (TOB) were obtained from Sigma-Aldrich (St. Louis, MO), arbekacin (ABK), dibekacin (DIB), and kanamycin A (KAN) were purchased from Meiji Seika Pharma Co. (Tokyo, Japan), aztreonam (ATM) was obtained from Eizai (Tokyo, Japan), cefepime (FEP) was obtained from Bristol-Myers Squibb Co. (New York, NY), gentamicin (GEN) and neomycin B and C mixtures (NEO) were obtained from Nacalai Tesque (Kyoto, Japan), imipenem (IPM) was obtained from Banyu Pharmaceutical Co. (Tokyo, Japan), isepamicin (ISP) was obtained from Nichi-Iko Co. (Toyama, Japan), meropenem (MEM) and netilmicin (NET) were obtained from Sumitomo Pharmaceutical Co. (Osaka, Japan), ofloxacin (OFX) was obtained from LKT laboratories (St. Paul, MN), and piperacillin-tazobactam (TZP) was obtained from Toyama Pure Chemical Industries (Tokyo, Japan).
In vitro susceptibility tests.
MICs were determined using a microdilution method according to the protocols recommended by the Clinical and Laboratory Standards Institute (13).
Serotyping.
The O serotypes of isolates were determined with a slide agglutination test kit (Denka Seiken Co., Tokyo, Japan) and sequence analysis of serotype-specific genes (14).
MLST.
Multilocus sequence typing (MLST) was performed according to the protocols described on the P. aeruginosa MLST database website (http://pubmlst.org/paeruginosa/). PCR and sequencing were performed for 7 chromosomal genes (acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE). The nucleotide sequences of these genes were compared with the sequences submitted to the MLST database to determine the allele numbers and sequence types (STs).
PCR amplification of a class 1 integron.
Genomic DNA was extracted using a Wizard Genomic DNA purification kit (Promega, Madison, WI). A class 1 integron was detected by PCR using 5′-CS and 3′-CS primers as described previously (9) and genetically mapped using the primers listed in Table 1. The Expand High-Fidelity PCR system (Roche Diagnostics GmbH, Penzberg, Germany) was used for PCR amplification. All PCR products were sequenced to identify genes and their orders in the integron.
Table 1.
PCR primers used in this study
| Primer | Sequencea (5′ to 3′) | Description |
|---|---|---|
| 5′-CS | GGCATCCAAGCAGCAAG | 5′-end common segment of class 1 integrons |
| 3′-CS | AAGCAGACTTGACCTGA | 3′-end common segment of class 1 integrons |
| intI-F | CTACCTCTCACTAGTGAGGGG | Positions 1–21 in intI1 |
| intI-R | TGCGTGTAAATCATCGTCGT | Positions 196–177 in intI1 |
| aac(6′)-Iaj-41F | ATCAGATCGATGCTGCAAGAATTC | Positions 41–64 in aac(6′)-Iaj |
| aac(6′)Iaj-543R | ACTTTTCCACATCCAAATATCGGG | Positions 543–520 in aac(6′)-Iaj |
| qacEdelta-F | TGAAAGGCTGGCTTTTTCTT | Positions 2–21 in qacEΔ1 |
| qacEdelta-R | GCAATTATGAGCCCCATACC | Positions 268–287 in qacEΔ1 |
| sul1-R | GGGTTTCCGAGAAGGTGATT | Positions 768–787 in sul1 |
| sul1-F | TCACCGAGGACTCCTTCTTC | Positions 29–48 in sul1 |
| IS6100-R | GGCTCTGTTGCAAAGATTGGC | Sequence 34–54 downstream of IS6100 |
| PstI-aac-F | aactgcagGGCTTGTTATGACTGTTTTT | Sequence in the 180- to 161-bp upstream region of aac(6′)-Iaj with PstI site |
| SalI-aac-R | ggtcgacTCAATTGAGTAGACTTTTCCAC | Positions 555–534 in aac(6′)-Iaj with SalI |
| SphI-aac-F | ccgcatgcgATGGAATATTCAATTATCAAT | Positions 1–21 in aac(6′)-Iaj with SphI |
| NotI-aac-R | gggcggccgcTCAATTGAGTAGACTTTTCC | Positions 555–536 in aac(6′)-Iaj with NotI |
| 23S-rRNA-F | CGAGGACAGTGTATGGTGGGCAGT | Positions 2207–2231 in 23S rRNA gene |
| 23S-rRNA-R | CTCAACGCCTCACAACGCTTACACA | Positions 2856–2832 in 23S rRNA gene |
Lowercase letters represent restriction enzyme recognition sites attached on the 5′ ends of primers.
