Abstract
Pathogenic Leptospira spp., the causative agents of leptospirosis, are slow-growing Gram-negative spirochetes. Isolation of Leptospira from clinical samples and testing of antimicrobial susceptibility are difficult and time-consuming. Here, we describe the development of a new solid medium that facilitates more-rapid growth of Leptospira spp. and the use of this medium to evaluate the Etest's performance in determining antimicrobial MICs to drugs in common use for leptospirosis. The medium was developed by evaluating the effects of numerous factors on the growth rate of Leptospira interrogans strain NR-20157. These included the type of base agar, the concentration of rabbit serum (RS), and the concentration and duration of CO2 incubation during the initial period of culture. The highest growth rate of NR-20157 was achieved using a Noble agar base supplemented with 10% RS (named LVW agar), with an initial incubation at 30°C in 5% CO2 for 2 days prior to continuous culture in air at 30°C. These conditions were used to develop the Etest for three species, L. interrogans (NR-20161), L. kirschnerii (NR-20327), and L. borgpetersenii (NR-20151). The MICs were read on day 7 for all samples. The Etest was then performed on 109 isolates of pathogenic Leptospira spp. The MIC90 values for penicillin G, doxycycline, cefotaxime, ceftriaxone, and chloramphenicol were 0.64 units/ml and 0.19, 0.047, 0.5, and 2 μg/ml, respectively. The use of LVW agar, which enables rapid growth, isolation of single colonies, and simple antimicrobial susceptibility testing for Leptospira spp., provides an opportunity for new areas of fundamental and applied research.
INTRODUCTION
Leptospirosis is a zoonotic disease caused by pathogenic members of the genus Leptospira (1, 2). This infection has a worldwide distribution but has the greatest impact on health in developing countries, where it is often grossly underrecognized (1–4). Making a diagnosis of leptospirosis in resource-limited settings is challenging because of the similarity between clinical features of this and other common infectious diseases that occur in the tropics such as scrub typhus, dengue, and malaria, combined with a lack of easy-to-use and affordable diagnostic tests (5).
In areas of the world where diagnostic microbiology capabilities are available, agar-based culture techniques continue to represent an effective and simple methodology for the isolation of a wide range of bacterial pathogens and testing of antimicrobial susceptibility. They are inexpensive relative to other diagnostic methodologies and often provide information on species and antimicrobial susceptibility within 24 to 48 h of a culture becoming positive. This is not the case for leptospirosis. Although Leptospira cells are present in blood during the first week of illness (6), these fastidious microorganisms do not grow readily in standard laboratory media. Culture can be performed but requires specific semisolid or liquid media and considerable expertise and may take several weeks or months before the organism is isolated (6). Furthermore, antimicrobial therapy is invariably empirical because of a lack of simple antimicrobial susceptibility testing methods for Leptospira (7). Broth macro- and microdilution methods have been described (8–16), but these are too time-consuming for routine diagnostic microbiology laboratories. Simple diffusion-based methods such as disk diffusion and Etest methodology are not described for these species. Drug resistance appears to be rare based on clinical response (7, 17, 18), but detection of the emergence of resistance would inevitably be slow, and suspicion for this would be based on an unexpectedly poor clinical outcome.
Here, we describe the development of a new agar that facilitates rapid growth of single colonies of pathogenic Leptospira and the use of this medium to develop the Etest to determine antimicrobial MICs to drugs in common use for leptospirosis.
MATERIALS AND METHODS
Bacterial isolates.
A total of 109 pathogenic Leptospira isolates were used during the development and evaluation of the new solid medium and Etest. This collection consisted of 100 clinical isolates from Asia, including Thailand (n = 63), Lao PDR (n = 28), and Sri Lanka (n = 9), together with 9 isolates from a reference collection that originated from Indonesia (n = 4), China (n = 1), Belgium (n = 1), Japan (n = 1), Denmark (n = 1), and the United States (n = 1). All organisms were maintained by continuous culture (stock culture) in semisolid EMJH medium containing 3% rabbit serum (RS) and 0.1% bacteriological agar.
Development of LVW agar.
The new solid medium, LVW agar, was developed by testing the effects of combinations of ingredients and incubation conditions on time to first visible colonies. All recipes contained three core ingredients and a variable concentration of two additional ingredients. The core ingredients were Leptospira Medium Base EMJH (2.3 g/liter; Difco), Leptospira Enrichment EMJH (100 ml/liter; Difco), and sodium pyruvate (10 mg/liter; Merck). The first two ingredients were chosen because they are present in a commonly used medium for Leptospira culture (EMJH), and sodium pyruvate was included because of a previous report that it enhanced growth of Leptospira when added to a solid medium (19). The variable ingredients were the agar base used, bacteriological agar (Agar No. 1; Oxoid, Hampshire, England) or Noble agar (Becton, Dickinson, Sparks, MD), and rabbit serum (Gibco, Invitrogen, NY) added to a final concentration of 3%, 5%, or 10%. Bacteriological agar was selected because it is commonly used in a semisolid Leptospira medium that is recommended by a WHO reference center (20), and Noble agar was selected because its successful use in solid and semisolid Leptospira media has been described previously (21, 22). Rabbit serum is a commonly used ingredient in Leptospira media, but the percentage used ranges from 3% to 10% (23). Agar was made up to a volume of 1 liter using distilled water and sterilized, and 12 ml of mixed agar was poured into a 55-mm-diameter petri dish to achieve a depth of 4 mm. After inoculation of Leptospira, agar plates were incubated at various concentrations of CO2 and durations before being switched to incubation in air. These incubations were included because of several reports showing that CO2 enhanced the growth of Leptospira (24, 25), but the optimal concentration of CO2 and duration of CO2 incubation are unknown.
