Abstract
A clinical isolate of Pseudomonas aeruginosa recovered from the lower respiratory tract of an 81-year-old patient hospitalized in Belgium was sent to the national reference center to determine its resistance mechanism. PCR sequencing identified a new GES variant, GES-18, which differs from the carbapenem-hydrolyzing enzyme GES-5 by a single amino acid substitution (Val80Ile, in the numbering according to Ambler) and from GES-1 by two substitutions (Val80Ile and Gly170Ser). Detailed kinetic characterization showed that GES-18 and GES-5 hydrolyze imipenem and cefoxitin with similar kinetic parameters and that GES-18 was less susceptible than GES-1 to classical β-lactamase inhibitors such as clavulanate and tazobactam. The overall structure of GES-18 is similar to the solved structures of GES-1 and GES-2, the Val80Ile and Gly170Ser substitutions causing only subtle local rearrangements. Notably, the hydrolytic water molecule and the Glu166 residue were slightly displaced compared to their counterparts in GES-1. Our kinetic and crystallographic data for GES-18 highlight the pivotal role of the Gly170Ser substitution which distinguishes GES-5 and GES-18 from GES-1.
INTRODUCTION
GES-type extended-spectrum β-lactamases (ESBLs) have been discovered in several Gram-negative bacteria, including Enterobacteriaceae and nonfermenters (1–4). Pseudomonas aeruginosa isolates that express GES-type β-lactamases have been reported from South America (5, 6), South Africa (7), and China (8). The nosocomial spread of isolates expressing GES-1 and GES-5 in Spain was reported in 2009 (9), and a new GES-13 variant was recently found in Greece (10).
GES-type β-lactamases form a distinct branch of the Ambler class A enzymes, but their origin is unknown. The first GES β-lactamase, GES-1, was reported in a Klebsiella pneumoniae isolate from France in 2000 and was found to be encoded by a plasmid-borne integron (11). Twenty-two GES variants have been discovered thus far, differing from each other by 1 to 3 amino acid substitutions (http://lahey.org/studies/other.asp) that affect substrate specificity. The GES-1 β-lactamase exhibits extended-spectrum characteristics, hydrolyzing oxyiminocephalosporins. However, unlike most ESBLs, GES-1 does not hydrolyze aztreonam. Moreover, although containing the disulfide bond characteristic of class A carbapenemases (Cys69-Cys238, using the standard numbering for class A β-lactamases [12]), GES-1 lacks the ability to significantly hydrolyze carbapenems and is even inhibited by low concentrations of imipenem due to its very strong affinity for this substrate (11, 13). Carbapenemase activity has been demonstrated in certain variants with a substitution (Asn or Ser) at the Gly170 residue, including GES-2, GES-4, GES-5, GES-6, and GES-14 (1, 2, 14–17). The residue at position 170 is located in the Ω-loop, which forms the bottom of the active site, and Asn is found at this position in most class A β-lactamases. In contrast to most class A β-lactamases, GES-4, GES-5, GES-6, and GES-14 also hydrolyze cefoxitin (1, 2, 14, 16, 17).
The blaGES-5 gene was detected for the first time in 2004 in an Escherichia coli isolate (E. coli 365-02) from a patient in Greece as part of a gene cassette located on a class 1 integron carried by a plasmid (16). The blaGES-5 gene was subsequently found to be carried by K. pneumoniae, Enterobacter cloacae, and P. aeruginosa in many parts of the world (4).
Here we report the detection of a new variant (GES-18), in a clinical isolate of P. aeruginosa collected from a patient hospitalized in Belgium in 2010. The blaGES-18 gene was cloned in E. coli, and we characterized the kinetic properties and crystallographic structure of the purified GES-18 protein. This variant differs from GES-5 by a single substitution (Val80Ile) and from GES-1 by two substitutions (Val80Ile and Gly170Ser).
MATERIALS AND METHODS
Antibiotics.
Imipenem was obtained from Merck Sharp and Dohme Research Laboratories (Rahway, NJ), nitrocefin was obtained from ProGenosis (Liège, Belgium), and temocillin (Negaban) was obtained from Eumedica Pharmaceuticals (Brussels, Belgium); all other antibiotics and inhibitors were from Sigma-Aldrich (St. Louis, MO).
Bacterial strains and antimicrobial susceptibility.
P. aeruginosa PA11122 is a blaGES-positive isolate identified during a screen for acquired carbapenemase resistance genes in multidrug-resistant P. aeruginosa and Acinetobacter baumannii isolates collected in 2010 in the setting of a national resistance survey. Bacterial species identification was confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) with a Microflex LT mass spectrometer (Bruker Daltonik GmbH, Leipzig, Germany). P. aeruginosa reference strain PAO1 (18), P. aeruginosa PU21 (19), and E. coli TOP10 (Invitrogen, Merelbeke, Belgium) and BL21 were used for plasmid transfer and/or cloning experiments. Antimicrobial susceptibility was determined by the disc diffusion method and by Etest (bioMérieux, Brussels, Belgium) on Mueller-Hinton (MH) agar plates (Bio-Rad, Nazareth, Belgium) according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (20) in the presence of 2 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) when testing pET28a/GES clones. Meropenem-dipicolinic acid (DPA) discs (Neo-Sensitabs KPC-MBL confirmation kit; Rosco Diagnostica, Taastrup, Denmark) were used to screen for metallo-β-lactamase production.
