Abstract
Seventy-seven porcine Enterococcus isolates with florfenicol MICs of ≥16 μg of were/ml screened for the presence of the multiresistance gene cfr, its location on plasmids, and its genetic environment. Three isolates—Enterococcus thailandicus 3-38 (from a porcine rectal swab collected at a pig farm), Enterococcus thailandicus W3, and Enterococcus faecalis W9-2 (the latter two from sewage at a different farm), carried the cfr gene. The SmaI pulsed-field gel electrophoresis patterns of the three isolates differed distinctly. In addition, E. faecalis W9-2 was assigned to a new multilocus sequence type ST469. Mating experiments and Southern blot analysis indicated that cfr is located on conjugative plasmids pW3 (∼75 kb) from E. thailandicus W3, p3-38 (∼72 kb) from E. thailandicus 3-38, and pW9-2 (∼55 kb) from E. faecalis W9-2; these plasmids differed in their sizes, additional resistance genes, and the analysis of the segments encompassing the cfr gene. Sequence analysis revealed that all plasmids harbored a 4,447-bp central region, in which cfr was bracketed by two copies of the novel insertion sequence ISEnfa4 located in the same orientation. The sequences flanking the central regions of these plasmids, including the partial tra gene regions and a ω-ε-ζ toxin-antitoxin module, exhibited >95% nucleotide sequence identity to the conjugative plasmid pAMβ1 from E. faecalis. Conjugative plasmids carrying cfr appear to play an important role in the dissemination and maintenance of the multiresistance gene cfr among enterococcal isolates and possibly other species of Gram-positive bacteria.
INTRODUCTION
Enterococci are Gram-positive bacteria that generally colonize the gastrointestinal tracts of animals. They are also found in food, water, and environmental samples. Enterococci were once considered harmless commensals of humans, but they have emerged as important nosocomial pathogens over the past 3 decades. Enterococcus spp., especially Enterococcus faecalis and Enterococcus faecium, are now the second major cause of surgical and urinary tract infections and the third major cause of bacteremia (1). The ability of Enterococcus spp. to acquire mobile genetic elements, such as plasmids and transposons carrying the determinants of antimicrobial resistance and virulence, has contributed to their emergence as often multiresistant pathogens. Antimicrobial multiresistance drastically limits the therapeutic options in the treatment of infections, and the emergence and rapid spread of glycopeptide-resistant enterococci has presented a particular challenge, since there are few remaining options for antimicrobial treatment (2). As donors and recipients of antimicrobial resistance genes, Enterococcus spp. play an important role in the dissemination of resistance genes by horizontal gene transfer. The transmission of the vanA gene cluster-carrying transposon Tn1546 from vancomycin-resistant enterococci (VRE) to methicillin-resistant Staphylococcus aureus (MRSA), creating a vancomycin- and methicillin-resistant S. aureus strain in a hospitalized patient, represents a dramatic example of this phenomenon (3).
As the first oxazolidinone introduced for clinical use in the United States since 2000 and in China since 2007, linezolid is currently the most efficient antimicrobial against VRE, MRSA, and penicillin-resistant pneumococci (4). As a last-resort antimicrobial agent, linezolid not only exhibits strong antibacterial activity but also has properties that mitigate against the development of drug resistance in bacteria (5). No evidence of a significant increase in the percentage of linezolid-resistant bacteria has been reported in the LEADER Program, whose results are available on an annual basis since 2006 (6). The most recent results from the LEADER Program for the year 2009 reported an overall percentage of linezolid resistance of 0.34% (6). The most common mechanism of linezolid resistance involves mutations in the central loop of domain V of 23S rRNA, with G2576T (Escherichia coli numbering) being the most frequent mutation (7), followed by mutations in the genes for the ribosomal proteins L3 and L4 (8). Transferable linezolid resistance is due to the gene cfr encoding an rRNA methyltransferase which methylates position 8 of A2503 (9) and inhibits ribose methylation at nucleotide C2498 in the 23S rRNA (10). Thereby, Cfr confers resistance to five chemically unrelated antimicrobial classes including phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A (11) and decreased susceptibility to the 16-membered macrolides spiramycin and josamycin (12).
The observation that the gene cfr is often located on plasmids underlines the potential for the spread of this gene (11). Although it was initially discovered on the 17.1-kb multiresistance plasmid, pSCFS1, in a bovine Staphylococcus sciuri isolate (13), the gene cfr has been detected in seven bacterial genera, including Staphylococcus, Bacillus, Enterococcus, Macrococcus, Jeotgalicoccus, Proteus, and Escherichia. Most of the corresponding isolates were derived from domestic animals (mainly pigs) (14–23), and plasmids seemed to play an important role in the interspecies and intergenus transfer of the cfr gene. To date, 17 different cfr-carrying plasmids have been reported (Table 1). Among these plasmids, only the staphylococcal plasmids pSCFS7 and pERGB and the enterococcal plasmid pHOU-cfr were characterized as conjugative plasmids (14, 25, 27). In the present study, we describe three transferable multiresistance plasmids obtained from Enterococcus spp. from domestic pigs and farm environments which carry the cfr gene in new genetic environments.
Table 1.