First, all agar plates were tested for time to growth (visible colonies) of Leptospira interrogans serovar Pyrogenes strain NR-20157, a clinical isolate cultured from a febrile rice farmer presenting to a hospital in northeast Thailand in 2001 (6, 26). The inoculum was prepared by adding 1 ml of stock culture to 4 ml of liquid culture medium (EMJH supplemented with 10% RS) and incubating the mixture at 30°C in air for 1 week. This was diluted to an optical density at 420 nm of 0.32 (approximately 108 CFU/ml), followed by serial dilution using EMJH medium to achieve final concentrations of 105, 104, 103, and 102 CFU/ml. Ten microliters of each concentration was drop inoculated onto one quarter of an agar plate. Agar plates were incubated at 30°C in air or initially incubated for 1, 2, 3, 4, or 5 days in either 1% or 5% CO2 at 30°C, followed by incubation at 30°C in air for a total of 28 days. Agar plates were observed with the naked eye each day, and the time to first visible Leptospira colonies was recorded. The result was recorded as “no growth” if there were no visible Leptospira colonies on day 28. The different combinations of ingredients, bacterial concentrations, and incubation conditions tested are provided in Table 1 in the supplemental material.
Table 1.
Factors associated with time to visible growth of L. interrogans strain NR-20157 on solid agara
| Culture condition | Univariable model |
Multivariable model |
||
|---|---|---|---|---|
| HRb (95% CI) | P value | HR (95% CI) | P value | |
| Concn (CFU/ml) of Leptospira inoculum | ||||
| 102 | 1.0 | <0.001 | 1.0 | <0.001 |
| 103 | 1.4 (0.9–2.0) | 1.6 (1.1–2.3) | ||
| 104 | 2.2 (1.5–3.2) | 3.3 (2.2–4.9) | ||
| 105 | 4.5 (3.0–6.6) | 11.3 (7.3–17.4) | ||
| Type of agar | ||||
| BA with 3% RS | 1.0 | <0.001 | 1.0 | <0.001 |
| BA with 5% RS | 1.2 (0.8–1.9) | 1.2 (0.8–1.9) | ||
| BA with 10% RS | 0.7 (0.4–1.1) | 0.5 (0.3–0.8) | ||
| Noble agar with 3% RS | 0.9 (0.6–1.4) | 0.8 (0.5–1.4) | ||
| Noble agar with 5% RS | 2.4 (1.6–3.8) | 3.7 (2.4–5.8) | ||
| Noble agar with 10% RS | 6.2 (3.9–9.9) | 15.2 (9.0–25.5) | ||
| No. of days of incubation in CO2c | ||||
| 0 | 1.0 | 0.15 | 1.0 | <0.001 |
| 1 | 1.7 (1.0–3.0) | 3.2 (1.8–5.8) | ||
| 2 | 2.0 (1.2–3.5) | 4.0 (2.3–7.0) | ||
| 3 | 1.6 (0.9–2.8) | 2.9 (1.6–5.1) | ||
| 4 | 1.4 (0.8–2.5) | 1.9 (1.1–3.4) | ||
| 5 | 1.7 (1.0.–3.0) | 2.3 (1.3–4.1) | ||
| Concn of CO2 (%)d | ||||
| 0 | 1.0 | 0.03 | NA | NA |
| 1 | 1.5 (0.9–2.5) | NA | ||
| 5 | 1.8 (1.1–3.0) | NA | ||
Abbreviations: BA, bacteriological agar base; HR, hazard ratio; RS, rabbit serum; NA, not applicable.
HR with 95% confidence interval (95% CI) represents the ratio of Leptospira growth rate compared to the baseline condition (for which HR = 1.0) over time. An HR value of more than 1.0 indicates that time to visible colonies for a given condition was shorter than the baseline condition, and vice versa.
Initial incubation in CO2 for 1 to 5 days was followed by incubation in air for a total of 28 days.
Concentration of CO2 was not a predictive factor in the multivariable model.
Development and evaluation of Leptospira Etest.