PCR screening, characterization of the genetic environment of blaGES-18, and plasmid extraction.
Detection of carbapenemases (blaVIM, blaIMP, blaNDM-1, blaKPC) was performed by DNA microarray (Check-MDR CT102; Check-Points, Wageningen, Netherlands) according to a previously described protocol (21). Genes encoding class A ESBL enzymes (BEL, GES, VEB, and PER) were sought by PCR as previously described (1). The genetic context of blaGES-18 was determined by PCR sequencing of the integron variable region with primers matching the 5′ and 3′ conserved segments of class 1 integrons (22) using an external sequencing service (Macrogen, Seoul, South Korea) and a primer walking strategy. Nucleotide sequences were analyzed with the NCBI BLASTN algorithm (http://www.ncbi.nlm.nih.gov/BLAST/). Plasmid DNA was extracted using the Kieser method (23) and the QIAfilter plasmid midikit (Qiagen, Venlo, Netherlands) according to the manufacturer′s instructions.
Cloning, expression, and purification of GES-1, GES-5, and GES-18 β-lactamases.
The genes encoding GES-1, GES-5, and GES-18 were cloned into pET28a using the NcoI and EcoRI restriction sites. The different vectors were then introduced into the E. coli BL21(DE3) strain (New England BioLabs, Ipswich, MA). Overexpression and purification of these three proteins were performed as described for the GES-11, GES-12, and GES-14 enzymes (24).
Determination of kinetic parameters.
All experiments were carried out at 30°C in 100 mM sodium phosphate buffer (pH 7.0). Kinetic measurements of purified GES β-lactamases were performed as described previously (24). Low and very high Km values were determined as Ki values using 100 μM nitrocefin as the reporter substrate. For low Km values, the kcat values were derived from the initial hydrolysis rates measured at saturating substrate concentrations, whereas for high Km values, kcat was derived directly from the kcat/Km ratio (25). Despite its low value, the Km parameter for imipenem was determined by initial rates method [since, according to the work of Frase et al., Km is >Ks(i) (26)].
Fifty percent inhibitory concentrations (IC50s) were determined using 100 μM benzylpenicillin as the substrate and clavulanic acid and tazobactam as inhibitors, as previously described (11, 15, 17).
GES-18 crystallization.
The GES-18 β-lactamase (13 mg/ml) was crystallized at 21°C from a 20 mM citric acid–80 mM Bis-Tris propane (pH 8.8)–10% (wt/vol) polyethylene glycol (PEG) 3350 solution, using the sitting drop method with crystallization plates from Taorad GmbH (Aachen, Germany). The reservoir solution (1 μl) was mixed with the protein solution (1 μl), and the mixture was left to equilibrate against the solution reservoir. Typically, prismatic GES-18 crystals grew within a few days to dimensions of 80 by 80 by 250 μm and represented space group P212121 (a = 74.184 Å, b = 80.506 Å, and c = 87.748 Å).
Data collection and processing.
For data collection, crystals were gradually transferred to a cryoprotectant solution comprising reservoir solution supplemented with 30% (vol/vol) glycerol. The mounted crystals were flash-frozen at 100 K in a liquid nitrogen stream. Near-complete X-ray data sets were collected using a Bruker FR591 rotating-anode X-ray generator and a Mar345dtb detector. Diffraction data were processed with XDS (27) and scaled with SCALA from the CCP4 suite (28).
Structure determination and refinement.
The structure of GES-18 was solved by molecular replacement using Phaser (29) and the structure of GES-1 (PDB file 2QPN) as a search model. The resulting model was rebuilt using ARP/wARP (30). The structures were iteratively refined by manual inspection of the electron density with Coot (31) and refinement with Refmac5 (32). Refinement to convergence was carried out with isotropic B values and using TLS parameters (33). Alternative conformations were modeled for a number of side chains, and occupancies were adjusted to yield similar B values for the alternative conformations.
Accession numbers.
The nucleotide sequence data in this article have been deposited in the EMBL/GenBank nucleotide sequence database under accession number JQ028729. Coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession number 3V3S.
RESULTS
Clinical isolate and resistance traits.