Overviewof the so far identified cfr-carryingplasmids in different bacteria
| Host bacteria | Plasmid | Size (kb) | Reference |
|---|---|---|---|
| Staphylococcus spp. | pSCFS1 | 17.1 | 24 |
| pSCFS3 | 35.7 | 16 | |
| pSCFS6 | ∼43 | 24 | |
| pSCFS7 | ∼45 | 25 | |
| p004-737X | ∼55 | 26 | |
| p426-3147L | ∼175 | 26 | |
| pSS-01 | ∼40 | 22 | |
| pSS-02 | ∼35.4 | 22 | |
| pSS-03 | 7.1 | 22 | |
| pERGB | ∼50 | 27 | |
| Bacillus spp. | pBS-01 | 16.5 | 28 |
| pBS-02 | 16.5 | 23 | |
| pBS-03 | 7.4 | 19 | |
| Jeotgalicoccus and Macrococcus spp. | pJP1 | ∼53 | 21 |
| E. faecalis | pEF-01 | 32.4 | 17 |
| pHOU-cfr | ∼97 | 14 | |
| E. coli | pEC-01 | ∼110 | 29 |
MATERIALS AND METHODS
Bacterial isolates and detection of florfenicol resistance genes.
A total of 77 Enterococcus isolates (E. faecalis [n = 49], E. faecium [n = 15], E. hirae [n = 6], E. thailandicus [n = 3], E. durans [n = 2], E. asini [n = 1], and Enterococcus sp. strain CIFRI D-TSB15 [n = 1]) with florfenicol MIC values of ≥16 μg of were/ml collected from individual pigs (n = 65; 57 from rectal swabs and 8 from nasal swabs), pig farm environments (n = 9; 8 from sewage and 1 from soil), and farm workers (n = 3; all from rectal swabs) in three pig farms and one slaughterhouse in Shandong province in 2011. Although no clinical breakpoints for florfenicol applicable to enterococci are currently available, isolates with an MIC of ≥16 μg of were/ml tentatively considered as florfenicol resistant. Whole-cell DNA from the Enterococcus isolates was extracted using a commercial kit (TianGen, Beijing, China) according to the manufacturer's instructions. The presence of the florfenicol resistance genes cfr, fexA, and fexB was investigated using previously described methods (28, 30).
Molecular typing.
Pulsed-field gel electrophoresis (PFGE) was performed to investigate the clonality of the cfr-positive enterococcus isolates. DNA preparation and subsequent restriction analysis were conducted as described previously (31). Whole-cell DNA was incubated with SmaI (TaKaRa, Dalian, China) for 6 h at 30°C. The resulting restriction fragments were separated using a CHEF-DRIII system (Bio-Rad) with a clamped homogeneous electric field of 6 V/cm, using a 120° switch angle for 16 h at 14°C, with the pulse time linearly ramped from 1 s to 20 s. The cfr-positive bovine E. faecalis isolate EF-01, which was the first Enterococcus isolate of animal origin carrying the cfr gene (17), was included for comparison. In addition, typing of the cfr-positive E. faecalis isolates was performed by multilocus sequence typing (MLST), using an established set of MLST primers described previously (32).
Filter mating.
Filter mating was performed with each of the three cfr-positive original isolates using E. faecalis JH2-2 (rifampin resistance) as the recipient, following the method described previously (33). The concentrations of antibiotics in brain heart infusion agar plates used for the selection of transconjugants were 25 μg/ml for rifampin and 10 μg/ml for florfenicol. Transconjugants were confirmed as cfr positive and fexA and fexB negative by PCR analysis; PFGE was also performed in all transconjugants using SmaI to confirm that they were derivatives of the recipient strain JH2-2.
Antimicrobial susceptibility testing.
The MICs of all of the cfr-positive original Enterococcus isolates, transconjugants, and E. faecalis JH2-2 were determined using broth microdilution according to the recommendations of documents M31-A3 (34) and M100-S21 (35) of the Clinical and Laboratory Standards Institute. The reference strain E. faecalis ATCC 29212 served as a quality control.
Analysis of cfr-carrying plasmids.
To estimate the size of the cfr-carrying plasmids, whole-cell DNA of the enterococcal isolates in agarose gel plugs were treated with S1 nuclease (TaKaRa) and then separated by PFGE as described previously (36, 37); gels were run for 14 h. Plasmid DNA was extracted using a plasmid extraction midi kit (Qiagen, Germany) using a modification described previously (17). The extracted plasmids and S1 nuclease-linearized PFGE-separated plasmid DNA fragments were transferred to Hybond N+ membranes (Amersham Biosciences, USA) and hybridized with digoxigenin (DIG)-labeled cfr-, fexA-, or fexB-specific probes. The DIG High Prime DNA Labeling and Detection Starter Kit I (Roche Applied Sciences, Germany) was used for detection. The regions flanking the cfr gene were sequenced by a modified random primer sequencing walking strategy, as previously described (38). To determine the stability of the ISEnfa4 flanked segments in plasmids in both the original strains and in the transconjugants, inverse PCR was performed using the primers cfrIF and cfrIR (these are complementary to locations inside the cfr gene) (22).
Nucleotide sequence accession numbers.
The nucleotide sequences of a 27,360-bp segment of plasmid pW3, a 21,116-bp fragment of p3-38, and a 25,761-bp segment of pW9-2 have been deposited in GenBank under accession numbers JQ911739 (pW3), JQ911740 (p3-38), and JQ911741 (pW9-2), respectively.
