Abstract
More than 2 decades of study support the hypothesis that alginate lyases are promising therapeutic candidates for treating mucoid Pseudomonas aeruginosa infections. In particular, the enzymes' ability to degrade alginate, a key component of mucoid biofilm matrix, has been the presumed mechanism by which they disrupt biofilms and enhance antibiotic efficacy. The systematic studies reported here show that, in an in vitro model, alginate lyase dispersion of P. aeruginosa biofilms and enzyme synergy with tobramycin are completely decoupled from catalytic activity. In fact, equivalent antibiofilm effects can be achieved with bovine serum albumin or simple amino acids. These results provide new insights into potential mechanisms of alginate lyase therapeutic activity, and they should motivate a careful reexamination of the fundamental assumptions underlying interest in enzymatic biofilm dispersion.
INTRODUCTION
Premature mortality among cystic fibrosis (CF) patients is typically associated with chronic airway infection by the opportunistic pathogen Pseudomonas aeruginosa (1). Upon colonization of the airway, P. aeruginosa causes a range of clinical complications, including stimulation of a hyperinflammatory response, obstruction of the pulmonary tract, and progressive loss of lung function (1–3). The bacterium's persistence within the CF lung is thought to result in large part from a biofilm mode of growth (2, 4), which can subvert both the host immune response and antibacterial therapies (5). In contrast to environmental niches, in the CF lung, P. aeruginosa generally transitions to a mucoid phenotype characterized by overproduction of the exopolysaccharide alginate (6). This copolymer of (1,4)-linked β-d-mannuronic acid and α-l-guluronic acid alters biofilm architecture and function (7) and thereby compounds P. aeruginosa's persistence in the chronically inflamed airway (5). Indeed, alginate is one of the most extensively studied P. aeruginosa virulence factors (8).
Given alginate's contribution to mucoid biofilm structure, its function in bacterial virulence, and its role in the persistent nature of lung infections, it has long been considered an attractive target for interventional therapies (6). In particular, biocatalytic degradation of mucoid P. aeruginosa biofilms using alginate lyase enzymes (EC 4.2.2.3) has been the subject of more than 20 years of research. Alginate lyase treatment has been shown to reduce viscosity in cultures of clinical isolates and in CF sputum (9, 10); it strips biofilms from abiotic surfaces (11, 12), it enhances phagocytosis and killing of P. aeruginosa by human immune cells (13–15), and it improves the efficacy of various antipseudomonal antibiotics (14, 16–19). In aggregate, these reports make a rather convincing case for the use of inhaled alginate lyases for treating chronic P. aeruginosa infections of the CF lung, although no clinical trials have been conducted to date.
In an effort to further bolster the rationale for therapeutic alginate lyases, we set out to conduct systematic studies analyzing the solution phase kinetics, biofilm-disrupting potential, and antibiotic synergy of two promising enzyme candidates. Sphingomonas sp. A1 alginate lyase (A1-III) has been shown previously to exhibit high levels of activity toward bacterial alginate (20) and has also been subjected to molecular engineering with an eye toward therapeutic applications (12, 21). P. aeruginosa alginate lyase (AlgL) is produced by the CF-associated pathogen itself, and its confirmed role in bacterial alginate biosynthesis (22, 23) suggests the enzyme is an obvious choice for developing alginate-degrading CF biotherapies. On a superficial level, our results are entirely consistent with the encouraging outcomes of prior alginate lyase studies. Our systematic approach, however, has for the first time revealed that enzyme-mediated biofilm disruption and antibiotic synergy are decoupled from catalytic activity. The results described here suggest the need to carefully reexamine the veracity of fundamental assumptions motivating interest in therapeutic alginate lyases.
MATERIALS AND METHODS
Bacterial strains.
The mucoid P. aeruginosa clinical isolate FRD1 and its nonmucoid derivative, SMC406, were obtained from the G. O'Toole laboratory (Geisel School of Medicine at Dartmouth, Hanover, NH). SMC406 varies from FRD1 only in the deletion of the algT gene, which effectively eliminates alginate biosynthesis (24). The Escherichia coli expression strain SHuffle T7 Express {fhuA2 lacZ::T7 gene1 [lon] ompT ahpC gal λatt::pNEB3-r1-cDsbC (Specr lacIq) ΔtrxB sulA11 R(mcr-73::miniTn10-Tets)2 [dcm] R(zgb-210::Tn10 –Tets) endA1 Δgor Δ(mcrC-mrr)114::IS10} was purchased from New England BioLabs (Ipswich, MA).
Cloning of the A1-III and AlgL alginate lyases.
The alginate lyase gene algL was amplified from the P. aeruginosa PAO1 genome using a 5′ primer with an ATG start codon, homology to base pairs 82 to 96 on the native gene, and a 3′ primer with either a TGA stop codon or a C-terminal hexahistidine tag followed by a TGA stop codon. The amplified AlgL genes were ligated into the pET28b expression vector and transformed into electrocompetent SHuffle T7 Express. Individual clones were sequence verified. Additionally, pET28b constructs containing either Sphingomonas sp. alginate lyase A1-III or a C-terminal hexahistidine-tagged variant (A1-III–His) (12) were transformed into SHuffle T7 Express, and individual clones were sequence verified.
The catalytically inactivated double point mutant of A1-III–His (A1-DM-His) was constructed by targeting two of the active-site residues involved in the enzyme's β-elimination mechanism (25). Mutation H188A consisted of switching the histidine 188 GTG codon to a CGC codon, while mutation Y242F replaced the tyrosine 242 GTA codon with a GAA codon. The mutated gene was then ligated into the pET28b expression vector and transformed into SHuffle T7 Express, and individual clones were sequence verified.
Expression and purification of alginate lyases.
Expression hosts were grown overnight in 8 ml of LB supplemented with 30 μg/ml kanamycin (LB-KAN30) at 30°C. The overnight cultures were then subcultured 1:100 into 500 ml of fresh LB-KAN30 and grown to mid-log phase at 30°C, after which the cells were equilibrated at 25°C for 30 min and then induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 21 h. Following induction, the cell cultures were centrifuged at 6,000 × g at 4°C for 10 min, and the total cell pellet for each construct was resuspended in 5 ml of buffer A (20 mM NaH2PO4, pH 6.5). The resuspended pellets were transferred to 10-ml Pyrex beakers, equilibrated on ice for 20 min, and disrupted by sonication with 24 cycles of 10 s on and 10 s off (Fisher 550 Sonic Dismembrator). Whole-cell lysate was centrifuged at 17,000 × g at 4°C for 20 min, and the soluble fraction was filtered through a 0.22-μm polyethersulfone (PES) membrane.