P. aeruginosa PA11122 was isolated from an endotracheal aspirate obtained from an 81-year-old patient hospitalized in Antwerp, Belgium. This patient developed a ventilator-associated pneumonia while hospitalized in the intensive care unit. Isolate PA11122 was resistant to most β-lactams, including imipenem and meropenem (Table 1), and it was also resistant to aminoglycosides (gentamicin, tobramycin, and amikacin) and to ciprofloxacin. Phenotypic screening tests for the presence of acquired extended-spectrum β-lactamases and of carbapenemases failed to demonstrate any synergistic effect in the presence of different inhibitors, including clavulanic acid and dipicolinic acid (DPA) or EDTA. PCR screening was negative for metallo-β-lactamases as well as for class D oxacillin-hydrolyzing β-lactamases, but it yielded positive results for blaGES ESBL only. PCR sequencing confirmed that blaGES was a new allele, with an 864-bp open reading frame encoding a 287-amino-acid preprotein (including signal peptide) designated GES-18. This variant differed from GES-5 by a single substitution (Val80Ile) and from GES-1 by two substitutions (Val80Ile and Gly170Ser).
Table 1.
MICs of antibiotics for P. aeruginosa strain PA11122 expressing GES-18 and for E. coli BL21 carrying plasmid pET28a-GES-18, -GES-5, or -GES-1a
| Antibiotic | MIC (μg/ml) |
||||
|---|---|---|---|---|---|
| P. aeruginosa PA11122 |
E. coli carrying plasmid: |
||||
| pET28a | pET28a-GES-1 | pET28a-GES-5 | pET28a-GES-18 | ||
| Ampicillin | ND | 1.5 | >256 | >256 | >256 |
| Amoxicillin-clavulanic acid | ND | 1.5 | 32 | 64 | 128 |
| Ticarcillin-clavulanic acid | >256 | ND | ND | ND | ND |
| Piperacillin | >256 | 0.75 | >256 | >256 | >256 |
| Piperacillin-tazobactam | >256 | 0.75 | 1.5 | 32 | 32 |
| Cefuroxime | ND | 0.064 | 128 | 8 | 16 |
| Ceftazidime | 32 | 0.064 | 16 | 1 | 1.5 |
| Cefotaxime | ND | 0.064 | 4 | 0.38 | 0.38 |
| Cefepime | 96 | 0.064 | 0.5 | 0.094 | 0.094 |
| Aztreonam | 8 | <0.016 | 0.25 | 0.023 | 0.047 |
| Imipenem | >32 | 0.25 | 0.25 | 0.75 | 0.75 |
| Meropenem | >32 | 0.032 | 0.047 | 0.125 | 0.125 |
| Cefoxitin | ND | 0.125 | 0.25 | 8 | 8 |
MICs were determined by Etest. ND, not determined.
Genetic context.
The blaGES-18 gene was shown to be part of a cassette inserted between two copies of aacA4 located upstream of blaPSE-1 and aadA2 on a new class 1 integron, assigned number In762 in the integron database INTEGRALL (34). More than 4,550 bp of the integron was sequenced as shown schematically in Fig. 1 (accession number JQ028729).
Fig 1.
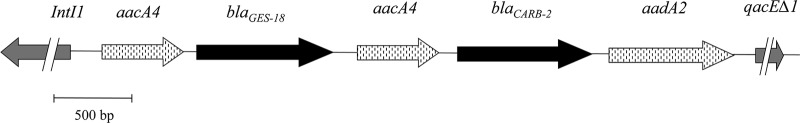
Schematic representation of In762 (accession number JQ028729) containing the blaGES-18 gene. Double slashes indicate the limit of sequencing, including a partial sequence of the integrase gene intI1 (5′-CS) and qacEΔ1 (3′-CS). Arrows indicate the direction of transcription of the coding regions.
Repeated plasmid extractions failed to visualize any plasmid by agarose gel electrophoresis. The nucleic acid extracts were nevertheless used for electroporation, but no transformants of the wild-type P. aeruginosa strain PAO1 were obtained, suggesting that blaGES-18 was possibly located on the chromosome in this isolate. Mating-out experiments between the clinical strain PA11122 as the donor and the rifampin-resistant P. aeruginosa strain PU21 as the recipient also failed to yield transconjugants.
Cloning of GES-18.
Compared to E. coli BL21 clone pET28a/GES-1, E. coli BL21 pET28a/GES-18 was more susceptible to oxyiminocephalosporins but more resistant to cefoxitin and to meropenem as well as to penicillin–β-lactamase inhibitor combinations such as amoxicillin-clavulanic acid and piperacillin-tazobactam (Table 1).
Production and purification.
After production of the GES-1, GES-5, and GES-18 enzymes in E. coli BL21(DE3) and their purification to homogeneity, a yield of 20 to 30 mg of β-lactamase per liter of culture was typically obtained. The masses of the different proteins were verified by electrospray ionization-mass spectrometry (ESI-MS). Within experimental errors, the variants were found to exhibit the expected masses (29,216 versus 29,217 Da for GES-1, 29,256 versus 29,257 Da for GES-5, and 29,260 versus 29,261 for GES-18).
Comparison of hydrolytic activities.
We compared the activities of GES-18 (Val80Ile, Gly170Ser), GES-5 (Gly170Ser), and GES-1 against compounds representing the four major families of β-lactam antibiotics, namely, penicillins, cephalosporins, carbapenems, and monobactams (Table 2).
Table 2.