RESULTS
Identification of the florfenicol resistance genes cfr, fexA, and fexB in Enterococcus isolates.
PCR analysis of whole-cell DNA with primers specific for the three florfenicol resistance genes—cfr, fexA, and fexB—thus far identified in Enterococcus demonstrated that these genes were present alone or in different combinations in most of the isolates included in the present study. Of the 77 enterococci studied, 68 isolates carried either fexA (n = 36), fexB (n = 14), or both genes (n = 18); only two isolates (E. thailandicus 3-38 and E. faecalis W9-2) carried both cfr and fexA, whereas only one isolate (E. thailandicus W3) carried both cfr and fexB. Interestingly, six isolates did not harbor any of these genes. Of the three cfr-positive isolates, E. thailandicus 3-38 was collected from a rectal swab of a pig, whereas E. thailandicus W3 and E. faecalis W9-2 were collected from sewage at a different pig farm. The nucleotide sequences of the amplified cfr genes in these three strains were 100% identical to the cfr gene of the S. sciuri plasmid pSCFS1 (accession number NC_005076). The 977-bp fexA amplicon from E. faecalis W9-2 was identical to the corresponding fexA sequence of the S. aureus plasmid pSCFS7 (FN995110), whereas a single nucleotide exchange (T→A) at position 551 was detected in the fexA amplicon of E. thailandicus 3-38R, which caused an amino acid change at position 71 of the phenicol exporter protein (Phe→Ile). Finally, the 786-bp fexB amplicon obtained from E. thailandicus W3 was identical to fexB from plasmid pEFM-1 (JN201336) of E. faecium.
Antimicrobial resistance, clonality, and plasmid profiles of the cfr-positive enterococci isolates.
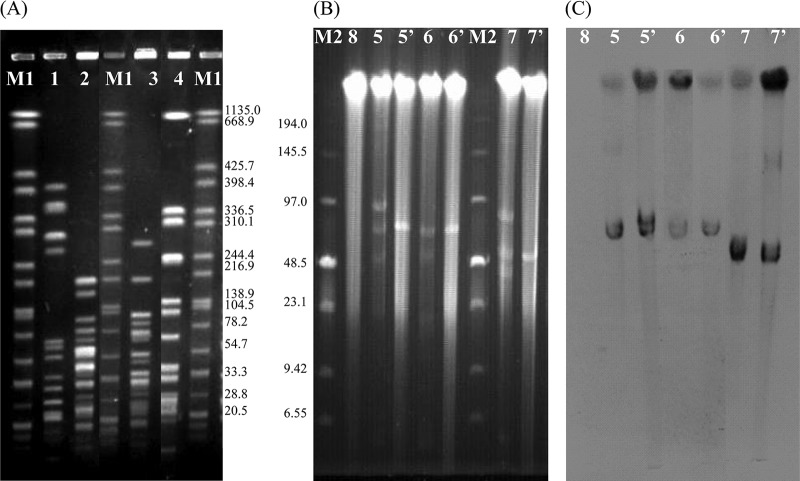
All three cfr-positive isolates exhibited resistance to chloramphenicol, erythromycin, tetracycline, ciprofloxacin, and rifampin and had elevated MICs against florfenicol (up to 64 to 128 μg/ml) but were susceptible to ampicillin and vancomycin. In addition, the MICs for linezolid were 8 μg/ml (E. thailandicus 3-38) and 4 μg/ml (E. thailandicus W3 and E. faecalis W9-2) (Table 2). PFGE analysis of the four cfr-positive Enterococcus isolates, including the previously described bovine cfr-carrying E. faecalis isolate EF-01, revealed marked genomic heterogeneity of less than 65% similarity in their SmaI patterns (Fig. 1A). MLST of E. faecalis EF-01 indicated that it belongs to ST21 within the clonal complex 21 (CC21) (39), whereas E. faecalis W9-2 represented the novel ST type ST469.
Table 2.
Antimicrobial susceptibility profiles of cfr-positive enterococci isolates W3, 3-38, and W9-2 and their transconjugants JHW3, JH3-38 and JHW9-2, for which E. faecalis JH2-2 served as recipient strain
| Isolate | Organism | Origin | MIC(μg/ml)a |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CHL | FFC | LIZ | ERY | TET | AMP | VAN | CIP | |||
| W3 | E. thailandicus | Sewage | 64 | 64 | 4 | >128 | 128 | ≤1 | ≤1 | >8 |
| 3-38 | E. thailandicus | Swine | 64 | 64 | 8 | >128 | 128 | 4 | ≤1 | >8 |
| W9-2 | E. faecalis | Sewage | 64 | 128 | 4 | >128 | 128 | ≤1 | ≤1 | >8 |
| JH2-2 | E. faecalis | NAb | 8 | 8 | 2 | 0.25 | 2 | ≤1 | ≤1 | 2 |
| JHW3 | E. faecalis | NA | 32 | 32 | 8 | 8 | 2 | ≤1 | ≤1 | 2 |
| JH3-38 | E. faecalis | NA | 32 | 32 | 8 | 8 | 2 | ≤1 | ≤1 | 2 |
| JHW9-2 | E. faecalis | NA | 32 | 32 | 8 | 8 | 2 | ≤1 | ≤1 | 2 |
CHL, chloramphenicol; FFC, florfenicol; LIZ, linezolid; ERY, erythromycin; TET, tetracycline; AMP, ampicillin; VAN, vancomycin; CIP, ciprofloxacin.