Filtered soluble cell extract containing AlgL was loaded onto a 30-ml hydroxyapatite column (Calbiochem, La Jolla, CA) equilibrated with buffer A. Buffer A continued to run for 10 ml at 2 ml/min before changing to a step gradient of 30% buffer A and 70% buffer B (500 mM NaH2PO4, pH 6.5) for 60 ml, followed by 100% buffer B for an additional 65 ml. At 100% buffer B, a single peak, containing AlgL and other proteins, was eluted. This peak was dialyzed against 50 mM KH2PO4, 150 mM NaCl, pH 7, at 4°C and concentrated using a 10,000-molecular-weight cutoff (MWCO) centrifugal filter. Concentrated sample (300 μl) was then loaded onto a 120-ml Superdex75 size exclusion chromatography (SEC) (GE Healthcare Life Sciences, Piscataway, NJ) column equilibrated with 50 mM KH2PO4, 150 mM NaCl, pH 7. Flow was maintained at 0.6 ml/min, and purified AlgL was eluted as a single peak at 60 ml. Samples from the SEC were concentrated using a 3,000-MWCO centrifugal filter and stored at 4°C. Enzyme purity was assessed by SDS-PAGE, and the final protein concentrations were measured by A280 (NanoDrop 1000; Thermo Scientific, Waltham, MA) using a molar absorptivity of 67,050 cm−1 M−1, estimated with the VectorNTI software package (Invitrogen, Carlsbad, CA).
The same hydroxyapatite column and buffers were used to purify A1-III enzyme from corresponding soluble cell extract. The column was washed with buffer A for 20 ml at 2 ml/min, then increased to 5% B for 30 ml, increased again to 12% B for 50 ml, and finally 100% B for another 80 ml. An unresolved peak at 100 ml containing A1-III was collected and dialyzed against 50 mM KH2PO4, 150 mM NaCl, pH 7 at 4°C before being concentrated with a 10,000-MWCO centrifugal filter. Two hundred microliters of the concentrated sample was then loaded onto the same Superdex75 SEC column, and purified A1-III was eluted at 60 ml under the same conditions described above. Samples did not require further concentration after SEC and were stored at 4°C. Enzyme purity was assessed by SDS-PAGE, and the protein concentration was measured by A280 (molar absorptivity = 63,350 cm−1 M−1).
In addition to the above-mentioned purification protocol, single-step immobilized metal ion affinity chromatography (IMAC) purification was also used as an alternative method for isolating the His-tagged variants. Briefly, 4 to 5 ml of filtered soluble cell extract was gently mixed with a 0.4-ml bed volume of nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen, Valencia, CA), which had been equilibrated with native lysis buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 8.0). Binding occurred at 4°C for 1 h, with gentle rocking, after which the column was drained and washed with 10 bed volumes of wash buffer (50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole, pH 8.0). Purified A1-III was eluted in a native elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0), dialyzed into storage buffer (20 mM NaH2PO4, pH 6.5), and kept at 4°C. Once again, enzyme purity was assessed by SDS-PAGE, and concentrations were determined by A280.
Enzyme kinetic analysis with soluble substrates.
Enzyme-mediated cleavage of the alginate carbohydrate backbone through beta elimination results in formation of a C-4–C-5 carbon-carbon π bond at the new nonreducing termini, causing a strong absorbance peak at 235 nm. Replicate kinetic analysis was conducted by monitoring product formation at 235 nm in UV-transparent 96-well plates (Costar Fisher number 3635). Using a multichannel pipette, 190-μl aliquots of medium-viscosity brown seaweed alginate (BSWA) (Sigma number A2033) were mixed thoroughly with 10 μl of enzyme residing in each well of the plate's upper row. BSWA dilutions were prepared in a reaction buffer (150 mM NaCl, 50 mM NaH2PO4, pH 7.0) and ranged from 0.01% to 0.1% (wt/vol). Upon completion of mixing, the plate was transferred to a UV-visible plate reader (SpectraMax 190; Molecular Devices, Sunnyvale, CA), and the formation of product was tracked every 15 s over the course of 10 min. Initial slopes from absorbance-versus-time plots were determined for each substrate concentration, and nonlinear regression of the initial reaction rates versus substrate concentration was used to determine apparent Vmax and Km values for each enzyme. Kinetics with bacterial alginate substrate, purified from P. aeruginosa FRD1 as described previously (26), followed the same procedure (bacterial alginate follows the same cleavage mechanism as BSWA).
Biofilm cell viability assays.
P. aeruginosa strains FRD1 and SMC406 were grown on LB agar plates at 37°C. Single colonies were selected and used to inoculate 3-ml cultures of tryptic soy broth (TSB). The cultures were placed on an orbital shaker for 21 h at 37°C before being diluted 1:1,000 in fresh TSB. Upon dilution, the cultures were aliquoted into 96-well minimum biofilm eradication concentration (MBEC) plates (Innovotech, Edmonton, AB, Canada) at a volume of 75 μl per well. The corresponding peg lid was then placed on top of the plate, and the sides of the plate were sealed with strips of Breathe-Easy membrane (Diversified Biotech, Dedham, MA), which eliminated evaporation but still allowed gas permeability. The plates were incubated statically at 37°C for 20 h before the peg lids were removed, rinsed in sterile deionized water for 10 s, and placed into black, clear-bottom, 96-well treatment plates (Corning, Lowell, MA). The treatment plates contained 125 μl of activity buffer (50 mM potassium phosphate, 150 mM NaCl, pH 7) only (untreated) or with one of the following agents: 100 or 500 μg/ml tobramycin (MP Biomedicals, Solon, OH), 1,000 μg/ml protein (A1-III–His, A1-DM-His, A53C-His-PEG [polyethylene glycol], AlgL-His, or bovine serum albumin [BSA]), the constituent amino acids of A1-III–His at the same molar concentrations contained in 1,000 μg/ml of intact enzyme, the arginine constituent of A1-III–His at the same molar concentration contained in 1,000 μg/ml of the intact enzyme, or arginine at the same molar concentration as the sum total of all amino acids in 1,000 μg/ml A1-III–His. Additionally, 100 or 500 μg/ml of tobramycin was combined with each of the protein and amino acid treatments to assess synergistic effects. The treatment plates were sealed with strips of Breathe-Easy membrane and statically incubated at 37°C for 12 h.
After treatment, the peg lids were removed from the treatment plates, rinsed in sterile deionized water for 10 s, and placed into black, clear-bottom, 96-well plates containing 125 μl per well of alamarBlue (Invitrogen, Carlsbad, CA) to measure biofilm cell viability. Concurrently, alamarBlue was added to the treatment plates to measure the viability of any detached cells. All alamarBlue plates were sealed with Breathe-Easy membranes and statically incubated for 12 h at 37°C. Membranes and peg lids were then removed, and reduction of the alamarBlue resazurin compound was measured by fluorescence at an excitation wavelength of 540 nm (550-nm cutoff) and an emission wavelength of 585 nm. Additionally, alamarBlue background signal was measured in the presence of all treatments without cells and subtracted from the cell viability signal.
SEM.
Polystyrene peg lids were prefixed in 0.1 M PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 7, containing 50 mM l-lysine, 0.075% ruthenium red, and 2.5% glutaraldehyde for 30 min at room temperature. The peg lids were then fixed in 0.1 M PIPES, pH 7, containing 0.075% ruthenium red and 2.5% glutaraldehyde for 3 h at room temperature before being rinsed with 0.1 M PIPES, pH 7, for 20 min (the rinse was repeated three times). Dehydration of the peg lids was performed in 30%, 50%, 70%, and 95% ethanol for 5 min each, with 2 final changes in 100% ethanol for 5 min each. The peg lids were then air dried in a desiccator for 72 h. Upon completion of air drying, individual pegs were snapped off from the lids with tweezers and mounted on scanning electron microscopy (SEM) stubs with a layer of melted apiezon wax. The mounted pegs were then sputter coated with gold and palladium and viewed by SEM (XL30 ESEM FEG; FEI, Hillsboro, OR).