Kinetic parameters for GES-18, GES-5, and GES-1 β-lactamasesa
| Antibiotic | GES-18 |
GES-5 |
GES-1 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (mM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (mM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (mM−1 s−1) | |
| Benzylpenicillin | 70 | 400 | 175 | 110 | 430 | 255 | 30 | 120 | 250 |
| Amoxicillin | 7 | 40 | 175 | 13 | 50 | 260 | 13 | 180 | 72 |
| Ticarcillin | 5 | 1,000 | 5 | 5 | 1,600 | 3 | 7 | 1,000 | 7 |
| Piperacillin | 75 | 700 | 100 | 160 | 1,000 | 160 | 8 | 900 | 9 |
| Temocillin | 0.004 | 3,500 | 0.001 | 0.003 | 4,500 | 0.0007 | 0.002 | 4,000 | 0.0005 |
| Cephalothin | 500 | 6,000 | 80 | 900 | 9,000 | 100 | 900 | 5,300 | 170 |
| Cephaloridine | 360 | 4,600 | 70 | 725 | 5,000 | 150 | 540 | 4,600 | 120 |
| Nitrocefin | 100 | 330 | 300 | 260 | 550 | 470 | 380 | 340 | 1100 |
| Cefoxitin | 3.4 | 1,000 | 3 | 4.5 | 860 | 5 | 0.08 | 60 | 1.3 |
| Cefotaxime | >20 | >20,000 | 1 | >28 | >20,000 | 1.4 | 370 | 9,000 | 40 |
| Ceftazidime | >2.5 | >20,000 | 0. 125 | >3 | >20,000 | 0. 15 | >56 | >20,000 | 2.8 |
| Cefepime | >1 | >20,000 | 0.05 | >2 | >20,000 | 0.1 | >16 | > 20,000 | 0. 8 |
| Aztreonam | 0.08 | 3,000 | 0.02 | 0.06 | 1,800 | 0.03 | 4.8 | 4,000 | 1.2 |
| Imipenem | 0.4 | 3.5 | 110 | 0.7 | 4.6 | 150 | 0.007 | 1.5 | 5 |
| Ertapenem | 0.06 | 1.3 | 40 | 0.09 | 1.5 | 60 | 0.003 | 0.4 | 7.5 |
| Meropenem | 0.04 | 1 | 40 | 0.06 | 1 | 60 | 0.0007 | 0.1 | 7 |
Data are the means of three independent experiments. Standard deviations were within 10% of the means (with the exception of imipenem, for which they were 30%).
The kinetic parameters of GES-18 and of GES-5 were similar for several substrates (e.g., ticarcillin, temocillin, cefotaxime, ceftazidime, aztreonam, cefoxitin, and carbapenems), indicating the conservation of many enzymatic properties (Table 2). However, the kcat values of GES-18 were about half of those of GES-5 for some penicillins (amoxicillin and piperacillin) and cephalosporins (cephalothin, nitrocefin, and cephaloridine). On the other hand, the Km values of GES-5 and GES-18 were almost in the same range for all β-lactam agents tested.
The carbapenemase activity of GES-18 was similar to that of GES-5 but higher than that of GES-1. The Gly170Ser mutation in GES-18 led to approximately 22-, 5-, and 6-fold increases in activity (kcat/Km) against imipenem, ertapenem, and meropenem, respectively (30-, 8-, and 8.5-fold increases for GES-5). This predominantly reflected the substantial increase in kcat values (approximately 60-, 20-, and 60-fold higher for GES-18 and approximately 100-, 30-, and 86-fold higher for GES-5, against imipenem, ertapenem, and meropenem, respectively) as well as the moderate increase in Km values (approximately 2-, 3-, and 10-fold higher for GES-18 and approximately 3-, 3.7-, and 10-fold higher for GES-5, respectively).
GES-18 and GES-5 were also active against cefoxitin (Table 2). The kcat values were approximately 42- and 56-fold higher than GES-1, but the Km values also increased (approximately 17- and 14-fold, respectively), resulting in kcat/Km ratios only 2.5- and 4-fold higher, respectively.
GES-18 and GES-5 had relatively poor efficiencies against ceftazidime (the kcat/Km ratios were 22- and 19-fold lower than that of GES-1). The kinetic parameters for cefotaxime hydrolysis indicated that GES-1 was the most efficient enzyme against this substrate (Table 2). Aztreonam was also a poor substrate for GES-5 and GES-18, with very low kcat values and relatively high Km values (Table 2).
Comparison of susceptibilities to inhibitors.
We tested two commercial β-lactamase inhibitors and found that tazobactam was the most active against all three GES β-lactamase variants. GES-1 was the most susceptible to inhibition by clavulanic acid and tazobactam (the IC50s of GES-1 were 5 and 3 μM, respectively). Higher apparent IC50s for both inhibitors were seen with the Ser170-containing GES-5 and GES-18 variants. GES-18 exhibited IC50s of 25 and 10 μM for clavulanic acid and tazobactam, respectively. For GES-5, these values were 20 and 10 μM, respectively.
Structure determination.