NA, not applicable.
Fig 1.
(A) SmaI-PFGE patterns from E. faecalis EF-01 and three cfr-positive isolates used in the present study. Lanes 1 to 4, E. faecalis EF-01, E. thailandicus W3, E. thailandicus 3-38, and E. faecalis W9-2, respectively; lanes M1 contain the XbaI pattern of Salmonella braenderup H9812 with the fragment sizes given in kilobases on the right-hand side. (B) Numbers and sizes of plasmids as determined by S1 nuclease PFGE. Lane M2, low-range PFG marker (NEB); lanes 5 and 5′, W3R and its transconjugant JHW3; lanes 6 and 6′, 3-38 and its transconjugant JH3-38; lanes 7 and 7′, W9-2 and its transconjugant JHW9-2; lane 8, the recipient E. faecalis JH2-2. (C) Southern hybridization with the cfr probe. The lane numbers correspond to those in panel B.
Conjugation proved successful using isolates W3, 3-38, and W9-2 as the donor strains and JH2-2 as the recipient strain; three transconjugants, designated JHW3, JH3-38 and JHW9-2, respectively, were obtained. The sizes of the plasmids present in the original strains and their transconjugants were determined by S1 nuclease-PFGE. Each of the original strains harbored at least three plasmids, but at least one visible plasmid band could be observed in each of the three transconjugants (Fig. 1B). Southern blot analysis showed that the cfr probe hybridized to a band of ∼75 kb in both W3 and JHW3, a band of ∼72 kb in both 3-38 and JH3-38, and a band of ∼55 kb in both W9-2 and JHW9-2 (Fig. 1C); these three cfr-carrying plasmids were designated pW3, p3-38 and pW9-2, respectively. In addition, we confirmed that the florfenicol exporter gene fexA was not associated with any of the plasmids in E. thailandicus 3-28 and located on an ∼40-kb plasmid in E. faecalis W9-2, while the fexB gene was located on an ∼50-kb plasmid in E. thailandicus W3 (data not shown).
MIC testing revealed that all transconjugants exhibited at least 4-fold-elevated MICs for chloramphenicol, florfenicol, and linezolid compared to the recipient JH2-2 strain. No observable MIC changes were found for tetracycline and ciprofloxacin. Interestingly, all transconjugants showed 32-fold elevated MICs for erythromycin, compared to JH2-2 (Table 2). PCR screening to detect erythromycin resistance genes using previously described primers (40, 41) revealed the presence of the erm(B) gene in transconjugants JHW3 and JHW9-2 and their original strains, while the erm(A) gene was detected in both JH3-38 and 3-38. The erm(B) gene in pW3 and pW9-2 encode a 245-amino-acid (aa) rRNA methylase that shares 100% identity with Erm(B) from Streptococcus suis D12 (AER19841), and the Erm(B) protein differs by only four aa exchanges (S100N, H163Y, H217Y, and N222D) from that from pAMβ1 (ACY79534). The 571-bp erm(A) amplicon from both 3-38 and JH3-38 was identical to the corresponding erm(A) sequence of the Streptococcus suis isolate B1 (EU047809).
Analysis of the genetic environment of cfr in the conjugative plasmids pW3, p3-38, and pW9-2.
To gain insight into the genetic environment of the cfr gene in the three newly identified conjugative plasmids, cfr-carrying segments of 27,360, 21,116, and 25,550 bp of the ∼75-kb plasmid pW3, the ∼72-kb plasmid p3-38, and the ∼55-kb pW9-2 plasmid, respectively, were sequenced. The corresponding maps are shown in Fig. 2.
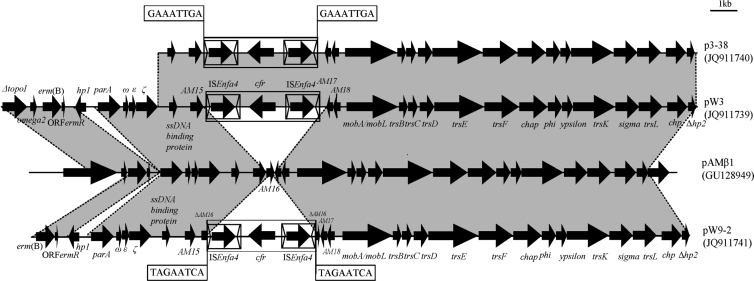
Fig 2.
Genetic environment of cfr gene in the conjugative plasmids p3-38, pW3, and pW9-2 and structural comparison with the conjugative plasmid pAMβ1 from E. faecalis strain DS5. Regions of >95% homology are marked by gray shading.
All three segments had an overall nucleotide sequence identity of >96% with respect to each other. The 4,447-bp central region, including cfr and the two 1,324-bp insertion sequences located in the same orientation, even displayed 100% nucleotide sequence identity. The further flanking sequences in pW3, p3-38, and pW9-2 exhibited a high nucleotide sequence identity (>95%) to the corresponding regions of the conjugative plasmid pAMβ1 (GU128949) from E. faecalis (Fig. 2).