Statistical analysis.
Statistical significance was assessed using one-way analysis of variance (ANOVA) and post hoc Tukey's multiple-comparison tests. Significance was determined at an α value of 0.05.
RESULTS
Selection, cloning, and production of alginate lyases.
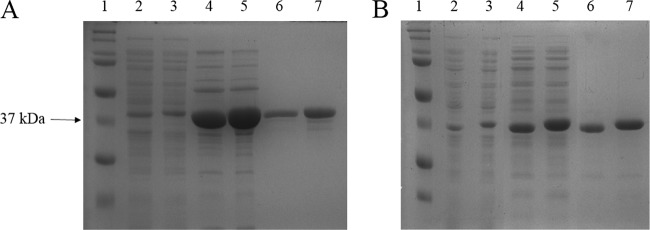
We selected two enzyme candidates for comparative analysis. Sphingomonas sp. A1-III has been reported to exhibit particularly high levels of activity toward bacterial alginate (20) and has been the subject of extensive study and preclinical development (9, 12, 21). P. aeruginosa AlgL has naturally evolved to degrade bacterial alginate, and the enzyme was shown previously to facilitate detachment of P. aeruginosa from biofilms (27). The algL gene was amplified from P. aeruginosa PAO1 genomic DNA, cloned into the pET28b expression vector, and transformed into electrocompetent E. coli SHuffle T7 Express. A gene encoding a variant of AlgL bearing a C-terminal hexahistidine tag (AlgL-His) was constructed in a similar manner. Shake flask cultures of the two expression strains were induced overnight, and the proteins were purified from whole-cell lysates via hydroxyapatite and then SEC. The yields were 0.39 mg/liter for native AlgL and 2.3 mg/liter for AlgL-His, with both proteins exhibiting >90% purity (Fig. 1A). The synthesis and cloning of genes encoding the native A1-III enzyme and its C-terminal His-tagged variant have been described previously (12). Expression and purification of A1-III and A1-III–His followed the same procedures described for AlgL above. The final yields for the two proteins were 9.3 mg per liter of bacterial culture and 15 mg per liter of culture, respectively, and again, both proteins were >90% pure following SEC (Fig. 1B).
Fig 1.
SDS-PAGE analysis of alginate lyase expression and purification. (A) P. aeruginosa AlgL. Lane 1, Bio-Rad Precision Plus Dual Color Standard; lane 2, AlgL soluble cell-free extract; lane 3, AlgL-His soluble cell-free extract; lane 4, AlgL eluted from hydroxyapatite; lane 5, AlgL-His eluted from hydroxyapatite; lane 6, AlgL following size exclusion chromatography; lane 7, AlgL-His following size exclusion chromatography. (B) Sphingomonas sp. A1-III. Samples containing the native and His-tagged proteins were loaded as for panel A.
In addition to the two-step hydroxyapatite and SEC purification described above, the His-tagged variants AlgL-His and A1-III–His were purified by one-step IMAC nickel chromatography. IMAC purification resulted in 7.6 mg AlgL-His per liter of bacterial culture and 13 mg A1-III–His per liter of culture. The purity of the proteins eluted from the IMAC resin was comparable to that obtained with the two-step purification (data not shown). It bears noting that the IMAC yields for AlgL-His were at least 3-fold greater than any previously reported purification of the enzyme from shake flask cultures (28, 29). Importantly, kinetic analysis (described below) showed that the IMAC-purified proteins were indistinguishable from samples prepared by two-step chromatography.
Solution phase enzyme kinetics.
Intuition suggests that the therapeutic potential of alginate lyase enzymes should be directly proportional to their catalytic firepower, and therefore, the solution phase kinetics of alginate degradation were used to compare the prospective utilities of the various enzymes (Table 1). Initial studies were conducted with commercially available BSWA, a standard substrate for analysis of alginate lyase activity. In comparing the two native enzyme constructs, A1-III exhibited a 10-fold greater Vmax than AlgL, but the difference in Km values was not statistically significant. With the BSWA substrate, the Vmax and Km values for the native A1-III enzyme were similar to those of the His-tagged analog A1-III–His, as were the respective values for AlgL and AlgL-His. Thus, appending a C-terminal hexahistidine tag onto either of the two alginate lyases did not substantially alter their solution phase kinetics.
Table 1.
Pseudo Michaelis-Menten kinetic parameters with BSWA and bacterial (FRD1) alginate substrates
| Enzyme | Substrate | Vmax [ΔA235 (min · mg)−1] | Km (μg/ml) | Vmax/Km |
|---|---|---|---|---|
| AlgL | BSWA | 26 ± 1 | 70 ± 10 | 0.36 ± 0.05 |
| AlgL-His | BSWA | 35 ± 2 | 40 ± 10 | 0.8 ± 0.2 |
| FRD1 | 127 ± 6 | 840 ± 70 | 0.15 ± 0.01 | |
| Al-III | BSWA | 290 ± 20 | 39 ± 9 | 8 ± 2 |
| Al-III–His | BSWA | 337 ± 9 | 100 ± 10 | 3.4 ± 0.4 |
| FRD1 | 310 ± 10 | 420 ± 40 | 0.74 ± 0.08 |
BSWA is a readily available commodity product commonly used for characterizing alginate lyase activity, but the ultimate therapeutic target is alginate produced by the bacterium P. aeruginosa. Bacterial alginate differs from BSWA in its relative mannuronic and guluronic acid content, as well as the presence of acetyl esters on C-2 and C-3 hydroxyl groups (30). To compare the activities of the two enzymes on a more clinically relevant substrate, the kinetics of A1-III–His and AlgL-His were determined with alginate purified from P. aeruginosa strain FRD1, a mucoid clinical isolate (Table 1). The Vmax value for A1-III–His was essentially unchanged compared to its value with the BSWA substrate, but the enzyme's Km for bacterial alginate was at least 4-fold higher. Thus, A1-III–His exhibited strong activity toward both BSWA and bacterial alginate, but it required substantially higher concentrations of the latter to achieve maximum rates of degradation. The AlgL-His enzyme, on the other hand, was found to have a significant increase in both Km (20-fold) and Vmax (3.6-fold) when comparing the bacterial substrate to BSWA. The P. aeruginosa enzyme, therefore, achieves higher maximum rates of degradation with its native bacterial substrate, but analogous to A1-III–His, it requires concentrated substrate solutions to do so. Despite the increased activity of AlgL-His toward its native substrate, the A1-III–His enzyme maintained a significant advantage with respect to the maximum degradation rates of bacterial alginate (>2-fold). In contrast, the catalytic efficiencies (Vmax/Km) of the two enzymes were not significantly different for the therapeutically relevant substrate.
Treatment of mucoid and nonmucoid biofilms with A1-III–His and tobramycin.