The agar that supported the earliest visible bacterial growth (named LVW agar [see Results]) was used to establish and evaluate the Etest for Leptospira spp. LVW agar was prepared, and 25 ml was poured into a 90-mm-diameter petri dish to reach a depth of 4 mm. A pilot experiment was performed using three clinical Leptospira isolates: L. interrogans serovar Autumnalis strain NR-20161 (isolated in 2003 from Lumpang, northern Thailand), L. borgpetersenii serovar Javanica strain NR-20151 (isolated in 2003 from Udon Thani, northeast Thailand), and L. kirschnerii serovar Grippotyphosa strain NR-20327 (isolated in 2002 from Udon Thani). The three isolates were grown in EMJH medium to a concentration of 108 CFU/ml as described above, and 300 μl of each culture was spread evenly across the surface of six LVW agar plates. These were preincubated at 30°C in 5% CO2 for 2 days (the optimal incubation conditions for LVW agar; see Results for more details). A single Etest strip of penicillin G, doxycycline, ceftriaxone, cefotaxime, or chloramphenicol (AB Biodisk, Solna, Sweden) was then applied to each of the five plates, the sixth plate without an Etest strip acting as a growth control. Plates were then incubated at 30°C in air and observed every day for a total of 14 days. The MIC was read on the first day that a demarcation of the inhibition ellipse within the bacterial lawn became clearly discernible to the naked eye (the MIC being the dissection point of the ellipse).
To test the reproducibility of the Etest, four technicians independently performed the assay for the three isolates (strains NR-20161, NR-20151, and NR-20327) and five antimicrobials (penicillin G, doxycycline, ceftriaxone, cefotaxime, and chloramphenicol). Each technician prepared their own Leptospira inoculum, used a different batch of LVW agar, and was blinded to the results of the other technicians.
Comparison between Etest and broth microdilution.
To evaluate agreement between MICs read by the Etest and the broth microdilution, broth microdilution susceptibility testing was performed for the three isolates (strains NR-20161, NR-20151, and NR-20327) and the five antimicrobials (penicillin G, doxycycline, ceftriaxone, cefotaxime, and chloramphenicol; Sigma-Aldrich, St. Louis, MO). Broth microdilution was performed as described by Murray et al. (13). In brief, one 96-well, round-bottomed microtiter plate was used per isolate. Each plate included a positive control (bacteria without an antimicrobial), a negative control (medium only), and serial 2-fold dilutions of each of the 5 antimicrobials with the final concentration of 25.0, 12.5, 6.25, 3.13, 1.56, 0.78, 0.39, 0.2, 0.1, 0.05, 0.025, or 0.012 μg/ml (units/ml for penicillin G). Leptospira cells were added to the positive-control and antibiotic-containing wells at a final concentration of 1 × 106 CFU/ml. After 3 days of incubation at 30°C in air, 20 μl of alamarBlue (catalogue no. DAL1025; Invitrogen, New York, NY) was added to every well. On the 5th day of incubation, the color of each well was noted, and the MIC was recorded as the lowest concentration that did not result in a blue-to-pink color change. The Etest MIC values deviate slightly from those of 2-fold dilutions, and for the purposes of making a direct comparison between Etest and broth MIC only, we rounded up the Etest results to the next broth microdilution value (e.g., 0.023 μg/ml was rounded to 0.025 μg/ml).
Antimicrobial susceptibilities of 109 pathogenic Leptospira isolates.
The validated Etest methodology was performed for 109 Leptospira isolates and five antimicrobial drugs (penicillin G, doxycycline, ceftriaxone, cefotaxime, and chloramphenicol).
Statistical analysis.
Univariable and multivariable Cox proportional hazard models were used to select the conditions associated with the fastest growth of Leptospira strain NR-20157. Analysis was based on time to event, with time measured from the inoculation of Leptospira. The outcome of interest was time to first detection of Leptospira colonies on the agar plates. Bacteriological agar with 3% RS and without CO2 incubation was selected as the baseline against which other conditions were compared (termed baseline condition). Hazard ratios (HR) were calculated based on the detection of Leptospira for a given condition compared to those observed for the baseline. An HR value of more than 1.0 indicates that time to visible colonies for a given condition was shorter than that for the baseline condition, and vice versa. The effects of the type of agar and percentage of RS used were presented as a combined variable because an interaction was observed between the two. All variables that were statistically significant in univariable analyses (at the 0.25 significance level) were included in multivariable analyses. A final multivariable model was developed using a purposeful selection method (27). Data were analyzed using Stata 12.0 (StataCorp, TX).
RESULTS
Development of LVW agar.
Development of a solid agar that supports the growth of Leptospira spp. was based on testing the effect of (i) two different base agars, (ii) three concentrations of rabbit serum (RS), and (iii) 11 different incubation atmospheres, in combination with other fixed components (2.3 g/liter Leptospira Medium Base EMJH, 100 ml/liter Leptospira Enrichment EMJH, and 10 mg/liter sodium pyruvate) that are used in alternative Leptospira media. Four different concentrations of bacterial inocula (105, 104, 103, and 102 CFU/ml of L. interrogans strain NR-20157) were used to represent a spectrum that may be present in clinical samples. These variables together gave rise to a total of 264 different conditions/samples (see Table 1 in the supplemental material). Of these, 35 (13%) had no visible growth after 28 days of incubation. The median time from Leptospira inoculation to first visible colonies was 18 days (interquartile range, 13 to 20 days). The shortest time was 10 days, which was observed for 10 different conditions. Colonies growing under all conditions were subsurface, the appearance of which is shown in Fig. 1A.