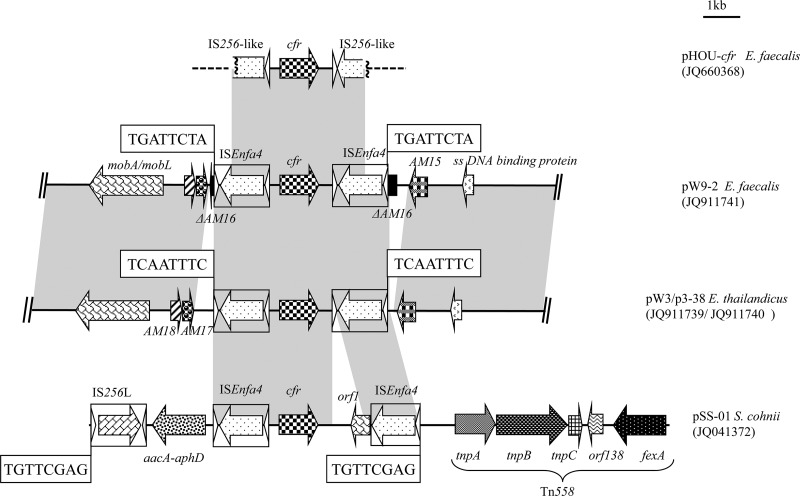
In the cfr upstream region, the putative promoter and the partly overlapping reading frames for peptides of 59 aa (ORF1) and 44 aa (ORF2) were present. However, in comparison to pSCFS1, a 51-bp deletion had occurred in ORF1 which resulted in the loss of the first 17 aa of ORF1. This deletion was also present in the cfr upstream region of pEF-01 from bovine E. faecalis (17) but differed from the 35-bp deletion in the region between the stop codon of ORF2 and the start codon of cfr in the staphylococcal plasmid pSCFS3 (16). Two copies of a 1,324-bp insertion sequence harboring a 1,173-bp transposase gene and imperfect 26-bp terminal inverted repeats were found. These insertion sequences are most closely related to IS1542 from E. faecium with an overall nucleotide sequence identity of 91.6% (1,213/1,324 bp) and 93.8% aa sequence identity (366/390 aa) in the transposase protein (42). This insertion sequence was submitted to the IS database (http://www-is.biotoul.fr/is.html) and received the designation ISEnfa4. Except for an 823-bp sequence, including the orf1 located downstream of the cfr gene on pSS-01, the 4,447-bp central region shared >99% nucleotide identity with the corresponding region of plasmid pSS-01 (accession no. JQ041372) present in S. cohnii and S. saprophyticus (Fig. 3).
Fig 3.
Schematic presentation of detailed genetic environment of the cfr gene in 4,447-bp center regions in conjugative plasmids pW9-2, pW3, and p3-38 and structure comparison with plasmid pHOU-cfr from E. faecalis 603-50427X of clinical human origin and pSS-01 from S. cohnii of swine origin. The arrows indicate the positions and direction of the transcription of the genes. The regions of >96% homology are marked using gray shading. The direct target site duplication is boxed. Δ, Truncated gene. A distance scale in kilobases is displayed in the upper right corner.
Immediately upstream of the righthand ISEnfa4 element and downstream of the left-hand ISEnfa4 element in the 4,447-bp central region, two different 8-bp direct repeats (DR) at the integration site were observed in the three conjugative plasmids. One DR (5′-TAGAATCA-3′), located in the open reading frame of AM16 (which exhibited >99% nucleotide [304/306 bp] identity to that in pAMβ1), was found in the E. faecalis plasmid pW9-2. The other DR (5′-GAAATTGA-3′), located in a noncoding region between the genes AM15 and AM17, was found in both E. thailandicus plasmids pW3-1 and p3-38. It should be noted that the AM16 gene could not be detected in these two plasmids (Fig. 3). To determine the stability of the ISEnfa4 flanked segment in the three conjugative plasmids, inverse PCR assays were performed using cfrIF and cfrIR primers. Amplicons 3,123 bp in size were obtained from each of the three parental strains and their transconjugants. Sequence analysis of these amplicons confirmed that they contained an intact cfr gene region and one complete ISEnfa4 element.
DISCUSSION
Since only little a information is currently available about the presence of the multiresistance gene cfr among enterococci of animal origin (17), we screened 77 enterococcal isolates for the presence of this gene. The detection of the cfr gene in 3 of 77 (3.9%) florfenicol-resistant enterococcus strains of swine and farm environmental origin suggested that this multiresistance gene is relatively rare among enterococci from pig farms in Shandong Province, China. To the best of our knowledge, this is the first report of the cfr gene in the species E. thailandicus. E. thailandicus is a newly discovered enterococcal species, which was initially isolated from fermented sausage in Thailand (43). To date, no clinical E. thailandicus isolate has been reported; however, two strains of Enterococcus sanguinicola, which in the meantime have been reclassified as E. thailandicus, were recovered from clinically relevant human sources (44). The presence of the cfr gene in E. thailandicus from swine and sewage origins points toward the presence of this multiresistance gene in enterococci from animal settings and the surrounding environment.
The predominant resistance gene among florfenicol-resistant enterococci was fexA, which was detected in 72.7% (56/77) of the isolates tested in the present study; this finding was similar to the percentage of fexA-positive isolates among florfenicol-resistant staphylococci of swine origin (73.2%, 109/149) (22). In addition, the recently identified florfenicol exporter gene fexB was detected in 41.6% (32/77) of the isolates tested in the present study. The coexistence of fexA and fexB in the same isolate was also observed in 18 (23.4%) of the strains tested. In addition, 7.8% (6/77) of the florfenicol-resistant isolates examined here did not harbor any of the three genes cfr, fexA, or fexB, which suggests that potential nonenzymatic or enzymatic mechanisms conferred by other, as-yet-identified resistance genes might be involved in florfenicol resistance in enterococci.