Within the hydrogel environment of bacterial biofilms, one may reasonably assume that the local concentrations of alginate will be quite high. Under conditions of high substrate concentration, Vmax, as opposed to Vmax/Km, is the most relevant kinetic parameter. We therefore elected to examine only A1-III–His in the initial biofilm assays. While we had previously validated the biofilm-disrupting potential of A1-III–His using lectins to quantify alginate in enzyme-linked immunosorbent assay (ELISA)-type assays (12), here, we sought to quantify bactericidal activity within established biofilms. The alamarBlue reagent had been validated previously as an accurate reporter of P. aeruginosa viability in both planktonic cultures and biofilms (31). Thus, alamarBlue was used to monitor relative viability in the subsequent studies.
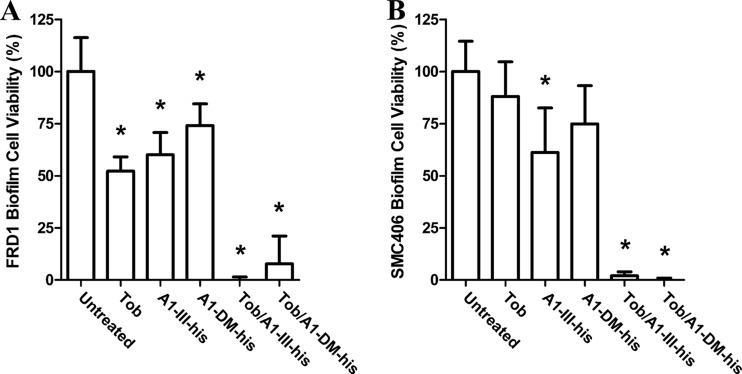
Treatment of mucoid FRD1 biofilms with 500 μg/ml of tobramycin, a clinically relevant concentration (32), resulted in a 48% reduction in cell viability compared to untreated controls (Fig. 2A). Standalone treatment with 1,000 μg/ml of A1-III–His showed a 40% reduction in alamarBlue signal, which likely resulted from enzyme-mediated cellular detachment as opposed to direct bactericidal action. In contrast, the combination of the two treatments (500 μg/ml tobramycin with 1,000 μg/ml of A1-III–His) repeatedly resulted in complete elimination of viable cell signal. Given the exceptional efficacy of the combined treatment on FRD1 biofilms, a simple control study was designed to confirm that the synergistic effect was a function of alginate degradation. Specifically, the treatment study with tobramycin and A1-III–His was repeated using the isogenic, nonmucoid P. aeruginosa strain SMC406 (Fig. 2B). The tobramycin treatment, lowered to 100 μg/ml to account for the greater sensitivity of nonmucoid strains (7), failed to reduce viable cells in the biofilm to a statistically significant degree. Surprisingly, the standalone A1-III–His treatment reduced alamarBlue signal by 40%, the same as observed with FRD1. Also similar to the results with FRD1, the combination treatment reduced SMC406 cell viability to background levels. These results were unanticipated, as the SMC406 strain does not produce alginate and should therefore be unaffected by alginate lyase treatment.
Fig 2.
Analysis of cell viability in P. aeruginosa biofilms following alginate lyase treatments. (A) Mucoid FRD1 biofilms were treated for 12 h with either 500 μg/ml tobramycin (Tob), 1,000 μg/ml alginate lyase, or a combination thereof. Subsequently, cell viability was quantified with alamarBlue, and the results were normalized to untreated biofilms. (B) Nonmucoid SMC406 biofilms were treated in the same manner as for panel A, except that tobramycin concentrations were lowered to 100 μg/ml. The error bars represent standard deviations from four replicates. Statistically significant (α = 0.05) differences relative to untreated biofilms are marked with asterisks.
Given the unexpected results with the nonmucoid SMC406 strain, studies were designed to more rigorously assess the role of alginate degradation in FRD1 biofilm disruption. Based upon an X-ray crystal structure and a hypothesized mechanism (25), an active-site double mutant (H188A and Y242F) of A1-III–His was constructed (here, the double mutant is referred to as A1-DM-His). The A1-DM-His protein was expressed and purified at levels comparable to A1-III–His, but the purified double mutant exhibited no detectable activity on BSWA, even at enzyme concentrations 40-fold higher than those used to characterize the active biocatalyst (data not shown). The inactive A1-DM-His enzyme was used to treat both mucoid FRD1 and nonmucoid SMC406 biofilms alone and in combination with tobramycin (Fig. 2). As a single-agent treatment, A1-DM-His reduced biofilm signals by 25%, whereas in combination with tobramycin, it reduced biofilm cell signals to background levels. The latter result was indistinguishable from those achieved with the wild-type enzyme and tobramycin combination. Thus, it was concluded that A1-III–His antibacterial synergy with tobramycin was decoupled from catalytic activity.
Treatment of mucoid biofilms with proteins lacking alginate-degrading activity.
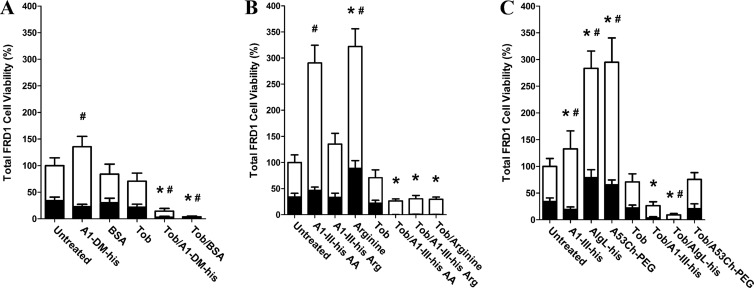
While the above-mentioned results demonstrated that catalytic activity was irrelevant to biofilm dispersion and tobramycin synergy, they did not rule out the possibility that the observed antibiofilm effects resulted from some other molecular feature specific to the A1-III alginate lyase protein. To test this hypothesis, the FRD1 biofilm studies were repeated with the inactive A1-DM-His enzyme, as well as BSA as a sham treatment (Fig. 3A). In addition to analyzing cell viability within the treated biofilms, viable cells were also quantified in the treatment supernatant itself. Thus, the new studies captured the treatment effects on both adherent and detached cell populations. BSA treatment affected neither biofilm nor planktonic alamarBlue signal. When used in combination with tobramycin, however, BSA essentially eradicated viable cells from both the planktonic and biofilm states. Likewise, A1-DM-His combined with tobramycin again eradicated the biofilm cells (consistent with the results shown in Fig. 2A) and left few viable cells in the planktonic state. In aggregate, these results indicated that the observed tobramycin synergy was not a function of any specific alginate lyase property but was more likely a nonspecific phenomenon stemming from biofilm exposure to proteins in general.
Fig 3.