To gain insight into the differences in substrate specificity between GES-1 and the GES-18 variant with the Ser170 substitution, the crystal structure of GES-18 was solved by molecular replacement using the structure of GES-1 (PDB code 2QPN) as a model. After refinement, almost all residues lay in the allowed or favored regions in the Ramachandran plot. Stereochemical parameters were calculated using PROCHECK (35) and fell within the range expected for structures with similar resolutions. The crystallographic and model statistics for the structure are summarized in Table 3. Attempts to generate good-quality GES-5 crystals have been unsuccessful so far.
Table 3.
X-ray data collection and structure refinement
| Parameter | GES-18 |
|---|---|
| Data collection statistics | |
| Unit cell parameters (Å) | |
| a | 74.184 |
| b | 80.506 |
| c | 87.748 |
| Space group | P212121 |
| Molecules/asymmetric unit | 2 |
| Resolution range (Å) | 25–1.87 |
| No. of observations | 336,536/45,819 |
| No. of unique reflections | 43,240/5,966 |
| Overall completeness (%) | 96.2/90.7 |
| I/σI | 32.8/5.9 |
| Multiplicity | 7.8/4.3 |
| Rsym | 0.051/0.267 |
| Refinement statistics | |
| Resolution range (Å) | 25–1.9 |
| No. of reflections | 41,653 |
| No. of atoms (non-H) | 4,741 |
| Rwork | 0.1576 |
| Rfree | 0.1955 |
| RMSD from ideal | |
| Bond (Å) | 0.009 |
| Angle (°) | 1.418 |
The crystal structure of GES-18 was refined to a resolution of 1.9 Å, yielding an Rwork value of 0.1576 and an Rfree value of 0.1955. The crystals belong to the P212121 space group, with two molecules of GES-18 in the asymmetric unit.
The final atomic resolution featured two independent structural models, model A comprising 269 amino acids (residues Ser24 to Lys295) and model B comprising 268 amino acids (residues Ser24 to Asp294). Furthermore, 440 water molecules were identified in the electron density map. Both models are related by a 2-fold noncrystallographic rotation axis and present an interface of approximately 890 Å2 dominated by hydrogen bonds and bridging water molecules. Superimposition of the individually refined models showed that they are essentially identical in structure, with a Cα root mean square deviation (RMSD) of 0.313 Å.
The structures of the GES-18 monomers show the typical class A serine β-lactamase fold comprising two domains, one α/β and the other all α (Fig. 2). The GES-18 dimer is essentially identical to the dimer in the P21 asymmetric unit of the GES-1 and GES-2 structures (13, 36). A superposition yields RMSD values of 0.454 Å and 0.336 Å compared to GES-1 and GES-2, respectively.
Fig 2.
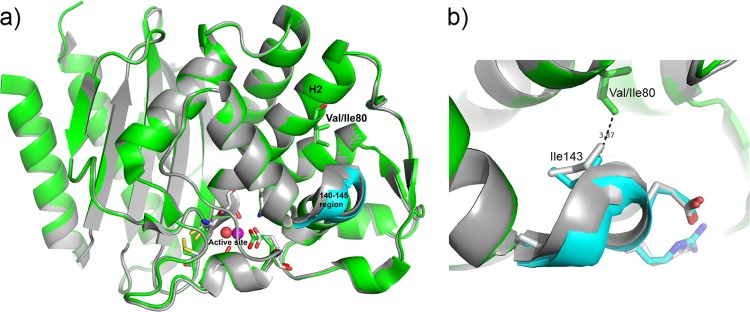
Superimposition of the crystallographic structures of GES-18 (in green) and GES-1 (in gray). (a) The active sites and residues (Val/Ile) at position 80 are represented by sticks. The respective hydrolytic water molecules are represented by spheres (in red for GES-18 and in magenta for GES-1). The 140-to-145 region which is slightly perturbed by the Val80Ile mutation is colored in cyan for GES-18. (b) Residues at position 80, Val or Ile, are represented by sticks (in green for GES-18 and in gray for GES-1). The 140-to-145 region which is slightly perturbed by the Val80Ile mutation is colored in cyan for GES-18. The residues 140 to 145 are also represented by sticks (cyan for GES-18 and gray for GES-1).
Residue 80 is located in a hydrophobic core within helix H2, which bears the catalytic Ser70 of the class A signature motif SXXK at its N terminus (Fig. 2). This residue is partially oriented to the protein surface. In GES-18, Val80 is replaced by Ile, which has a larger side chain (Fig. 2). Residues 140 to 145 of the neighboring region are situated at a typical distance for hydrophobic contacts from Ile80 and are marginally displaced in the GES-18 model compared to GES-1. The distance between Ile80:CD and Ile143:CG2 is 3.87 Å (Fig. 2b). The substitution with Ile possibly allows the protein to close a small hydrophobic cavity and thus reduces its hydrophobic surface area, perhaps influencing the stability and also the flexibility of the protein structure. Due to its location on the catalytically important SXXK-bearing helix H2, the mutation might indirectly influence the catalytic behavior.