DNA sequencing.
DNA sequences were determined using an ABI PRISM3130 sequencer (Applied Biosystems, Foster City, CA). Homology searches of nucleotide and translated protein sequences were performed using BLAST. Multiple-sequence alignments, searches for open reading frames (ORFs), and dendrograms for AACs were performed using Genetyx software (Genetyx, Tokyo, Japan).
PFGE and Southern hybridization.
DNA plugs were prepared as described previously (5) and digested overnight at 37°C with SpeI (TaKaRa Bio) or I-CeuI (New England BioLabs, Ipswich, MA). Pulsed-field gel electrophoresis (PFGE) analysis was performed as described previously (9). Southern hybridization was performed using an enhanced chemiluminescence direct nucleic acid-labeling and detection system according to the manufacturer's instructions (GE Healthcare, Tokyo, Japan), as described previously (11), to determine whether the novel class 1 integron identified in the P. aeruginosa isolates has a chromosomal location. Probes for aac(6′)-Iaj and 23S rRNA genes from NCGM1588 were amplified by PCR using the primer sets aac(6′)-Iaj-41F/aac(6′)Iaj-543R and 23S-rRNA-F/23S-rRNA-R, respectively (Table 1).
Cloning of aac(6′)-Iaj gene.
The ORF of aac(6′)-Iaj and 180 bp of the upstream region of the gene, including the promoter, were amplified by PCR from P. aeruginosa NCGM1558 using the primer set PstI-aac-F and SalI-aac-R (Table 1). The PCR products were digested with PstI and SalI and ligated into the PstI and SalI sites of pSTV28. The plasmids were used to transform DH5α, and transformants were selected on LB agar containing 100 μg/ml of chloramphenicol. To determine the MICs of aminoglycosides, E. coli DH5α was transformed with pSTV28-aac(6′)-Iaj.
Purification of recombinant AAC(6′)-Iaj.
The aac(6′)-Iaj gene from P. aeruginosa NCGM1588 was amplified by PCR using the primer set SphI-aac-F and NotI-aac-R (Table 1), and the product was digested with SphI and NotI and ligated into pQE2 (Invitrogen, Carlsbad, CA), which had been digested with the same restriction enzymes. The plasmid was used to transform DH5α, and the transformants were selected on LB agar containing 100 μg/ml of ampicillin. The resulting plasmid, pQE-aac(6′)-Iaj, was used to transform BL21-CodonPlus (DE3)-RIP (Agilent Technologies), which was used for recombinant protein purification. BL21-CodonPlus (DE3)-RIP carrying plasmid pQE2-aac(6′)-Iaj was grown in LB medium containing 200 μg/ml ampicillin at 37°C until the A600 reached 0.3. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a concentration of 0.1 mM to induce expression of AAC(6′)-Iaj, and the culture was incubated for 4 h at 37°C. The soluble fraction of six-histidine-tagged AAC(6′)-Iaj was obtained from the bacterial cells lysed by sonication in buffer A (20 mM Tris, 300 mM NaCl, and 10 mM imidazole, pH 8.0). The AAC(6′)-Iaj was purified from the soluble fraction using Ni-NTA agarose according to the manufacturer's instruction (Qiagen, Tokyo, Japan).
TLC analysis of acetylated aminoglycosides.