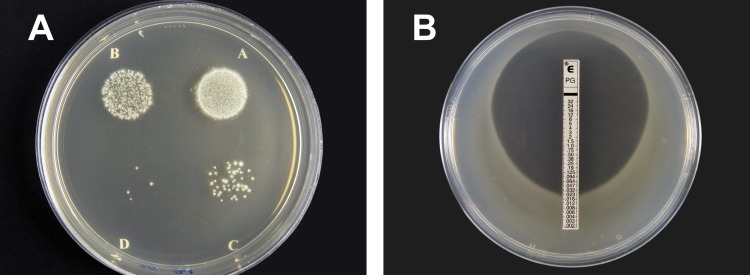
Fig 1.
(A) Colonies of Leptospira interrogans serovar Pyrogenes strain NR-20157 on LVW agar. Plates were drop inoculated with 10 μl of bacteria at an approximate concentration of 105, 104, 103, or 102 CFU/ml (A, B, C, and D, respectively) and incubated at 30°C in 5% CO2 for 2 days followed by 30°C in air for a total of 28 days. (B) Penicillin G Etest on LVW agar for Leptospira interrogans serovar Autumnalis strain NR-20161. The plate was prepared by spreading 300 μl of 108 CFU/ml and incubated at 30°C in 5% CO2 for 2 days followed by 30°C in air for a total of 7 days.
Data were analyzed using a two-step process. The optimal combination of medium and incubation conditions was initially evaluated using a univariable Cox proportional hazard model, in which the time to first visible colonies was compared with the time to first visible colonies for a baseline condition (bacteriological agar as the base agar, with 3% RS and no CO2 incubation) (Table 1). This analysis demonstrated that Noble agar base with 10% RS was associated with a 6.2 times higher rate of Leptospira growth (time to first detection) compared with bacteriological agar base with 3% RS (hazard ratio, 6.2; 95% confidence interval [CI], 3.9 to 9.9; P < 0.001 [Table 1]). Initial incubation in 5% or 1% CO2 (for any length of duration) was associated with a 1.8 or 1.5 times higher rate of Leptospira growth compared to no initial incubation in CO2, respectively (P = 0.03). There was a trend toward the highest rate of Leptospira growth for media incubated in CO2 for 2 days (P = 0.15).
Data were then analyzed using a multivariable Cox proportional hazard model to determine which of the variables had an independent effect. In the final model, we found that a higher Leptospira inoculum, Noble agar base with 10% RS, and use of an initial CO2 incubation for 2 days were independent factors associated with less time to visible Leptospira growth (Table 1). The model suggested that using the same duration of initial CO2 incubation, the concentration of CO2 (1% or 5%) was not an independent factor associated with the rate of Leptospira growth (P = 0.25; log likelihood ratio test). However, 5% CO2 was chosen based on the trend observed in the univariable model (Table 1). We concluded that the optimal solid medium contained 1% Noble agar base with 10% RS, and this medium was named LVW agar.
Development and validation of Etest for Leptospira.
Three Leptospira species (L. interrogans strain NR-20161, L. borgpetersenii strain NR-20151, and L. kirschnerii strain NR-20327) were evaluated using Etest strips for five drugs (penicillin G, doxycycline, ceftriaxone, cefotaxime, and chloramphenicol). Leptospira colonies were visible on day 4 or 5 after inoculation for all samples, but the zone of inhibition was not clear-cut until day 7 (Fig. 1B). The MICs were read on day 7 and each day for a further 7 days; no changes were observed in the MICs during this period, and so day 7 was selected for further analysis. There was good reproducibility between the results generated by four technicians who independently performed the assay for the three isolates (strains NR-20161, NR-20151, and NR-20327) and the five antimicrobials, with no more than a two-dilution difference observed for any given antimicrobial (Table 2).
Table 2.