Hydrogen bonding between Glu166, Lys73, and Ser70 is crucial in the activation of the Ser70 hydroxyl group for the nucleophilic attack of the β-lactam ring (acylation) and that of the hydrolytic water for the hydrolysis of the acyl enzyme intermediate (deacylation) (37–40). In GES-18, the presence of the Ser170 residue modifies the network of hydrogen bonds in the active site. In particular, the side chain of the Ser170 residue can form a hydrogen bond with the hydrolytic water molecule and may therefore stabilize the water molecule in the active site (Fig. 3a). In GES-2, the hydrolytic water molecule is also hydrogen bonded to the side chain of Asn170 (PDB code 3NI9) (36). Moreover, in GES-18, the hydrolytic water molecule is shifted by 2.15 Å compared to its position in GES-1 (Fig. 3b) and by 2.67 Å compared to its position in GES-2. The Ser170 side chain therefore potentially stabilizes the network of three catalytic residues (Ser70, Lys73, and Glu166) by forming an additional hydrogen bond with the carboxylate group of Glu166 (Fig. 3a). Furthermore, the distance between Ser70 and Glu166 is shorter in GES-18 (3.06 Å) than in GES-1 (3.96 Å) and in GES-2 (4.38 Å). Consequently, the hydroxyl moiety of Ser70 can form a direct hydrogen bond with the Glu166 carboxyl group (Fig. 3a), an interaction that could enhance the nucleophilicity of the Ser70 hydroxyl group for the initial attack on the carbonyl C atom of the β-lactam ring.
Fig 3.
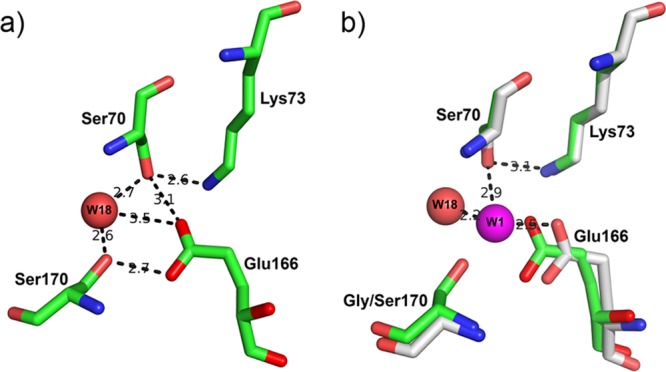
(a) Representation of the active site of GES-18. The network of hydrogen bonds is represented. (b) Superimposition of the respective active sites of GES-1 (in gray) and GES-18 (in green). The hydrolytic water molecule is represented in magenta for GES-1 (W1) and in red for GES-18 (W18). W18 is shifted by 2.15 Å compared to its position in GES-1. The hydrogen bond distances between W1 and Ser70, Lys73, and Glu166 are also represented.
DISCUSSION
We have characterized GES-18, a novel carbapenem-hydrolyzing GES-type β-lactamase from P. aeruginosa. GES-18 differs from GES-5 by a single Val80Ile substitution located on helix H2 in a hydrophobic core distant from the catalytic center. The Val80 residue in CTX-M-1 is known to be involved in cefotaxime hydrolysis, and a Val80Ala substitution may affect the dynamics and flexibility of this enzyme (41). We have shown that the Val80Ile substitution in GES-18 does not significantly modify the substrate profile in comparison to GES-5, but the kcat value was modified for some substrates.
As previously shown for GES-5 (39), the presence of Ser170 increased the carbapenemase and cephamycinase activities of the enzyme compared to those of GES-1. Cefoxitin is a poor substrate for most class A β-lactamases, and cephamycinase activity is typically associated with class C β-lactamases (42). The ability of GES variants containing Ser170 to use cefoxitin and imipenem as the substrates may be associated with the different positioning of the 7α-methoxy group or the equivalent 6α-hydroxyethyl substituent of the substrate (39). Furthermore, the Gly170Ser substitution markedly reduces the activity of the enzyme against ceftazidime and aztreonam. These antibiotics share the same (C-3/C-7) side chain even though the latter is attached to a different nucleus. In agreement with a recent study (43), we also found that the Gly170Ser substitution affected resistance to β-lactamase inhibitors. From a structural perspective, the Gly170Ser substitution induces subtle local rearrangements in the active site (Fig. 3). Ser70 and Ser170 form hydrogen bonds with Glu166, and the hydrolytic water molecule presents an additional H bond with Ser170, yielding a more rigid structure. The flexibility of the protein backbone is also reduced through the replacement of a glycine by a serine residue. Molecular dynamic simulations of GES apoenzymes have also demonstrated that in GES-5, the Ω loop and the catalytic Ser70, Lys73, and Glu166 residues are less mobile than the corresponding regions of GES-1 (39). Furthermore, our structure demonstrates that the hydrolytic water molecule and Glu166 were displaced compared to their counterparts in GES-1, potentially achieving positioning that is catalytically more favorable for the hydrolysis of carbapenems or cephamycins.