Mixtures containing 2 mM aminoglycoside, 2 mM acetyl coenzyme A (acetyl-CoA), and 50 μg/ml AAC(6′)-Iaj in 20 μl of phosphate buffer (pH 7.4) were incubated for 16 h at 37°C. Aliquots of 3 μl of each aminoglycoside mixture were spotted onto the surface of a Silica Gel 60 thin-layer chromatography (TLC) plate containing a fluorescence indicator with an excitation wavelength of 254 nm (Merck, Darmstadt, Germany), and the results were developed with a 5% phosphate potassium solution. The aminoglycosides and their acetylated products were detected with 0.2% ninhydrin in acetone.
Nucleotide sequence accession number.
The nucleotide sequence of In151 determined in this study has been deposited in the EMBL and GenBank databases and the DDBJ and assigned the accession number AB709942.
RESULTS AND DISCUSSION
Characterization of P. aeruginosa NCGM1588.
The MICs of antibiotics for NCGM1588 were as follows: PIP, 32 μg/ml; TZP, 32 μg/ml; CAZ, 8 μg/ml; FEP, 16 μg/ml; IPM, 32 μg/ml; MEM, 16 μg/ml; ATM, 32 μg/ml; AMK, 128 μg/ml; ABK, 32 μg/ml; GEN, 8 μg/ml; CIP, 16 μg/ml; OFX, 32 μg/ml; PMB, 4 μg/ml; and CST, 4 μg/ml. NCGM1588 showed high levels of AMK resistance. In particular, it showed high levels of ABK resistance, whereas the representative epidemic strain of MDR P. aeruginosa IMCJ2.S1 in Japan was susceptible to ABK (9, 15). The serotype of NCGM1588 was O7, and the MLST was ST560. NCGM1588 showed different PFGE patterns from that of MDR P. aeruginosa IMCJ2.S1, with similarity of 46.2%.
P. aeruginosa NCGM1588 is an emerging MDR pathogen in Japan. Therefore, it is necessary to carefully investigate whether the NCGM1588 will expand in medical settings. NCGM1588 seems to be quite different from the epidemic strain of MDR P. aeruginosa IMCJ2.S1, which is widespread in Japan (9, 10, 12), because of different PFGE patterns, MLSTs (ST560 versus ST235), and serotypes (O7 versus O11). IMCJ2.S1 causes mainly urinary tract infections (9), whereas NCGM1588 caused respiratory infection. During our surveillance from 2009 to 2010, we found 16 isolates of MDR P. aeruginosa, including a novel MDR P. aeruginosa strain, NCGM1179 (16), which had identical PFGE patterns and were ST991 and serotype O18; all of these isolates were isolated from the respiratory tract (17). To date, 14 strains of P. aeruginosa showing ST560, including NCGM1588, have been reported—8 in Australia, 3 in China, 1 in the Netherlands, and 1 in Spain (http://pubmlst.org/paeruginosa/). This is the first report of the isolation of P. aeruginosa showing ST560 in Japan.
aac(6′)-Iaj in the class 1 integron.
To identify the drug resistance genes of NCGM1588, the variable regions of class 1 integrons were amplified with the primers 5′-CS and 3′-CS (Table 1). PCR products of 1.1 kb were generated from this strain. DNA sequence analysis revealed a variable region containing a cassette of a novel aac(6′) gene. Based on the standard nomenclature (18), we named this ORF aac(6′)-Iaj. The novel gene consisted of an ORF of 555 bp, and its sequence showed 70% identity to that of aac(6′)-Ia from Corynebacterium resistens (accession number FN825254) (19). The aac(6′)-Iaj gene had a G+C content of 31.2%.
We designated the gene aac(6′)-Iaj according to a system of nomenclature proposed by Shaw et al. (1), which is easy to understand and indicates the functional properties of the enzymes in a straightforward manner as follows: numbers in parentheses, e.g., (1), (2), (3), and (6′), etc., for the site of modification; roman numerals, e.g., I, II, IV, etc., for unique resistance profiles; and lowercase letters, e.g., a, b, c, etc., for unique protein designations (1, 20).