Analysis of P. aeruginosa viability following biofilm treatments. Mucoid FRD1 biofilms were treated for 12 h with a panel of biologic reagents, tobramycin, or combinations thereof. alamarBlue was then used to quantify cell viability in the remaining adherent biofilm (black bars), as well as in the treatment supernatant containing detached planktonic cells (white bars). (A) A1-DM-His alginate lyase and BSA were used to assess the effects of protein treatments that lacked alginate-degrading activity. (B) Treatments consisting of individual amino acids were used to assess whether higher-order protein structure was required for tobramycin synergy. The treatments were composed of (i) A1-III–His AA, with each individual amino acid at the same concentration as its specific molar contribution to 1,000 μg/ml intact A1-III–His; (ii) A1-III–His Arg, with arginine at the same concentration as its specific molar contribution to 1,000 μg/ml intact A1-III–His; and (iii) arginine, with arginine at the same molar concentration as the total of all individual amino acids in 1,000 μg/ml intact A1-III–His. (C) Treatment with diverse alginate lyase enzymes was used to assess the effects of varied catalytic activity and proteolytic sensitivity. A1-III–His was repeated as a positive control. AlgL-His was included as a reduced-activity enzyme evolved to function specifically on bacterial alginate. A53C-His-PEG is a genetically engineered and PEGylated variant of A1-III–His that exhibits nearly identical solution phase kinetics but should have greater resistance to proteolytic degradation. The error bars represent standard deviations from six replicates. For biofilm cells, statistically significant (α = 0.05) differences relative to untreated biofilm are marked with asterisks. For planktonic cells, statistically significant (α = 0.05) differences relative to untreated biofilm are marked with pound signs.
Amino acid treatment of mucoid biofilms.
In an effort to assess whether intact primary/secondary/tertiary protein structure was required for antibiofilm synergy with tobramycin, a panel of individual amino acid mixtures was examined next. Each treatment was based on the molar concentration of the individual constituent amino acids in 1,000 μg/ml intact A1-III–His. Treatment with either the full complement of constituent amino acids (A1-III–His AA) or just the constituent arginines of A1-III–His (A1-III–His Arg) did not affect the biofilm cell signal but did increase the planktonic cell signal (Fig. 3B). Treatment with arginine at a molar concentration equal to that of the sum total of all individual constituent amino acids (arginine) yielded a 2-fold increase in the biofilm signal and a 5-fold increase in the planktonic cell signal. These observations suggest that the amino acids were used as a nutrient source in the otherwise nutrient-depleted buffer, and they are also consistent with the prior use of arginine as a growth supplement for P. aeruginosa biofilms (33). As observed in the earlier trials, tobramycin alone yielded only a moderate reduction in cell viability (36% for biofilm and 26% for planktonic cells). In combination with tobramycin, however, each of the amino acid treatments resulted in complete knockdown of the biofilm cell signal. The combined treatments were found to retain some level of planktonic cell viability, but the levels were at least 55% lower than those observed with tobramycin treatment alone. Thus, amino acid mixtures were as good as or even better than intact proteins with respect to bactericidal synergy toward biofilm cells.
Alginate lyase proteolytic sensitivity as a parameter in mucoid biofilm treatments.
The above-mentioned results suggested that the alginate lyase treatments manifested antibiotic synergy by virtue of their consumption as a nutrient source, which would necessitate proteolytic degradation of the enzymes. As a preliminary test of this hypothesis, mucoid biofilms were treated with an active alginate lyase that had been conjugated to a 20-kDa polyethylene glycol chain, i.e., PEGylated. PEGylation is a well-validated method of reducing the proteolytic susceptibility of proteins (34, 35), and the PEGlyated alginate lyase (A53C-His-PEG) had been shown previously to exhibit wild-type solution phase kinetics and better than wild-type biofilm disruption (12). Treatment of mucoid biofilms with A1-III–His once again showed a decreased biofilm cell signal and at the same time an increased planktonic cell signal, which is consistent with enzyme-mediated detachment of biofilm cells (Fig. 3C). Similar to earlier results, A1-III–His combined with tobramycin essentially eradicated biofilm cells. Interestingly, treatment with AlgL-His alone caused an increase in both the biofilm cell signal and the planktonic cell signal. A combination of AlgL-His and tobramycin, however, yielded the expected knockdown of bacterial viability. Similar to AlgL alone, treatment with only A53C-His-PEG resulted in both increased biofilm cell signal and planktonic cell signal. In sharp contrast to all other combination treatments, however, A53C-His-PEG with tobramycin was no different than tobramycin alone. Thus, the PEGylated alginate lyase was the only biological treatment that failed to exhibit synergy with tobramycin.
Analysis of treatments by SEM.
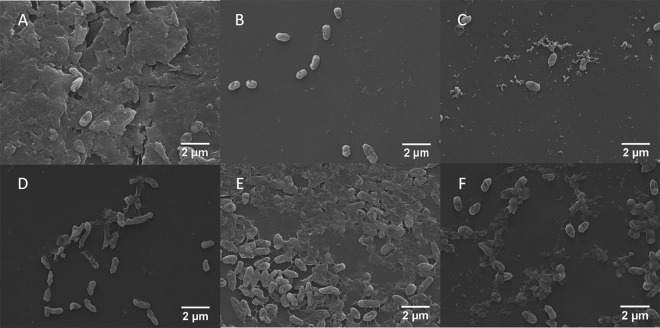
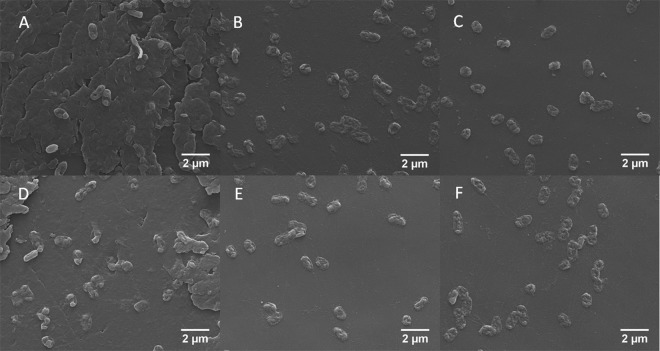
The alamarBlue assays of cell viability consistently pointed to a catalysis-independent mode of action for the alginate lyases, but to corroborate these results, the biofilm treatment studies were repeated and SEM was used to visualize treated biofilms in a direct fashion. Importantly, pre-SEM alamarBlue testing of these biofilms gave results matching those from the initial studies described above (data not shown). Representative images at ×8,000 magnification (Fig. 4) highlight the differential effects of various treatments with respect to biofilm architecture, total biomass, and vitality of adherent cells. Blank buffer treatment of biofilms resulted in bacterial cells embedded within robust and copious extracellular matrix. In the absence of tobramycin, treatment with A1-III–His or A1-DM-His caused the nearly complete removal of extracellular matrix. As single agents, A53C-His-PEG and BSA also appeared to remove the majority of biofilm matrix, but each appeared to subtly increase the number of attached cells compared to biofilms treated with the wild-type A1-III–His enzyme. Importantly, the qualitative SEM analysis of matrix removal by A1-III–His and A53C-His-PEG showed good agreement with prior quantitative analysis based on binding of an alginate-sensitive lectin (12). In contrast to the intact proteins, treatment with the constituent amino acids resulted in only partial removal of the biofilm matrix and a dramatic increase in the density of attached cells. Following treatment with biological agents, the bacteria generally appeared to have intact structure and regular shape, although some deviations from normal morphology were seen with the A53-His-PEG standalone treatment (Fig. 4D).
Fig 4.