Reproducibility of Etest MIC for Leptospira speciesa
| Antimicrobial | Strain | MIC determined by Etest |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.004 | 0.006 | 0.008 | 0.012 | 0.016 | 0.023 | 0.032 | 0.047 | 0.064 | 0.094 | 0.125 | 0.19 | 0.25 | 0.38 | 0.5 | 0.75 | 1 | ||
| Penicillin G | NR-20161 | 1 | 2 | 1 | ||||||||||||||
| NR-20151 | 2 | 2 | ||||||||||||||||
| NR-20327 | 2 | 2 | ||||||||||||||||
| Doxycycline | NR-20161 | 2 | 2 | |||||||||||||||
| NR-20151 | 1 | 3 | ||||||||||||||||
| NR-20327 | 2 | 2 | ||||||||||||||||
| Cefotaxime | NR-20161 | 1 | 1 | 2 | ||||||||||||||
| NR-20151 | 2 | 2 | ||||||||||||||||
| NR-20327 | 4 | |||||||||||||||||
| Ceftriaxone | NR-20161 | 1 | 3 | |||||||||||||||
| NR-20151 | 3 | 1 | ||||||||||||||||
| NR-20327 | 4 | |||||||||||||||||
| Chloramphenicol | NR-20161 | 1 | 3 | |||||||||||||||
| NR-20151 | 2 | 2 | ||||||||||||||||
| NR-20327 | 1 | 3 | ||||||||||||||||
Values are in μg/ml except for penicillin G values, which are in units/ml. Four technicians independently performed the assay for three isolates (strains NR-20161, NR-20151, and NR-20327) and five antimicrobial drugs. Each technician prepared their own Leptospira inoculum, used a different batch of LVW agar, and was blinded to the results of the other technicians.
Comparison between Etest and broth microdilution.
Three Leptospira species (L. interrogans strain NR-20161, L. borgpetersenii strain NR-20151, and L. kirschnerii strain NR-20327) were evaluated using a broth microdilution assay for all five drugs. Of 15 MICs, 7 (47%) were the same for both methods, 5 (33%) differed by no more than 2 dilutions, and 3 (20%) were different by 3 dilutions (Table 3).
Table 3.
Comparison of MIC determined by Etest and broth microdilution test
| Antimicrobial | Strain | MIC (μg/ml)a determined by: |
|
|---|---|---|---|
| Etestb | Broth microdilutionc | ||
| Penicillin G | NR-20161 | 0.025 | 0.025 |
| NR-20151 | 0.012 | 0.012 | |
| NR-20327 | 0.025 | 0.1 | |
| Doxycycline | NR-20161 | 0.1 | 0.39 |
| NR-20151 | 0.2 | 0.78 | |
| NR-20327 | 0.1 | 0.78 | |
| Cefotaxime | NR-20161 | 0.012 | 0.012 |
| NR-20151 | 0.012 | 0.012 | |
| NR-20327 | 0.05 | 0.05 | |
| Ceftriaxone | NR-20161 | 0.2 | 0.025 |
| NR-20151 | 0.05 | 0.025 | |
| NR-20327 | 0.2 | 0.2 | |
| Chloramphenicol | NR-20161 | 1.56 | 1.56 |
| NR-20151 | 0.78 | 6.25 | |
| NR-20327 | 1.56 | 6.25 | |
MIC values are given in units/ml for penicillin G.
The Etest MICs deviate slightly from 2-fold dilutions, and for the purposes of making a direct comparison between Etest and broth MIC only we rounded up the Etest results to the next broth microdilution value (e.g., 0.023 μg/ml was rounded to 0.025 μg/ml).
The 2-fold dilution series for the broth microdilution method included concentrations of 25.0, 12.5, 6.25, 3.13, 1.56, 0.78, 0.39, 0.2, 0.1, 0.05, 0.025, and 0.012 μg/ml.
Antimicrobial susceptibilities of 109 pathogenic Leptospira isolates by Etest.
The MICs to the five test antimicrobial drugs were determined for a total of 109 isolates using the Etest (see Table S2 in the supplemental material). The MIC90 values were as follows: penicillin G, 0.064 units/ml; doxycycline, 0.19 μg/ml; cefotaxime, 0.047 μg/ml; ceftriaxone, 0.5 μg/ml; and chloramphenicol, 2 μg/ml (Table 4). There was no difference in MIC90 values for isolates originating from Thailand, Lao PDR, Sri Lanka, and a set of reference strains from other countries (Table 4). The exception was the MIC90 of penicillin G in Sri Lanka, which was higher than for isolates from elsewhere (1 unit/ml versus ≤0.064 unit/ml), an observation that could be explained by a single outlier with an MIC of 1 unit/ml in Sri Lanka.
Table 4.
MICs of five antimicrobial agents for 109 pathogenic Leptospira isolates
| Origin of isolates (n) | MIC90 (range) (μg/ml)a |
||||
|---|---|---|---|---|---|
| Penicillin G | Doxycycline | Cefotaxime | Ceftriaxone | Chloramphenicol | |
| All origins (109) | 0.064 (≤0.002–1) | 0.19 (0.023–0.5) | 0.047 (≤0.002–0.125) | 0.5 (0.006–2) | 2 (0.094–3) |
| Thailand (63) | 0.047 (≤0.002–0.75) | 0.19 (0.023–0.5) | 0.032 (≤0.002–0.094) | 0.38 (0.006–1) | 2 (0.094–3) |
| Lao PDR (28) | 0.094 (≤0.002–0.125) | 0.25 (0.032–0.25) | 0.047 (≤0.002–0.064) | 0.5 (0.008–2) | 1.5 (0.125–2) |
| Sri Lanka (9) | 1 (0.006–1) | 0.38 (0.047–0.38) | 0.032 (0.003–0.032) | 0.38 (0.012–0.38) | 1.5 (0.094–1.5) |
| Othersb (9) | 0.064 (≤0.002–0.064) | 0.19 (0.047–0.19) | 0.125 (≤0.002–0.125) | 1.5 (0.047–1.5) | 2 (0.25–2) |
MIC values are given in units/ml for penicillin G.