The different SmaI PFGE patterns observed suggested that the three porcine and the single bovine cfr-positive enterococcal isolates are not related. Furthermore, MLST of two cfr-carrying E. faecalis strains originating from swine and cattle in China revealed that one of these isolates (EF-01) belongs to ST21, and the other (W9-2) represents the novel type ST469. The E. faecalis EF-01 isolate, which belonged to the clonal complex CC21, is mainly found in animals and in the community, while clinical isolates of this CC have rarely been detected (45). Concerning the W9-2 isolates, the ST469 belongs to a singleton, which did not have single or double locus variants by performing an eBURST analysis (data not shown).
In the present study, three different types of large cfr-carrying conjugative enterococcal plasmids—pW3, p3-38, and pW9-2—could be differentiated on the basis of their sizes, their additional erythromycin resistance genes, and the sequences surrounding the cfr gene. Despite these differences, high structural similarities between these three plasmids and plasmid pAMβ1 were observed. Plasmid pAMβ1, a member of a family of low-copy-number conjugative plasmids (46), was originally identified in a E. faecalis DS5 clinical isolate that harbored the MLSB resistance gene erm(B) (47) and could be transferred to, and maintained in, a wide range of Gram-positive bacteria (e.g., enterococci, streptococci, S. aureus, Bacillus subtilis, and some Lactobacillus casei strains), indicating a broad host range (48). In the present study, the MLSB resistance genes erm(A) and erm(B), the partial nucleotide sequence of the tra gene regions, and an ω-ε-ζ toxin-antitoxin module were observed in all three partially sequenced conjugative plasmids. This suggested that the 4,447-bp central region with the cfr gene and two ISEnfa4 elements had integrated into a pAMβ1-like backbone. A closer look at the sequences identified two integration sites. We propose that the 4,447-bp central region in pW9-2 has integrated into the coding sequence of AM16 via sample transposition; while in pW3 and p3-38, this element probably integrated into the region between the AM15 and AM17 genes, most likely by homologous recombination. The PCR-based stability tests also revealed that the cfr gene and one ISEnfa4 copy can easily be excised, suggesting that this multiresistance gene may also be transferable via ISEnfa4-mediated recombination.
With the exception of the multiresistance gene cfr, the MLSB genes erm(A) or erm(B) have also been found in three conjugative plasmids in our study. The coexistence of MLSB genes and cfr in the same plasmid will allow for persistence and coselection of the cfr gene under the positive selective pressure imposed by the use of macrolides, such as tylosin and tilmicosin. The macrolides, as well as a number of other classes of antimicrobials, including ampicillin, florfenicol, trimethoprim-sulfamethoxazole, lincomycin, and tiamulin, have been used for curing or preventing bacterial infections in three of the pig farms investigated here, according to the antibiotic usage records of these farms. The persistence of the cfr-carrying plasmids among enterococci identified here may also be attributed to the coexistence of ω-ε-ζ toxin-antitoxin module, which is known to promote the persistence of plasmids by encoding a system that kills or prevents the growth of plasmid-free cells (49). Therefore, the potential threat that the accumulation of multiple resistance genes including cfr on a plasmid with a toxin-antitoxin module poses is of great concern.
In conclusion, the data presented here described three conjugative plasmids carrying the multiresistance gene cfr in enterococci from pigs and their surrounding environments. The cfr-carrying center region flanked by the ISEnfa4 elements in these conjugative plasmids harboring tra regions and toxin-antitoxin systems is a potential risk factor for dissemination of this multiresistance gene among enterococci isolates and possibly other species of Gram-positive bacteria, which many believe cause a threat to public health.
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (grants 31001087 and U1031004) and by the Special Fund for Agro-Scientific Research in the Public Interest (grant 201203040).