Representative SEM images of FRD1 biofilms treated with buffer only (A), A1-III–His (B), A1-DM-His (C), A53C-His-PEG (D), A1-III–His AA (E), or BSA (F). The distinct architecture of biofilm matrix versus simple adherent cells is clear in panel A compared to panel B, C, D, or F.
SEM analysis of biofilms treated with combinations of tobramycin and biologics again showed that the biological agents contributed to a reduction in extracellular matrix. Compared to the results from biological agents alone, the only differences were that the amino acid treatment with tobramycin left no identifiable matrix (compare Fig. 4E and 5E) while A53C-His-PEG with tobramycin yielded more matrix than the enzyme alone (compare Fig. 4D and 5D). More striking was the effect of tobramycin treatments on cellular integrity. As a single agent, the antibiotic yielded a subpopulation of adherent cells with atypical membrane morphology (Fig. 5A). The combination of tobramycin with A53C-His-PEG yielded largely similar results (Fig. 5D). In contrast, few if any intact adherent cells could be identified from biofilms treated with combinations of tobramycin and A1-III–His, A1-DM-His, BSA, or A1-III–His AA; only ghost cells were observed (Fig. 5B, C, E, and F).
Fig 5.
SEM images of FRD1 biofilms treated with tobramycin (A), tobramycin and A1-III–His (B), tobramycin and A1-DM-His (C), tobramycin and A53C-His-PEG (D), tobramycin and A1-III–His AA (E), or tobramycin and BSA (F).
Overall, SEM imaging correlated well with the results of alamarBlue viability analysis. One apparent discrepancy was the A53C-His-PEG treatment versus the PBS control. The quantitative alamarBlue studies showed that biofilms treated with the PEGylated enzyme exhibited almost 2-fold-higher signals than those treated with PBS. The SEM images, on the other hand, suggest a dramatic reduction in total biomass upon treatment with A53C-His-PEG. It is important to note that the alamarBlue reagent measures cellular metabolic activity, and it is therefore insensitive to biofilm matrix components, which constitute a substantial fraction of the biomass seen in PBS-treated biofilms (Fig. 4A). Moreover, as noted above, some fraction of cells from A53C-His-PEG-treated biofilms exhibited anomalous morphology (Fig. 4D). This atypical morphology might correlate with altered cellular metabolic activity, which could in turn impact the alamarBlue assay. Together, these observations might explain the apparent discrepancy between alamarBlue and SEM analyses of A53C-His-PEG- versus PBS-treated biofilms.
DISCUSSION
P. aeruginosa's persistence in the CF airway can, to a large extent, be attributed to the bacterium's biofilm mode of growth (36). Additionally, P. aeruginosa's transition to a mucoid phenotype in the majority of CF patients (1) leads to alginate-rich biofilms and enhanced antibiotic resistance (7). Therefore, disruption of the mucoid biofilm architecture with alginate lyases has long been considered a promising approach for enhancing antibiotic efficacy (16), and recent studies support this hypothesis (17, 19). In contrast, at least one report showed that exogenously added Azotobacter vinelandii alginate lyase failed to remove mucoid P. aeruginosa biofilms despite the enzyme's confirmed activity toward the alginate substrate (37). One explanation for such an outcome is that the alginate biopolymer has a negligible contribution to mucoid biofilm adhesion (38). Regardless, the apparent disconnect between reports of alginate lyase antibiotic synergy versus enzyme-mediated biofilm disruption motivated the design of systematic studies to more fully assess the antibacterial potential of these biocatalysts.
Alginate lyase A1-III–His was selected over AlgL-His for the initial biofilm cell viability assays as a result of the enzyme's superior alginate degradation kinetics. SEM analysis showed that A1-III–His drastically reduced extracellular matrix compared to untreated and tobramycin-treated biofilms. Additionally, cotreatment of biofilms with A1-III–His and tobramycin demonstrated a vast improvement in cell killing compared to no treatment or tobramycin-only treatment. These results are consistent with a long series of prior reports demonstrating cooperativity between alginate lyases and antibiotics (14, 16–18). Surprisingly, equivalent results were obtained in controlled studies with an isogenic nonmucoid strain that does not produce alginate exopolysaccharide. Although not peer reviewed, a recent report from the University of British Columbia found similar results with the nonmucoid P. aeruginosa strain PAO1 (39), which also fails to produce alginate under typical growth conditions (40). These observations suggested that alginate lyase catalytic activity was not a requisite factor for biofilm disruption or synergy with the antibiotic tobramycin. To test this hypothesis, we repeated our biofilm cell viability assays with the genetically inactivated alginate lyase A1-DM-His, as well as BSA sham treatments. Both catalytically inactive proteins yielded SEM evidence of biofilm dispersion and exhibited enhanced killing of biofilm cells when used in combination with tobramycin. Thus, biofilm dispersal and increased antibiotic efficacy appear to be nonspecific effects manifested by proteins in general. While early reports showed that heat-treated alginate lyase improved macrophage uptake of P. aeruginosa (15), the results reported here represent the first direct evidence that alginate lyase catalytic activity is superfluous to biofilm disruption and antibiotic synergy.
An alternative explanation for the antibiofilm effects of alginate lyases could be related to enzyme-mediated changes in cell physiology. Indeed, there is evidence that the growth behavior of P. aeruginosa and other biofilm-associated cells is a greater contributor to antibiotic resistance than any physical barrier the biofilm matrix might provide (41, 42). Nutrient limitation within biofilm communities leads to a general reduction in cellular metabolism (43), as well as specific response mechanisms that increase antibiotic tolerance (44). Another key piece of the puzzle is that P. aeruginosa biofilms can be dispersed by sudden increases in carbon source availability (45), and it has been shown that this effect depends upon protein synthesis (46). Additionally, arginine treatment is thought to upregulate the metabolism of P. aeruginosa in anaerobic biofilms, and it thereby increases antibiotic efficacy by 10- to 100-fold (47). Thus, even simple nutrients have the potential to profoundly influence adherent cell behavior and/or modulate antibiotic efficacy toward biofilms.
These insights motivated a hypothesis that, as opposed to actively degrading biofilm matrix, alginate lyases were acting as a nutrient source, modulating cellular metabolism and thus inducing cellular detachment and enhancing tobramycin efficacy. To test this hypothesis, biofilms were treated with the individual amino acid components of A1-III–His alginate lyase. With respect to enhancing tobramycin efficacy, the amino acid cotreatments matched the effectiveness of the intact enzyme. In the absence of tobramycin, however, SEM imaging showed the amino acid treatments failed to completely remove biofilm matrix surrounding the adherent cells. This contrasted with the intact A1-III–His, A1-DM-His, or BSA treatment, each of which individually resulted in complete erosion of the biofilm matrix. A potential explanation for the observed differences in the dispersive potential of intact proteins versus constituent amino acids relates to bioavailability. During infection, bacteria encounter nutrient-depleted environments and secrete proteases that can, through various mechanisms, facilitate degradation of host proteins for use as a nutrient source (48, 49). At the same time, several secreted pseudomonal proteases have been implicated in the degradation of structural proteins or adhesins that contribute to biofilm formation (50, 51). Together, these facts suggest a nonintuitive mechanism for the antibiofilm effects of alginate lyases: (i) biofilm bacteria sense the intact protein treatments and in response secrete proteases, (ii) subsequent assimilation of the freed amino acids upregulates cellular metabolism and renders the bacteria more susceptible to killing by tobramycin, and (iii) the secreted proteases destabilize key components of the biofilm matrix and initiate biofilm dissolution. Although this mechanism is speculative in nature, the literature supports each postulate independently, and their proposed connectivity is consistent with the observations reported here.