The structure of the class 1 integron harboring aac(6′)-Iaj was determined using external primers (Table 1). The sequence of the integron was not found in any database; therefore, it was named In151. In151 had a structure similar to that of In4 integron (accession number U12338) except for the gene cassette array (21). Between the 5′-CS and 3′-CS, In151 had one gene cassette that contained aac(6′)-Iaj and a 60-nt 59-base element, which is known as attC, located 11 bp downstream of aac(6′)-Iaj (22).
In151 could be derived from the same origin as In4 of P. aeruginosa plasmid R1033. In4 was found in plasmid R1033 of a P. aeruginosa strain isolated in 1975 (23). The In151 backbone differed from that of In4 by the presence of a partial copy of IS6100; i.e., In151 had the 5′-CS, 3′-CS, and a complete copy of IS6100 located downstream of the 3′-CS.
Amino acid sequence of AAC(6′)-Iaj enzyme.
AAC(6′)-Iaj consists of 184 amino acids. Multiple sequence alignments among AAC(6′) enzymes revealed that AAC(6′)-Iaj had 70% identity to AAC(6′)-Ia from Shigella sonnei (24), 66% to AAC(6′)-Iaf from P. aeruginosa (11), 65% to AAC(6′)-Iq from Klebsiella pneumoniae (25), 64% to AAC(6′)-Im from Citrobacter freundii (26), 63% to AAC(6′)-33 from P. aeruginosa (27), and 63% to AAC(6′)-I30 from Salmonella enterica (28). Based on the work of Neuwald and Landsman (29), four motifs in the amino acid sequences of the subfamily proteins belonging to AAC(6′)-Iaj were designated motifs C, D, A, and B (Fig. 1). Comparison of amino acid sequences of members of the AAC(6′)-I subfamily with that of AAC(6′)-Iaj revealed that motifs C, D, A, and B, which are found in most GCN5-related N-acetyltransferases (GNATs) (29, 30), were conserved in AAC(6′)-Iaj (Fig. 1). A large motif at the C terminus, motif B (30), was 77.6% identical between AAC(6′)-Ia (24) and AAC(6′)-Iaj.
Fig 1.
Alignment of the AAC(6′)-Iaj amino acid sequence with those of six members of AAC(6′)-I subfamily. Identical residues are marked with black boxes. Four motifs, C, D, A, and B, are indicated by a dotted line, a gray line, double lines, and a black line, respectively.
Localization of the aac(6′)-Iaj gene.
PFGE analysis and Southern hybridization using NCGM1588 genomic DNA digested by I-CeuI revealed that the probes specific for the 23S rRNA and aac(6′)-Iaj were detected in a chromosomal fragment of about 4.4 Mb (Fig. 2). These results indicate that aac(6′)-Iaj was located on the chromosomal DNA. Lower DNA bands were observed in lanes 2 to 4 of Fig. 2. They were probably due to nonspecific cleavage during DNA preparation and enzyme digestion. However, we cannot exclude the possibility that NCGM1588 has multiple copies of aac(6′)-Iaj.
Fig 2.

Localization of the aac(6′)-Iaj gene on I-CeuI-digested total DNA of P. aeruginosa strain NCGM1588 separated by PFGE. P. aeruginosa NCGM1588 genomic DNA digested by I-CeuI was done as previously described (11). Lane 1, molecular standard of Schizosaccharomyces pombe chromosomal DNA; lane 2, I-CeuI-digested total DNA of P. aeruginosa strain NCGM1588 with ethidium bromide; lane 3, Southern hybridization was performed with probes for 23S rRNA gene; lane 4, Southern hybridization was performed with probes for aac(6′)-Iaj.
Drug resistance mediated by AAC(6′)-Iaj enzyme.
P. aeruginosa NCGM1588 was resistant to all aminoglycosides tested except GEN (Table 2). A vector control of E. coli DH5α/pSTV28 was susceptible to all aminoglycosides tested, whereas E. coli DH5α/pSTV28-aac(6′)-Iaj was resistant to all aminoglycosides, including ABK, except GEN, with 4- to 128-fold-higher MIC values than those of the vector control (Table 2). The MIC of GEN in E. coli DH5α/pSTV28-aac(6′)-Iaj was the same as that in the vector control.