Footnotes
Published ahead of print 15 October 2012
REFERENCES
- 1. Ogier JC, Serror P. 2008. Safety assessment of dairy microorganisms: the Enterococcus genus. Int. J. Food. Microbiol. 126:291–301 [DOI] [PubMed] [Google Scholar]
- 2. Werner G, Coque TM, Hammerum AM, Hope R, Hryniewicz W, Johnson A, Klare I, Kristinsson KG, Leclercq R, Lester CH, Lillie M, Novais C, Olsson-Liljequist B, Peixe LV, Sadowy E, Simonsen GS, Top J, Vuopio-Varkila J, Willems RJ, Witte W, Woodford N. 2008. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. 13:19046. [PubMed] [Google Scholar]
- 3. Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569–1571 [DOI] [PubMed] [Google Scholar]
- 4. Bozdogan B, Appelbaum PC. 2004. Oxazolidinones: activity, mode of action, and mechanism of resistance. Int. J. Antimicrob. Agents 23:113–119 [DOI] [PubMed] [Google Scholar]
- 5. Meka VG, Gold HS. 2004. Antimicrobial resistance to linezolid. Clin. Infect. Dis. 39:1010–1015 [DOI] [PubMed] [Google Scholar]
- 6. Farrell DJ, Mendes RE, Ross JE, Sader HS, Jones RN. 2011. LEADER Program results for 2009: an activity and spectrum analysis of linezolid using 6,414 clinical isolates from 56 medical centers in the United States. Antimicrob. Agents Chemother. 55:3684–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsiodras S, Gold HS, Sakoulas G, Eliopoulos GM, Wennersten C, Venkataraman L, Moellering RC, Ferraro MJ. 2001. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 358:207–208 [DOI] [PubMed] [Google Scholar]
- 8. Locke JB, Hilgers M, Shaw KJ. 2009. Novel ribosomal mutations in Staphylococcus aureus strains identified through selection with the oxazolidinones linezolid and torezolid (TR-700). Antimicrob. Agents Chemother. 53:5265–5274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaminska KH, Purta E, Hansen LH, Bujnicki JM, Vester B, Long KS. 2010. Insights into the structure, function and evolution of the radical-SAM 23S rRNA methyltransferase Cfr that confers antibiotic resistance in bacteria. Nucleic. Acids Res. 38:1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. 2005. A new mechanism for chloramphenicol, florfenicol, and clindamycin resistance: methylation of 23S rRNA at A2503. Mol. Microbiol. 57:1064–1073 [DOI] [PubMed] [Google Scholar]
- 11. Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smith LK, Mankin AS. 2008. Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob. Agents Chemother. 52:1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schwarz S, Werckenthin C, Kehrenberg C. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44:2530–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Diaz L, Kiratisin P, Mendes R, Panesso D, Singh KV, Arias CA. 2012. Transferable plasmid-mediated resistance to linezolid due to cfr in a human clinical isolate of Enterococcus faecalis. Antimicrob. Agents Chemother. 56:3917–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kehrenberg C, Cuny C, Strommenger B, Schwarz S, Witte W. 2009. Methicillin-resistant and -susceptible Staphylococcus aureus strains of clonal lineages ST398 and ST9 from swine carry the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 53:779–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kehrenberg C, Schwarz S. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50:1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu Y, Wang Y, Wu C, Shen Z, Schwarz S, Du XD, Dai L, Zhang W, Zhang Q, Shen J. 2012. First report of the multidrug resistance gene cfr in Enterococcus faecalis of animal origin. Antimicrob. Agents Chemother. 56:1650–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Toh SM, Xiong L, Arias CA, Villegas MV, Lolans K, Quinn J, Mankin AS. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64:1506–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y, Schwarz S, Shen Z, Zhang W, Qi J, Liu Y, He T, Shen J, Wu C. 2012. Co-location of the multiresistance gene cfr and the novel streptomycin resistance gene aadY on a small plasmid in a porcine Bacillus strain. J. Antimicrob. Chemother. 67:1547–1549 [DOI] [PubMed] [Google Scholar]
- 20. Wang Y, Wang Y, Schwarz S, Shen Z, Zhou N, Lin J, Wu C, Shen J. 2012. Detection of the staphylococcal multiresistance gene cfr in Macrococcus caseolyticus and Jeotgalicoccus pinnipedialis. J. Antimicrob. Chemother. 67:1824–1827 [DOI] [PubMed] [Google Scholar]
- 21. Wang Y, Wang Y, Wu C, Schwarz S, Shen Z, Zhang W, Zhang Q, Shen J. 2011. Detection of the staphylococcal multiresistance gene cfr in Proteus vulgaris of food animal origin. J. Antimicrob. Chemother. 66:2521–2526 [DOI] [PubMed] [Google Scholar]
- 22. Wang Y, Zhang W, Wang J, Wu C, Shen Z, Fu X, Yan Y, Zhang Q, Schwarz S, Shen J. 2012. Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob. Agents Chemother. 56:1485–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang W, Wu C, Wang Y, Shen Z, Dai L, Han J, Foley SL, Shen J, Zhang Q. 2011. The new genetic environment of cfr on plasmid pBS-02 in a Bacillus strain. J. Antimicrob. Chemother. 66:1174–1175 [DOI] [PubMed] [Google Scholar]