Although the solved structure of the GES-18 apoenzyme provides insight into the differences in substrate specificity induced by the Gly170Ser substitution, it does not explain the strong positive effect on the acylation and deacylation rates for imipenem (e.g., GES-5 acylates imipenem 5,000 times faster than does GES-1 and deacylates this substrate 15 times faster [26, 39]). This challenge can be addressed by solving the crystal structures of enzyme complexes with carbapenem substrates.
In conclusion, the Val80Ile substitution does not significantly alter the substrate profile of the enzyme compared to that of GES-5. The Gly170Ser substitution in GES-18 affects not only cephamycin and carbapenem hydrolysis but also inhibitor resistance. The crystallographic structure of GES-18 is similar to other solved GES structures (GES-1 and GES-2), the Val80Ile and Gly170Ser substitution inducing only minor local rearrangements, but these included the displacements of the hydrolytic water molecule and the catalytically important Glu166 residue, which altogether could provide the structural basis for partially explaining differences in substrate specificity and performance.
ACKNOWLEDGMENTS
We thank Jean-Marie Frère for helpful discussions about kinetics and for reviewing the manuscript.
The work in Aachen (Germany) was supported by the European Regional Development Fund (ERDF), the European Union (Die Europäische Kommission investiert in Ihre Zukunft), and the Alma in Silico project financed by the Interreg IV Program. C. Bebrone was a fellow of the Alexander von Humboldt Foundation (Bonn, Germany). The work in Belgium was supported by EU grant FP7-HEALTH-2009-SINGLE-STAGE TEMPOtest-QC, project 241742, and the INAMI-RIZIV/WIV-ISP funded national reference center for antibiotic-resistant Pseudomonas and Acinetobacter.
Footnotes
Published ahead of print 31 October 2012
REFERENCES
- 1. Bogaerts P, Naas T, El Garch F, Cuzon G, Deplano A, Delaire T, Huang TD, Lissoir B, Nordmann P, Glupczynski Y. 2010. GES extended-spectrum β-lactamases in Acinetobacter baumannii isolates in Belgium. Antimicrob. Agents Chemother. 54:4872–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bonnin RA, Nordmann P, Potron A, Lecuyer H, Zahar JR, Poirel L. 2011. Carbapenem-hydrolyzing GES-type extended spectrum β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55:349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moubareck C, Brémont S, Conroy MC, Courvalin P, Lambert T. 2009. GES-11, a novel integron-associated GES variant in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:3579–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Naas T, Poirel L, Nordmann P. 2008. Minor extended-spectrum β-lactamases. Clin. Microbiol. Infect. 14:42–52 [DOI] [PubMed] [Google Scholar]
- 5. Pasteran F, Faccone D, Petroni A, Rapoport M, Galas M, Vázquez M, Procopio A. 2005. Novel variant (bla(VIM-11)) of the metallo-β-lactamase bla(VIM) family in a GES-1 extended-spectrum-β-lactamase-producing Pseudomonas aeruginosa clinical isolate in Argentina. Antimicrob. Agents Chemother. 49:474–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Picão RC, Poirel L, Gales AC, Nordmann P. 2009. Diversity of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolates causing bloodstream infections in Brazil. Antimicrob. Agents Chemother. 53:3908–3913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Poirel L, Weldhagen GF, De Champs C, Nordmann P. 2002. A nosocomial outbreak of Pseudomonas aeruginosa isolates expressing the extended-spectrum β-lactamase GES-2 in South Africa. J. Antimicrob. Chemother. 49:561–565 [DOI] [PubMed] [Google Scholar]
- 8. Wang J, Zhou JY, Qu TT, Shen P, Wei ZQ, Yu YS, Li LJ. 2010. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Chinese hospitals. Int. J. Antimicrob. Agents 35:486–491 [DOI] [PubMed] [Google Scholar]
- 9. Viedma E, Juan C, Acosta J, Zamorano L, Otero JR, Sanz F, Chaves F, Oliver A. 2009. Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum β-lactamases GES-1 and GES-5 in Spain. Antimicrob. Agents Chemother. 53:4930–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kotsakis SD, Papagiannitsis CC, Tzelepi E, Legakis NJ, Miriagou V, Tzouvelekis LS. 2010. GES-13, a β-lactamase variant possessing Lys-104 and Asn-170 in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 54:1331–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ambler RP, Coulson AF, Frère JM, Ghuysen JM, Joris B, Forsman M, Levesque RC, Tiraby G, Waley SG. 1991. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276:269–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smith CA, Caccamo M, Kantardjieff KA, Vakulenko S. 2007. Structure of GES-1 at atomic resolution: insights into the evolution of carbapenemase activity in the class A extended-spectrum β-lactamases. Acta Crystallogr. Sect. D 63:982–992 [DOI] [PubMed] [Google Scholar]
- 14. Bae IK, Lee YN, Jeong SH, Hong SG, Lee JH, Lee SH, Kim HJ, Youn H. 2007. Genetic and biochemical characterization of GES-5, an extended-spectrum class A β-lactamase from Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 58:465–468 [DOI] [PubMed] [Google Scholar]