Table 2.
MICs of various aminoglycosides for P. aeruginosa NCGM1588 and E. coli strains transformed with aac(6′)-Iaj
| Straina | MICb (μg/ml) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMK | ABK | DIB | GEN | ISP | KAN | NEO | NET | SIS | TOB | |
| NCGM1588 | 128 | 32 | 1,024 | 8 | 512 | 1,024 | 256 | >1,024 | 1,024 | 128 |
| E. coli DH5α/pSTV28 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 2 | 0.25 | 1 | 0.25 |
| E. coli DH5α/pSTV28-aac(6′)-Iaj | 16 | 4 | 16 | 0.5 | 4 | 32 | 8 | 32 | 4 | 16 |
The MICs for E. coli strains were determined with Mueller-Hinton broth preparations containing chloramphenicol (30 μg/ml) and individual aminoglycosides.
AMK, amikacin; ABK, arbekacin; DIB, dibekacin; GEN, gentamicin; ISP, isepamicin; KAN, kanamycin; NEO, neomycin; NET, netilmicin; SIS, sisomicin; TOB, tobramycin.
To examine the acetylase activity of AAC(6′)-Iaj to aminoglycosides, we performed thin-layer chromatography using the purified recombinant AAC(6′)-Iaj. LIV, an aminoglycoside compound, was used as a negative control. LIV has a hydroxyl group instead of an amino group at the 6′ position and therefore cannot be acetylated by AAC(6′). As shown in Fig. 3, all of these aminoglycosides, except GEN, were acetylated by AAC(6′)-Iaj. The acetylation rates were only 2% for GEN when estimated with the ImageJ analyzer (http://rsbweb.nih.gov/ij/index.html). The TLC data for GEN were consistent with the MICs of GEN for E. coli DH5α/pSTV28-aac(6′)-Iaj and E. coli DH5α/pSTV28 (Table 2). The reason for the incomplete acetylation is that commercially available gentamicin is a mixture of its derivatives; some of them have a methyl group on N-6′ and are refractory to AAC(6′)-I enzymes (1).
Fig 3.
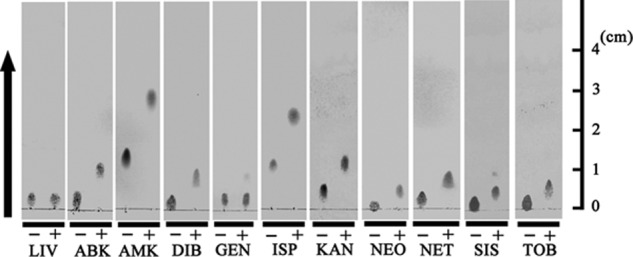
Analysis of acetylated aminoglycosides by TLC. AAC(6′)-Iaj and various aminoglycosides were incubated in the absence (−) or presence (+) of acetyl coenzyme A. The arrow indicates the direction of development.
AAC(6′)-Iaj-producing P. aeruginosa NCGM1588 was more resistant to ABK (MIC, 32 μg/ml) than AAC(6′)-Iae-producing IMCJ2.S1 (MIC, 2 μg/ml) (see Table 2 in reference 9), a representative epidemic MDR P. aeruginosa strain in Japan, indicating that AAC(6′)-Iaj could inactivate ABK more effectively than AAC(6′)-Iae. E. coli DH5α producing AAC(6′)-Iaj was relatively resistant to ABK compared to E. coli DH5α producing AAC(6′)-Iae (compare Table 2 in this paper and Table 3 in reference 9). As demonstrated by TCL analyses, both AAC(6′)-Iaj and AAC(6′)-Iae catalyzed inactivation of ABK (compare Fig. 3 in this paper and Fig. 6 in reference 9). The enzymatic activity of AAC(6′)-Iaj against ABK may be stronger than that of AAC(6′)-Iae, although further kinetic studies of both enzymes and chemical analysis of the products of acetylation by both enzymes will be necessary. The chemical structures of AMK and ABK are nearly identical, with only a few differences at the 2′, 3′, and 4′ positions in ring I; that is, AMK has 2′-, 3′-, and 4′-hydroxyl groups, whereas ABK has a 2′-amino group (31). The different substitutions at the 2′, 3′, and 4′ positions in ring I would be responsible for the different levels of ABK resistance between NCGM1179 and IMCJ2.S1.