- 24. Kehrenberg C, Aarestrup FM, Schwarz S. 2007. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob. Agents Chemother. 51:483–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shore AC, Brennan OM, Ehricht R, Monecke S, Schwarz S, Slickers P, Coleman DC. 2010. Identification and characterization of the multidrug resistance gene cfr in a Panton-Valentine leukocidin-positive sequence type 8 methicillin-resistant Staphylococcus aureus IVa (USA300) isolate. Antimicrob. Agents Chemother. 54:4978–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mendes RE, Deshpande LM, Castanheira M, DiPersio J, Saubolle MA, Jones RN. 2008. First report of cfr-mediated resistance to linezolid in human staphylococcal clinical isolates recovered in the United States. Antimicrob. Agents Chemother. 52:2244–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ruiz de Gopegui E, Juan C, Zamorano L, Perez JL, Oliver A. 2012. Transferable multidrug resistance plasmid carrying cfr associated with tet(L), ant(4′)-Ia and dfrK genes from a clinical methicillin-resistant Staphylococcus aureus ST125 strain. Antimicrob. Agents Chemother. 56:2139–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dai L, Wu CM, Wang MG, Wang Y, Huang SY, Xia LN, Li BB, Shen JZ. 2010. First report of the multidrug resistance gene cfr and the phenicol resistance gene fexA in a Bacillus strain from swine feces. Antimicrob. Agents Chemother. 54:3953–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Y, He T, Schwarz S, Zhou D, Shen Z, Wu C, Ma L, Zhang Q, Shen J. 2012. Detection of the staphylococcal multiresistance gene cfr in Escherichia coli of domestic-animal origin. J. Antimicrob. Chemother. 67:1094–1098 [DOI] [PubMed] [Google Scholar]
- 30. Liu H, Wang Y, Wu C, Schwarz S, Shen Z, Jeon B, Ding S, Zhang QJ, Shen J. 2012. A novel phenicol exporter gene, fexB, found in enterococci of animal origin. J. Antimicrob. Chemother. 67:322–325 [DOI] [PubMed] [Google Scholar]
- 31. Kim SY, Lee JE, Lee S, Lee HT, Hur HG, Ko G. 2010. Characterization of Enterococcus spp. from human and animal feces using 16S rRNA sequences, the esp gene, and PFGE for microbial source tracking in Korea. Environ. Sci. Technol. 44:3423–3428 [DOI] [PubMed] [Google Scholar]
- 32. Aanensen DM, Spratt BG. 2005. The multilocus sequence typing network: mlst.net. Nucleic. Acids Res. 33:W728–W733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huys G, D'Haene K, Collard JM, Swings J. 2004. Prevalence and molecular characterization of tetracycline resistance in Enterococcus isolates from food. Appl. Environ. Microbiol. 70:1555–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clinical and Laboratory Standards Institute 2008. Performance standards for antimicrobial disk and dilution susceptibility test for bacteria isolated from animals; approved standard M31-A3, 3rd ed CLSI, Wayne, PA [Google Scholar]
- 35. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. Approved standard M100-S21. CLSI, Wayne, PA [Google Scholar]
- 36. Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235–240 [DOI] [PubMed] [Google Scholar]
- 37. Rosvoll TC, Pedersen T, Sletvold H, Johnsen PJ, Sollid JE, Simonsen GS, Jensen LB, Nielsen KM, Sundsfjord A. 2010. PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501- and pHTβ-related replicons associated with glycopeptide resistance and stabilizing toxin-antitoxin systems. FEMS Immunol. Med. Microbiol. 58:254–268 [DOI] [PubMed] [Google Scholar]
- 38. Zhang K, McClure JA, Elsayed S, Conly JM. 2009. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Freitas AR, Novais C, Ruiz-Garbajosa P, Coque TM, Peixe L. 2009. Clonal expansion within clonal complex 2 and spread of vancomycin-resistant plasmids among different genetic lineages of Enterococcus faecalis from Portugal. J. Antimicrob. Chemother. 63:1104–1111 [DOI] [PubMed] [Google Scholar]
- 40. Khan SA, Nawaz MS, Khan AA, Cerniglia CE. 1999. Simultaneous detection of erythromycin-resistant methylase genes ermA and ermC from Staphylococcus spp. by multiplex-PCR. Mol. Cell Probe 13:381–387 [DOI] [PubMed] [Google Scholar]
- 41. Malhotra-Kumar S, Lammens C, Piessens J, Goossens H. 2005. Multiplex PCR for simultaneous detection of macrolide and tetracycline resistance determinants in streptococci. Antimicrob. Agents Chemother. 49:4798–4800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Darini AL, Palepou MF, Woodford N. 1999. Nucleotide sequence of IS1542, an insertion sequence identified within VanA glycopeptide resistance elements of enterococci. FEMS Microbiol. Lett. 173:341–346 [DOI] [PubMed] [Google Scholar]
- 43. Tanasupawat S, Sukontasing S, Lee JS. 2008. Enterococcus thailandicus sp. nov., isolated from fermented sausage (‘mum’) in Thailand. Int. J. Syst. Evol. Microbiol. 58:1630–1634 [DOI] [PubMed] [Google Scholar]
- 44. Shewmaker PL, Steigerwalt AG, Nicholson AC, Carvalho Mda G, Facklam RR, Whitney AM, Teixeira LM. 2011. Reevaluation of the taxonomic status of recently described species of Enterococcus: evidence that E. thailandicus is a senior subjective synonym of “E. sanguinicola” and confirmation of E. caccae as a species distinct from E. silesiacus. J. Clin. Microbiol. 49:2676–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ruiz-Garbajosa P, Bonten MJ, Robinson DA, Top Nallapareddy JSR, Torres C, Coque TM, Canton R, Baquero F, Murray BE, del Campo R, Willems RJ. 2006. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 44:2220–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. d'Alencon E, Ehrlich SD. 2000. A study of the CopF repressor of plasmid pAMbeta1 by phage display. J. Bacteriol. 182:2973–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Clewell DB, Yagi Y, Dunny GM, Schultz SK. 1974. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J. Bacteriol. 117:283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marcina FL, Archer GL. 1993. Conjugation and broad host range plasmids in streptococci and staphylococci, p 313–329 In Clewell DB. (ed), Bacterial conjugation. Plenum Press, Inc, New York, NY [Google Scholar]
- 49. Meinhart A, Alonso JC, Strater N, Saenger W. 2003. Crystal structure of the plasmid maintenance system ω/ζ: functional mechanism of toxin ζ and inactivation by ω2 ζ2 complex formation. Proc. Natl. Acad. Sci. U. S. A. 100:1661–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]