- 15. Poirel L, Weldhagen GF, Naas T, De Champs C, Dove MG, Nordmann P. 2001. GES-2, a class A β-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob. Agents Chemother. 45:2598–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vourli S, Giakkoupi P, Miriagou V, Tzelepi E, Vatopoulos AC, Tzouvelekis LS. 2004. Novel GES/IBC extended-spectrum β-lactamase variants with carbapenemase activity in clinical enterobacteria. FEMS Microbiol. Lett. 234:209–213 [DOI] [PubMed] [Google Scholar]
- 17. Wachino J, Doi Y, Yamane K, Shibata N, Yagi T, Kubota T, Arakawa Y. 2004. Molecular characterization of a cephamycin-hydrolyzing and inhibitor-resistant class A β-lactamase, GES-4, possessing a single G170S substitution in the omega-loop. Antimicrob. Agents Chemother. 48:2905–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Riezer J, Saier MH, Hancock RE, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 19. Jacoby GA. 1974. Properties of R plasmids determining gentamicin resistance by acetylation in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 6:239–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing. CLSI M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 21. Naas T, Cuzon G, Bogaerts P, Glupczynski Y, Nordmann P. 2011. Evaluation of a DNA microarray (Check-MDR CT102) for rapid detection of TEM, SHV, and CTX-M extended-spectrum β-lactamases and of KPC, OXA-48, VIM, IMP, and NDM-1 carbapenemases. J. Clin. Microbiol. 49:1608–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lévesque C, Piché L, Larose C, Roy PH. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kieser T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19–36 [DOI] [PubMed] [Google Scholar]
- 24. Delbrück H, Bogaerts P, Kupper MB, Rezende de Castro R, Bennink S, Glupczynski Y, Galleni M, Hoffmann KM, Bebrone C. 2012. Kinetic and crystallographic studies of extended spectrum GES-11, GES-12 and GES-14 β-lactamases. Antimicrob. Agents Chemother. 56:5618–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cornish-Bowden A. 2001. Fundamentals of enzyme kinetics. Portland Press Ltd., London, United Kingdom [Google Scholar]
- 26. Frase H, Shi Q, Testero SA, Mobashery S, Vakulenko SB. 2009. Mechanistic basis for the emergence of catalytic competence against carbapenem antibiotics by the GES family of β-lactamases. J. Biol. Chem. 284:29509–29513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kabsch W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795–800 [Google Scholar]
- 28. Collaborative Computational Project Number 4 1994. CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D 50:760–763 [DOI] [PubMed] [Google Scholar]
- 29. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perrakis A, Morris R, Lamzin VS. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458–463 [DOI] [PubMed] [Google Scholar]
- 31. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D 53:240–255 [DOI] [PubMed] [Google Scholar]
- 33. Painter J, Merritt EA. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. Sect. D 62:439–450 [DOI] [PubMed] [Google Scholar]
- 34. Moura A, Soares M, Pereira C, Leitao N, Henriques I, Correia A. 2009. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25:1096–1098 [DOI] [PubMed] [Google Scholar]
- 35. Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 36. Frase H, Smith CA, Toth M, Champion MM, Mobashery S, Vakulenko SB. 2011. Identification of products of inhibition of GES-2 β-lactamase by tazobactam by X-ray crystallography and spectrometry. J. Biol. Chem. 286:14396–14409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Drawz SM, Bonomo RA. 2010. Three decades of beta-lactamase inhibitors. Clin. Microbiol. Rev. 23:160–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Golemi-Kotra D, Meroueh SO, Kim C, Vakulenko SB, Bulychev A, Stemmler AJ, Stemmler TL, Mobashery S. 2004. The importance of a critical protonation state and the fate of the catalytic steps in class A β-lactamases and penicillin-binding proteins. J. Biol. Chem. 279:34665–34673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kotsakis SD, Miriagou V, Tzelepi E, Tzouvelekis LS. 2010. Comparative biochemical and computational study of the role of naturally occurring mutations at Ambler positions 104 and 170 in GES β-lactamases. Antimicrob. Agents Chemother. 54:4864–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meroueh SO, Fisher JF, Schlegel HB, Mobashery S. 2005. Ab initio QM/MM study of class A β-lactamase acylation: dual participation of Glu166 and Lys73 in a concerted base promotion of Ser70. J. Am. Chem. Soc. 127:15397–15407 [DOI] [PubMed] [Google Scholar]
- 41. Pérez-Llarena FJ, Kerff F, Abián O, Mallo S, Fernández MC, Galleni M, Sancho J, Bou G. 2011. Distant and new mutations in CTX-M-1 β-lactamase affect cefotaxime hydrolysis. Antimicrob. Agents Chemother. 55:4361–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Philippon A, Arlet G, Jacoby GA. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Frase H, Toth M, Champion MM, Antunes NT, Vakulenko SB. 2011. Importance of position 170 in the inhibition of GES-type β-lactamases by clavulanic acid. Antimicrob. Agents Chemother. 55:1556–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]