Others were reference isolates from Indonesia (4 isolates), China (1 isolate), Belgium (1 isolate), Japan (1 isolate), Denmark (1 isolate), and the United States (1 isolate).
DISCUSSION
In regions of the world where diagnostic microbiology is available, bacteriological agar plays a central role in diagnosing bacterial diseases and evaluating antimicrobial resistance of the causative pathogens. In this study, we developed a solid agar that supports the rapid and visible growth of Leptospira colonies. Using LVW agar, we also developed diffusion-based antimicrobial susceptibility testing of Leptospira spp. on solid medium. The Etest was selected because it is a rapid, simple, and well-established method for antimicrobial resistance testing with easy-to-read MICs. Rapid growth and clear MICs could be read within 7 days after bacterial inoculation for all 109 Leptospira isolates tested. The ingredients of LVW agar are commercially available (1% Noble agar base, 10% RS, 2.3 g/liter Leptospira Medium Base EMJH, 100 ml/liter Leptospira Enrichment EMJH, and 10 mg/liter sodium pyruvate), and preparation is straightforward.
Several previous studies have described the growth of Leptospira on solid agar (24, 28, 29), although growth on these usually took more than 2 weeks. The first report in 1956 by Cox and Larson described a medium containing 1% agar (Difco), 10% RS, rabbit erythrocyte lysate, and 0.2% tryptose phosphate broth (Difco) (30). Colonies of the 10 Leptospira isolates evaluated were observed within 15 days of incubation in air at 30°C (30). In 1963, Yanagawa et al. used the Cox and Larson agar and observed colonies of 24 pathogenic Leptospira isolates within 21 days of incubation in 1% CO2 at 30°C (25). In 1973, Johnson et al. used solid medium containing 1% agar, 100 mg/liter pyruvate, and Tween 80-albumin medium and observed colonies of 2 pathogenic isolates (Leptospira interrogans serovars Canicola and Hardjo) within 30 days (19). In 1979, Bromley and Charon used solid medium containing 1% Noble agar and EMJH medium, on which colonies of L. interrogans serovar Illini were observed within 10 days of incubation in air at 30°C (21). In 1986, Rule and Alexander used solid medium containing gellan gum (Gelrite; Merck Co.) and EMJH medium, on which colonies of 11 pathogenic Leptospira isolates were observed within 20 days of incubation in air at 30°C (31). Using LVW agar and an inoculum of 108 CFU/ml, we showed that colonies of all 109 pathogenic Leptospira isolates tested could be observed within 7 days after bacterial inoculation.
The rapid growth of Leptospira on LVW agar is probably associated with several factors. We observed that a combination of Noble agar with 10% RS provided the fastest growth of Leptospira compared to the baseline condition (bacteriological agar with 3% RS). Noble agar has a lower percentage of ash, calcium, magnesium, potassium, sodium, sulfate, and sulfur than bacteriological agar, and one or more of these variables may facilitate the growth of Leptospira. The requirement for a high percentage of RS is consistent with previous studies, suggesting that the nitrogen source and thiamine in RS can stimulate growth of Leptospira (32). We found that an initial CO2 incubation for 2 days improved the rate of growth of Leptospira, although paradoxically the growth rate was decreased if the duration of CO2 incubation was longer than 2 days, with inhibition of growth if exposed to CO2 for the entire incubation period of 28 days (data not shown). Growth stimulation by CO2 is consistent with previous reports (24, 25). Unsurprisingly, a high initial bacterial inoculum was also associated with a shorter duration to culture positivity.
Leptospira colonies observed on LVW agar are very similar to those described previously on solid medium (30, 31) and are subsurface. One study on the growth of two isolates of L. interrogans (serovar Ponoma and serovar Icterohaemorrhagiae) on solid medium containing 1% agar, 100 mg/liter pyruvate, 10% RS, and EMJH medium reported the presence of both surface and subsurface colonies (29). This was not observed for LVW agar in our study, which also included L. interrogans serovar Ponoma and serovar Icterohaemorrhagiae.
Rapid, simple, and easy-to-read antimicrobial susceptibility testing methodology is needed for Leptospira spp. Broth macro- and microdilution methods have been developed and used to determine the MICs of small collections of Leptospira isolates (8, 9, 11–16). However, broth dilution techniques are rarely used in tropical settings because of cost and demands on time and technical skill. The Etest method developed here is based on a widely used diffusion-based methodology. It provides clear MIC results within 7 days with good reproducibility between technicians and batch tests and is comparable overall with the MIC results determined by broth microdilution.