The above hypothesis was tested in a preliminary fashion by treating mucoid biofilms with the A53C-His-PEG alginate lyase, whose pendent 20-kDa PEG chain should at least partly shield the protein from proteases. Consistent with prior reports of the enzyme's ability to disrupt mucoid biofilms (12), SEM imaging revealed that A53C-His-PEG itself caused nearly complete dissolution of the biofilm matrix. The above-described mechanism would predict this outcome, if indeed the bacteria sensed the enzyme and subsequently secreted proteases. In contrast to all other intact protein treatments, however, a combination of A53C-His-PEG with tobramycin was no more effective at killing than treatment with tobramycin alone. Again, this is consistent with the proposed mechanism, if tobramycin synergy is ultimately dependent on the presence of individual amino acids, which are less readily liberated from the PEGylated enzyme. As a corollary, direct treatment of biofilms with constituent amino acids manifests a less pronounced degradation of biofilm matrix but fully replicates the intact proteins' synergy with tobramycin. Regardless of the mechanistic details, the studies reported here demonstrate that protein treatments nonspecifically promote P. aeruginosa biofilm dispersal, and they may enhance tobramycin efficacy through the action of their constituent amino acids.
Taken together, the above results reveal that complete biofilm dispersal is neither sufficient nor required for tobramycin synergy with biological cotreatments. Importantly, the results disclosed here do not contradict prior reports of therapeutically relevant activity among alginate lyases. Indeed, they reinforce the widely published conclusions that this class of enzymes manifests biofilm-dispersive properties and synergy with antibiotics, yet the current studies are the first to probe the mechanism underlying these effects. In the biofilm model used here, alginate lyase catalytic activity is irrelevant to biofilm dispersion and tobramycin synergy. In fact, equivalent disruption of the biofilm matrix is achieved with the commodity protein BSA, and equivalent tobramycin synergy is achieved with either BSA or simple amino acids. While further work is required to extend the current studies to more clinically relevant models, it is clear that the long history of alginate lyase enzyme therapies should be reexamined with a more critical eye. In particular, the motivating assumption that enzymatic degradation of alginate would drive therapeutic effects may well be unfounded.
ACKNOWLEDGMENTS
This work was supported by grants from the National Center for Research Resources (5P20RR018787-10) and the National Institute of General Medical Sciences (8 P20 GM103413-10) of the National Institutes of Health. John W. Lamppa was also supported in part by a Renal Function and Disease Training Grant (T32 DK007301) from the NIH.
We thank Charles Daghlian and Louisa Howard for their assistance with the SEM imaging, as well as George O'Toole for the kind gift of strain SMC406 and critical comments on the manuscript.
Footnotes
Published ahead of print 15 October 2012
REFERENCES
- 1. Elkin S, Geddes D. 2003. Pseudomonal infection in cystic fibrosis: the battle continues. Expert Rev. Anti Infect. Ther. 1:609–618 [DOI] [PubMed] [Google Scholar]
- 2. Hoiby N, Krogh Johansen H, Moser C, Song Z, Ciofu O, Kharazmi A. 2001. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microbes Infect. 3:23–35 [DOI] [PubMed] [Google Scholar]
- 3. Smedley YM, Marriott Hodges CN, James SL. 1986. Rheological interactions of cystic fibrosis tracheal mucin and Pseudomonas aeruginosa extracellular alginate. J. Pharm. Pharmacol. 38:54P2869128 [Google Scholar]
- 4. Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762–764 [DOI] [PubMed] [Google Scholar]
- 5. Davies JC, Bilton D. 2009. Bugs, biofilms, and resistance in cystic fibrosis. Respir. Care 54:628–638 [DOI] [PubMed] [Google Scholar]
- 6. Ramsey DM, Wozniak DJ. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 56:309–322 [DOI] [PubMed] [Google Scholar]
- 7. Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. May TB, Shinabarger D, Maharaj R, Kato J, Chu L, DeVault JD, Roychoudhury S, Zielinski NA, Berry A, Rothmel RK. 1991. Alginate synthesis by Pseudomonas aeruginosa: a key pathogenic factor in chronic pulmonary infections of cystic fibrosis patients. Clin. Microbiol. Rev. 4:191–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Murata K, Inose T, Hisano T, Abe S, Yonemoto Y, Yamashita T, Takagi M, Sakaguchi K, Kimura A, Imanaka T. 1993. Bacterial alginate lyase: enzymology, genetics and application. J. Ferment. Bioeng. 76:427–437 [Google Scholar]
- 10. Mrsny RJ, Lazazzera BA, Daugherty AL, Schiller NL, Patapoff TW. 1994. Addition of a bacterial alginate lyase to purulent CF sputum in vitro can. Result in the disruption of alginate and modification of sputum viscoelasticity. Pulm. Pharmacol. 7:357–366 [DOI] [PubMed] [Google Scholar]
- 11. Strathmann M, Wingender J, Flemming H-C. 2002. Application of fluorescently labelled lectins for the visualization and biochemical characterization of polysaccharides in biofilms of Pseudomonas aeruginosa. J. Microbiol. Methods 50:237–248 [DOI] [PubMed] [Google Scholar]
- 12. Lamppa JW, Ackerman ME, Lai JI, Scanlon TC, Griswold KE. 2011. Genetically engineered alginate lyase-PEG conjugates exhibit enhanced catalytic function and reduced immunoreactivity. PLoS One 6:e17042 doi:10.1371/journal.pone.0017042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mai GT, Seow WK, Pier GB, McCormack JG, Thong YH. 1993. Suppression of lymphocyte and neutrophil functions by Pseudomonas aeruginosa mucoid exopolysaccharide (alginate): reversal by physicochemical, alginase, and specific monoclonal antibody treatments. Infect. Immun. 61:559–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bayer AS, Speert DP, Park S, Tu J, Witt M, Nast CC, Norman DC. 1991. Functional role of mucoid exopolysaccharide (alginate) in antibiotic-induced and polymorphonuclear leukocyte-mediated killing of Pseudomonas aeruginosa. Infect. Immun. 59:302–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eftekhar F, Speert DP. 1988. Alginase treatment of mucoid Pseudomonas aeruginosa enhances phagocytosis by human monocyte-derived macrophages. Infect. Immun. 56:2788–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bayer AS, Park S, Ramos MC, Nast CC, Eftekhar F, Schiller NL. 1992. Effects of alginase on the natural history and antibiotic therapy of experimental endocarditis caused by mucoid Pseudomonas aeruginosa. Infect. Immun. 10:3979–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alkawash MA, Soothill JS, Schiller NL. 2006. Alginate lyase enhances antibiotic killing of mucoid Pseudomonas aeruginosa in biofilms. APMIS 114:131–138 [DOI] [PubMed] [Google Scholar]