ACKNOWLEDGMENTS
This study was supported by a grant (H24-Shinko-ippan-010) from the Ministry of Health, Labor and Welfare of Japan and JSPS KAKENHI Grant number 24790432.
Footnotes
Published ahead of print 15 October 2012
REFERENCES
- 1. Shaw KJ, Rather PN, Hare RS, Miller GH. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rather PN, Munayyer H, Mann PA, Hare RS, Miller GH, Shaw KJ. 1992. Genetic analysis of bacterial acetyltransferases: identification of amino acids determining the specificities of the aminoglycoside 6′-N-acetyltransferase Ib and IIa proteins. J. Bacteriol. 174:3196–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vakulenko SB, Mobashery S. 2003. Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 16:430–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hall RM, Collis CM. 1998. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updat. 1:109–119 [DOI] [PubMed] [Google Scholar]
- 5. Grundmann H, Schneider C, Hartung D, Daschner FD, Pitt TL. 1995. Discriminatory power of three DNA-based typing techniques for Pseudomonas aeruginosa. J. Clin. Microbiol. 33:528–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Laraki N, Galleni M, Thamm I, Riccio ML, Amicosante G, Frere JM, Rossolini GM. 1999. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob. Agents Chemother. 43:890–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee K, Lim JB, Yum JH, Yong D, Chong Y, Kim JM, Livermore DM. 2002. bla(VIM-2) cassette-containing novel integrons in metallo-beta-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob. Agents Chemother. 46:1053–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Partridge SR, Hall RM. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47:342–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sekiguchi J, Asagi T, Miyoshi-Akiyama T, Fujino T, Kobayashi I, Morita K, Kikuchi Y, Kuratsuji T, Kirikae T. 2005. Multidrug-resistant Pseudomonas aeruginosa strain that caused an outbreak in a neurosurgery ward and its aac(6′)-Iae gene cassette encoding a novel aminoglycoside acetyltransferase. Antimicrob. Agents Chemother. 49:3734–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sekiguchi J, Asagi T, Miyoshi-Akiyama T, Kasai A, Mizuguchi Y, Araake M, Fujino T, Kikuchi H, Sasaki S, Watari H, Kojima T, Miki H, Kanemitsu K, Kunishima H, Kikuchi Y, Kaku M, Yoshikura H, Kuratsuji T, Kirikae T. 2007. Outbreaks of multidrug-resistant Pseudomonas aeruginosa in community hospitals in Japan. J. Clin. Microbiol. 45:979–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kitao T, Miyoshi-Akiyama T, Kirikae T. 2009. AAC(6′)-Iaf, a novel aminoglycoside 6′-N-acetyltransferase from multidrug-resistant Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 53:2327–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miyoshi-Akiyama T, Kuwahara T, Tada T, Kitao T, Kirikae T. 2011. Complete genome sequence of highly multidrug-resistant Pseudomonas aeruginosa NCGM2.S1, a representative strain of a cluster endemic to Japan. J. Bacteriol. 193:7010 doi:10.1128/JB.06312-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clinical and Laboratory Standards Institute 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 9th ed Approved standard M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 14. Raymond CK, Sims EH, Kas A, Spencer DH, Kutyavin TV, Ivey RG, Zhou Y, Kaul R, Clendenning JB, Olson MV. 2002. Genetic variation at the O-antigen biosynthetic locus in Pseudomonas aeruginosa. J. Bacteriol. 184:3614–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kitao T, Miyoshi-Akiyama T, Shimada K, Tanaka M, Narahara K, Saito N, Kirikae T. 2010. Development of an immunochromatographic assay for the rapid detection of AAC(6′)-Iae-producing multidrug-resistant Pseudomonas aeruginosa. J. Antimicrob. Chemother. 65:1382–1386 [DOI] [PubMed] [Google Scholar]