The MIC90s of penicillin G, doxycycline, and cefotaxime defined for the 109 pathogenic isolates tested in this study were within 2 dilutions of the MIC90 results reported in 2008 by Murray et al. using the broth microdilution technique for 13 pathogenic isolates (16), while the MIC90 of cefotaxime was within 3 dilutions (16). In addition, the MIC90 of chloramphenicol reported in our study was within 2 dilutions of the MIC90 previously reported in a study by Murray et al. in 2004, in which 26 Leptospira isolates were included (13, 14). In general, we did not observe regional differences in antimicrobial susceptibility in Thailand, Lao PDR, and Sri Lanka, and our findings support the continued use of penicillin, cephalosporin, and doxycycline for the treatment of leptospirosis in these areas.
Rapid, sensitive, and easy-to-perform methods to isolate Leptospira from clinical and environmental specimens are required in rural tropical settings where leptospirosis is endemic (3, 5, 24, 33). Further investigations of the utility of LVW agar for such purposes are under way. We speculate that LVW agar, which enables rapid growth, isolation of single colonies, and simple antimicrobial susceptibility testing for Leptospira spp., will become widely used in diagnostic microbiology and during fundamental and applied research.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following people who provided Leptospira isolates: Thareerat Kalambaheti, Department of Microbiology and Immunology, Faculty of Tropical Medicine, Mahidol University; Paul Newton, LOMWRU, Mahosot hospital, Vientiane, Lao PDR; Hitanadura Janika De Silva, University of Kelaniya, Colombo, Sri Lanka; and Lee Smythe, WHO/FAO/OIE Collaborating Centre for Reference & Research on Leptospirosis, Centre for Public Health Sciences, Queensland Health Scientific Services, Brisbane, Australia. We thank Sunee Chueasuwanchai, Amornwadee Sangkakam, Wilarat Jedsadapanpong, and the staff of the Mahidol-Oxford Tropical Medicine Research Unit for their assistance. We thank Prapass Wannapinij for computational support.
D.H.P. was supported by a Wellcome Trust Clinical Research Training Fellowship (078990/Z/06/Z), D.L. was supported by a project grant awarded by the Wellcome Trust (090219/Z/09/Z), and S.J.P. was supported by the NIHR Cambridge Biomedical Research Centre. This study was funded by the Wellcome Trust.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 31 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01812-12.
REFERENCES
- 1. Ko AI, Goarant C, Picardeau M. 2009. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 7:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Levett PN. 2001. Leptospirosis. Clin. Microbiol. Rev. 14:296–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adler B, de la Pena Moctezuma A. 2010. Leptospira and leptospirosis. Vet. Microbiol. 140:287–296 [DOI] [PubMed] [Google Scholar]
- 4. Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, Levett PN, Gilman RH, Willig MR, Gotuzzo E, Vinetz JM. 2003. Leptospirosis: a zoonotic disease of global importance. Lancet Infect. Dis. 3:757–771 [DOI] [PubMed] [Google Scholar]
- 5. Suttinont C, Losuwanaluk K, Niwatayakul K, Hoontrakul S, Intaranongpai W, Silpasakorn S, Suwancharoen D, Panlar P, Saisongkorh W, Rolain JM, Raoult D, Suputtamongkol Y. 2006. Causes of acute, undifferentiated, febrile illness in rural Thailand: results of a prospective observational study. Ann. Trop. Med. Parasitol. 100:363–370 [DOI] [PubMed] [Google Scholar]
- 6. Wuthiekanun V, Chierakul W, Limmathurotsakul D, Smythe LD, Symonds ML, Dohnt MF, Slack AT, Limpaiboon R, Suputtamongkol Y, White NJ, Day NP, Peacock SJ. 2007. Optimization of culture of Leptospira from humans with leptospirosis. J. Clin. Microbiol. 45:1363–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Suputtamongkol Y, Niwattayakul K, Suttinont C, Losuwanaluk K, Limpaiboon R, Chierakul W, Wuthiekanun V, Triengrim S, Chenchittikul M, White NJ. 2004. An open, randomized, controlled trial of penicillin, doxycycline, and cefotaxime for patients with severe leptospirosis. Clin. Infect. Dis. 39:1417–1424 [DOI] [PubMed] [Google Scholar]
- 8. Harris BM, Blatz PJ, Hinkle MK, McCall S, Beckius ML, Mende K, Robertson JL, Griffith ME, Murray CK, Hospenthal DR. 2011. In vitro and in vivo activity of first generation cephalosporins against Leptospira. Am. J. Trop. Med. Hyg. 85:905–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hospenthal DR, Murray CK. 2003. In vitro susceptibilities of seven Leptospira species to traditional and newer antibiotics. Antimicrob. Agents Chemother. 47:2646–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim D, Kordick D, Divers T, Chang YF. 2006. In vitro susceptibilities of Leptospira spp. and Borrelia burgdorferi isolates to amoxicillin, tilmicosin, and enrofloxacin. J. Vet. Sci. 7:355–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murgia R, Cinco M. 2001. Sensitivity of Borrelia and Leptospira to quinupristin-dalfopristin (Synercid) in vitro. New Microbiol. 24:193–196 [PubMed] [Google Scholar]