- 18. Hatch RA, Schiller NL. 1998. Alginate lyase promotes diffusion of aminoglycosides through the extracellular polysaccharide of mucoid Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:974–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alipour M, Suntres ZE, Omri A. 2009. Importance of DNase and alginate lyase for enhancing free and liposome encapsulated aminoglycoside activity against Pseudomonas aeruginosa. J. Antimicrob. Chemother. 64:317–325 [DOI] [PubMed] [Google Scholar]
- 20. Hisano T, Nishimura M, Yonemoto Y, Abe S, Yamashita T, Sakaguchi K, Kimura A, Murata K. 1993. Bacterial alginate lyase highly active on acetylated alginates. J. Ferment. Bioeng. 75:332–335 [Google Scholar]
- 21. Sakakibara H, Tamura T, Suzuki TH, Abe TS, Murata K. 2002. Preparation and properties of alginate lyase modified with poly(ethylene glycol). J. Pharm. Sci. 91:1191–1199 [DOI] [PubMed] [Google Scholar]
- 22. Albrecht MT, Schiller NL. 2005. Alginate lyase (AlgL) activity is required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 187:3869–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jain S, Ohman DE. 2005. Role of an alginate lyase for alginate transport in mucoid Pseudomonas aeruginosa. Infect. Immun. 73:6429–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hershberger CD, Ye RW, Parsek MR, Xie ZD, Chakrabarty AM. 1995. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative sigma factor (sigma E). Proc. Natl. Acad. Sci. U. S. A. 92:7941–7945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoon H-J, Hashimoto W, Miyake O, Murata K, Mikami B. 2001. Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 A resolution. J. Mol. Biol. 307:9–16 [DOI] [PubMed] [Google Scholar]
- 26. Wingender J, Strathmann M, Rode A, Leis A, Flemming HC. 2001. Isolation and biochemical characterization of extracellular polymeric substances from Pseudomonas aeruginosa, p 302–314 In Doyle RJ. (ed), Microbial growth in biofilms, part A. Academic Press Inc., San Diego, CA: [DOI] [PubMed] [Google Scholar]
- 27. Boyd A, Chakrabarty AM. 1994. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 60:2355–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schiller NL, Monday SR, Boyd CM, Keen NT, Ohman DE. 1993. Characterization of the Pseudomonas aeruginosa alginate lyase gene (algL): cloning, sequencing, and expression in Escherichia coli. J. Bacteriol. 175:4780–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yamasaki M, Moriwaki S, Miyake O, Hashimoto W, Murata K, Mikami B. 2004. Structure and function of a hypothetical Pseudomonas aeruginosa protein PA1167 classified into family PL-7. J. Biol. Chem. 279:31863–31872 [DOI] [PubMed] [Google Scholar]
- 30. Pedersen SS, Espersen F, Hoiby N, Shand GH. 1989. Purification, characterization, and immunological cross-reactivity of alginates produced by mucoid Pseudomonas aeruginosa from patients with cystic fibrosis. J. Clin. Microbiol. 27:691–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Elkhatib WF, Noreddin AM. 2009. A new fluorogenic assay for monitoring and determining planktonic and biofilm forms of Pseudomonas aeruginosa viable count in vitro. J. Rapid Methods Autom. Microbiol. 17:304–314 [Google Scholar]
- 32. Novartis Pharmaceuticals 2010. Prescribing information for PrTobi* (tobraymycin solution for inhalation) 300 mg/5 ml tobramycin (as sulfate) respiratory antibiotic. Novartis Pharmaceuticals, Dorval, Quebec, Canada [Google Scholar]
- 33. Caiazza NC, O'Toole GA. 2004. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J. Bacteriol. 186:4476–4485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kodera Y, Matsushima A, Hiroto M, Nishimura H, Ishii A, Ueno T, Inada Y. 1998. Pegylation of proteins and bioactive substances for medical and technical applications. Progr. Polymer Sci. 23:1233–1271 [Google Scholar]
- 35. Harris JM, Chess RB. 2003. Effect of pegylation on pharmaceuticals. Nat. Rev. Drug Discov. 2:214–221 [DOI] [PubMed] [Google Scholar]
- 36. Høiby N, Ciofu O, Bjarnsholt T. 2010. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 5:1663–1674 [DOI] [PubMed] [Google Scholar]
- 37. Christensen BE, Ertesvag H, Beyenal H, Lewandowski Z. 2001. Resistance of biofilms containing alginate-producing bacteria to disintegration by an alginate degrading enzyme (AlgL). Biofouling 17:203–210 [Google Scholar]
- 38. Xavier JB, Picioreanu C, Rani SA, van Loosdrecht MCM, Stewart PS. 2005. Biofilm-control strategies based on enzymic disruption of the extracellular polymeric substance matrix: a modelling study. Microbiology 151:3817–3832 [DOI] [PubMed] [Google Scholar]
- 39. Cotton LA, Graham RJ, Lee RJ. 2009. The role of alginate in P. aeruginosa PAO1 biofilm structural resistance to gentamicin and ciprofloxacin. J. Exp. Microbiol. Immunol. 13:58–62 [Google Scholar]
- 40. Wozniak DJ, Wyckoff TJO, Starkey M, Keyser R, Azadi P, O'Toole GA, Parsek MR. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. U. S. A. 100:7907–7912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walters MC, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47:317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qu Y, Daley AJ, Istivan TS, Rouch DA, Deighton MA. 2010. Densely adherent growth mode, rather than extracellular polymer substance matrix build-up ability, contributes to high resistance of Staphylococcus epidermidis biofilms to antibiotics; authors' response. J. Antimicrob. Chemother. 65:2055–2056 [DOI] [PubMed] [Google Scholar]
- 43. Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6:199–210 [DOI] [PubMed] [Google Scholar]
- 44. Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sauer K, Cullen MC, Rickard AH, Zeef LAH, Davies DG, Gilbert P. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 186:7312–7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roy AB, Petrova OE, Sauer K. 2012. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J. Bacteriol. 194:2904–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Borriello G, Richards L, Ehrlich GD, Stewart PS. 2006. Arginine or nitrate enhances antibiotic susceptibility of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 50:382–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Travis J, Potempa J. 2000. Bacterial proteinases as targets for the development of second-generation antibiotics. Biochim. Biophys. Acta 1477:35–50 [DOI] [PubMed] [Google Scholar]
- 49. Supuran CT, Scozzafava A, Clare BW. 2002. Bacterial protease inhibitors. Med. Res. Rev. 22:329–372 [DOI] [PubMed] [Google Scholar]
- 50. Nikolaev YA, Panikov NS. 2002. Extracellular protease as a reversible adhesion regulator in Pseudomonas fluorescens. Microbiology 71:541–545 [PubMed] [Google Scholar]
- 51. Tielen P, Rosenau F, Wilhelm S, Jaeger K-E, Flemming Wingender H-CJ. 2010. Extracellular enzymes affect biofilm formation of mucoid Pseudomonas aeruginosa. Microbiology 156:2239–2252 [DOI] [PubMed] [Google Scholar]