- 16. Tada T, Kitao T, Miyoshi-Akiyama T, Kirikae T. 2011. Genome sequence of multidrug-resistant Pseudomonas aeruginosa NCGM1179. J. Bacteriol. 193:6397 doi:10.1128/JB.06129-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kitao T, Tada T, Tanaka M, Narahara K, Shimojima M, Shimada K, Miyoshi-Akiyama T, Kirikae T. 2012. Emergence of a novel multidrug-resistant Pseudomonas aeruginosa strain producing IMP-type metallo-beta-lactamases and AAC(6′)-Iae in Japan. Int. J. Antimicrob. Agents 39:518–521 [DOI] [PubMed] [Google Scholar]
- 18. Vanhoof R, Hannecart-Pokorni E, Content J. 1998. Nomenclature of genes encoding aminoglycoside-modifying enzymes. Antimicrob. Agents Chemother. 42:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schroder J, Maus I, Meyer K, Wordemann S, Blom J, Jaenicke S, Schneider J, Trost E, Tauch A. 2012. Complete genome sequence, lifestyle, and multi-drug resistance of the human pathogen Corynebacterium resistens DSM 45100 isolated from blood samples of a leukemia patient. BMC Genomics 13:141 doi:10.1186/1471-2164-13-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist. Updat. 13:151–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Partridge SR, Brown HJ, Stokes HW, Hall RM. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stokes HW, O'Gorman DB, Recchia GD, Parsekhian M, Hall RM. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731–745 [DOI] [PubMed] [Google Scholar]
- 23. Smith DI, Lus RG, Rubio Calvo MC, Datta N, Jacob AE, Hedges RW. 1975. Third type of plasmid conferring gentamicin resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 8:227–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tenover FC, Filpula D, Phillips KL, Plorde JJ. 1988. Cloning and sequencing of a gene encoding an aminoglycoside 6′-N-acetyltransferase from an R factor of Citrobacter diversus. J. Bacteriol. 170:471–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Centron D, Roy PH. 1998. Characterization of the 6′-N-aminoglycoside acetyltransferase gene aac(6′)-Iq from the integron of a natural multiresistance plasmid. Antimicrob. Agents Chemother. 42:1506–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hannecart-Pokorni E, Depuydt F, de Wit L, van Bossuyt E, Content J, Vanhoof R. 1997. Characterization of the 6′-N-aminoglycoside acetyltransferase gene aac(6′)-Im [corrected] associated with a sulI-type integron. Antimicrob. Agents Chemother. 41:314–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Viedma E, Juan C, Acosta J, Zamorano L, Otero JR, Sanz F, Chaves F, Oliver A. 2009. Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum beta-lactamases GES-1 and GES-5 in Spain. Antimicrob. Agents Chemother. 53:4930–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mulvey MR, Boyd DA, Baker L, Mykytczuk O, Reis EM, Asensi MD, Rodrigues DP, Ng LK. 2004. Characterization of a Salmonella enterica serovar Agona strain harbouring a class 1 integron containing novel OXA-type beta-lactamase (blaOXA-53) and 6′-N-aminoglycoside acetyltransferase genes [aac(6′)-I30. J. Antimicrob. Chemother. 54:354–359 [DOI] [PubMed] [Google Scholar]
- 29. Neuwald AF, Landsman D. 1997. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22:154–155 [DOI] [PubMed] [Google Scholar]
- 30. Dyda F, Klein DC, Hickman AB. 2000. GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 29:81–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mingeot-Leclercq MP, Glupczynski Y, Tulkens PM. 1999. Aminoglycosides: activity and resistance. Antimicrob. Agents Chemother. 43:727–737 [DOI] [PMC free article] [PubMed] [Google Scholar]