- 12. Murray CK, Ellis MW, Hospenthal DR. 2004. Susceptibility of Leptospira serovars to antimalarial agents. Am. J. Trop. Med. Hyg. 71:685–686 [PubMed] [Google Scholar]
- 13. Murray CK, Hospenthal DR. 2004. Broth microdilution susceptibility testing for Leptospira spp. Antimicrob. Agents Chemother. 48:1548–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Murray CK, Hospenthal DR. 2004. Determination of susceptibilities of 26 Leptospira sp. serovars to 24 antimicrobial agents by a broth microdilution technique. Antimicrob. Agents Chemother. 48:4002–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oie S, Hironaga K, Koshiro A, Konishi H, Yoshii Z. 1983. In vitro susceptibilities of five Leptospira strains to 16 antimicrobial agents. Antimicrob. Agents Chemother. 24:905–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ressner RA, Griffith ME, Beckius ML, Pimentel G, Miller RS, Mende K, Fraser SL, Galloway RL, Hospenthal DR, Murray CK. 2008. Antimicrobial susceptibilities of geographically diverse clinical human isolates of Leptospira. Antimicrob. Agents Chemother. 52:2750–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Panaphut T, Domrongkitchaiporn S, Vibhagool A, Thinkamrop B, Susaengrat W. 2003. Ceftriaxone compared with sodium penicillin G for treatment of severe leptospirosis. Clin. Infect. Dis. 36:1507–1513 [DOI] [PubMed] [Google Scholar]
- 18. Watt G, Padre LP, Tuazon ML, Calubaquib C, Santiago E, Ranoa CP, Laughlin LW. 1988. Placebo-controlled trial of intravenous penicillin for severe and late leptospirosis. Lancet i:433–435 [DOI] [PubMed] [Google Scholar]
- 19. Johnson RC, Walby J, Henry RA, Auran NE. 1973. Cultivation of parasitic leptospires: effect of pyruvate. Appl. Microbiol. 26:118–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smythe LD. 2012. EMJH agar. Quality Information Systems Document number 14340, vol 3.0 WHO/FAO/OIE Collaborating Center for Reference and Research on Leptospirosis, Queensland Health Forensic and Scientific Services, Queensland Health, Queensland, Australia [Google Scholar]
- 21. Bromley DB, Charon NW. 1979. Axial filament involvement in the motility of Leptospira interrogans. J. Bacteriol. 137:1406–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Orr HS, Little TW. 1979. Isolation of Leptospira of the serotype hardjo from bovine kidneys. Res. Vet. Sci. 27:343–346 [PubMed] [Google Scholar]
- 23. Freitas JC, Silva FG, Oliveira RC, Delbem ACB, Muller EE, Alves LA, Teles PS. 2004. Isolation of Leptospira spp from dogs, bovine and swine naturally infected. Cien. Rur. Santa Maria 34:853–856 [Google Scholar]
- 24. Faine S, Adler B, Bolin C, Perolat P. 1999. Leptospira and leptospirosis. MediSci, Melbourne, Australia [Google Scholar]
- 25. Yanagawa R, Hiramune T, Fujita J. 1963. Effect of carbon dioxide on the colonial growth of pathogenic leptospirae. J. Bacteriol. 85:875–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thaipadungpanit J, Wuthiekanun V, Chierakul W, Smythe LD, Petkanchanapong W, Limpaiboon R, Apiwatanaporn A, Slack AT, Suputtamongkol Y, White NJ, Feil EJ, Day NP, Peacock SJ. 2007. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS Negl. Trop. Dis. 1:e56 doi:10.1371/journal.pntd.0000056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bursac Z, Gauss CH, Williams DK, Hosmer DW. 2008. Purposeful selection of variables in logistic regression. Source Code Biol. Med. 3:17 doi:10.1186/1751-0473-3-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stalheim OH, Wilson JB. 1963. Leptospiral colonial morphology. J. Bacteriol. 86:482–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wood J, Johnson RC, Palin K. 1981. Surface colonies of Leptospira interrogans. J. Clin. Microbiol. 13:102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cox CD, Larson AD. 1957. Colonial growth of leptospirae. J. Bacteriol. 73:587–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rule PL, Alexander AD. 1986. Gellan gum as a substitute for agar in leptospiral media. J. Clin. Microbiol. 23:500–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson RC, Gary ND. 1962. Nutrition of Leptospira pomona. 1. A chemically defined substitute for rabbit serum ultrafiltrate. J. Bacteriol. 83:668–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Limmathurotsakul D, Turner EL, Wuthiekanun V, Thaipadungpanit J, Suputtamongkol Y, Chierakul W, Smythe LD, Day NP, Cooper B, Peacock SJ. 2012. Fool's gold: why imperfect reference tests are undermining the evaluation of novel diagnostics: a reevaluation of 5 diagnostic tests for leptospirosis. Clin. Infect. Dis. 55:322–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.