Abstract
Vancomycin-resistant Staphylococcus aureus (VRSA) is thought to result from the in vivo conjugative transfer of a vanA plasmid from an Enterococcus sp. to S. aureus. We studied bacterial isolates from VRSA cases that occurred in the United States to identify microbiological factors which may contribute to this plasmid transfer. First, vancomycin-susceptible, methicillin-resistant S. aureus (MRSA) isolates from five VRSA cases were tested for their ability to accept foreign DNA by conjugation in mating experiments with Enterococcus faecalis JH2-2 containing pAM378, a pheromone-response conjugative plasmid. All of the MRSA isolates accepted the plasmid DNA with similar transfer efficiencies (∼10−7/donor CFU) except for one isolate, MRSA8, for which conjugation was not successful. The MRSA isolates were also tested as recipients in mating experiments between an E. faecalis isolate with an Inc18-like vanA plasmid that was isolated from a VRSA case patient. Conjugative transfer was successful for 3/5 MRSA isolates. Successful MRSA recipients carried a pSK41-like plasmid, a staphylococcal conjugative plasmid, whereas the two unsuccessful MRSA recipients did not carry pSK41. The transfer of a pSK41-like plasmid from a successful MRSA recipient to the two unsuccessful recipients resulted in conjugal transfer of the Inc18-like vanA plasmid from E. faecalis at a frequency of 10−7/recipient CFU. In addition, conjugal transfer could be achieved for pSK41-negative MRSA in the presence of a cell-free culture filtrate from S. aureus carrying a pSK41-like plasmid at a frequency of 10−8/recipient CFU. These results indicated that a pSK41-like plasmid can facilitate the transfer of an Inc18-like vanA plasmid from E. faecalis to S. aureus, possibly via an extracellular factor produced by pSK41-carrying isolates.
INTRODUCTION
Vancomycin-resistant Staphylococcus aureus (VRSA) was first reported in 2002. To date, there have been 13 cases in the United States (1, 2; http://www.cdc.gov/HAI/settings/lab/vrsa_lab_search_containment.html), several reports of isolates in India (3, 4), and unconfirmed cases in Iran (5). The occurrence of vancomycin resistance in S. aureus is a concern from a patient management and public health perspective. Vancomycin is the treatment of choice for serious infections caused by MRSA, and all of the VRSA isolates have been MRSA that acquired plasmid-borne, vanA-mediated vancomycin resistance. Often, the presence of a mobile genetic determinant conferring resistance to a primary therapeutic agent in a common bacterial pathogen suggests an impending public health crisis. However, there is no evidence of VRSA transmission between patients, no patients have died of a VRSA infection, and there are newer antimicrobial agents that are active against VRSA (6; Centers for Disease Control and Prevention, unpublished data). In addition, the expression of vanA appears to have a fitness cost for MRSA. As a result, some have questioned whether VRSA really matters (7). It is difficult to predict the extent to which VRSA may become a public health threat. Nevertheless, understanding how S. aureus acquired vanA plasmids may provide important insight into the plasmid transfer mechanisms between Gram-positive bacteria of different genera, namely, Enterococcus spp. and S. aureus. If the plasmid transfer that occurred in the VRSA case becomes a more frequent event, then the potential for transfer of resistance determinants to S. aureus could also increase.
Bacterial conjugation is an important mechanism for the horizontal spread of genetic material, including antimicrobial resistance genes, between bacteria. Most VRSA have occurred in polymicrobial infections, and in many cases a vancomycin-resistant enterococcus (VRE) isolate and a vancomycin-susceptible MRSA isolate were recovered, along with the VRSA isolate from the case patient. Previously, we reported an Inc18-like vanA plasmid that was associated with at least five of seven VRSA cases in Michigan (1), and VRE strains carrying this plasmid were found more frequently in Michigan than in other states. This finding may at least partially explain why most of the cases VRSA infection occurred in Michigan. The VRE strains isolated from VRSA case patients are the likely donors of vanA to S. aureus, resulting in a VRSA strain (1, 8, 9).
Inc18 incompatibility plasmids are a family of broad-host-range conjugative plasmids that occur naturally in isolates of Enterococcus and Streptococcus spp. The streptococcal plasmid pIP501 and E. faecalis plasmids pAMβ1 and pRE25 are well-characterized examples of this group (10). These plasmids often carry antimicrobial resistance determinants, including ermB, which confers resistance to macrolides. In the laboratory, Inc18-like plasmids can be transferred by conjugation to a wide variety of bacteria, including streptococci (11), lactococci (12), staphylococci (13), and enterococci (14), but with the exception of a few VRSA strains that maintained the VRE Inc18-like vanA plasmid, these plasmids have not been identified in naturally occurring staphylococci (15).
The first U.S. VRSA isolate carried the vanA transposon, Tn1546, on a pSK41-like plasmid (8). It was hypothesized that vanA was transferred to S. aureus by conjugative transfer of the vanA plasmid from a VRE to S. aureus and then moved by the transposition of Tn1546 to the pSK41-like plasmid of S. aureus, and the Inc18-like vanA plasmid was then lost during replication (1, 8).
The pSK41-like plasmids are a family of multiresistant conjugative plasmids of staphylococci (16, 17). The members of this plasmid family include pSK41, pG01, and pJE1 (17). These plasmids were initially observed in studies characterizing gentamicin resistance in S. aureus (17–19). Conjugative transfer of these plasmids was thought to be limited to Staphylococcus spp. (18, 20). In addition to mediating their own conjugative transfer, pSK41-like plasmids are able to mobilize coresident plasmids (21–23). Conjugation by pSK41 family plasmids is mediated by a transfer system composed of ∼15 genes (10). The transfer mechanism of pSK41-like plasmids is related to that of Inc18-like plasmids. Specifically, the transfer mechanisms demonstrate similar genetic organization and tra product amino acid sequences (10), especially for the first six genes of the plasmid transfer regions. The transfer mechanisms of pSK41-like plasmids and Inc18-like plasmids have been categorized into the same superfamily of macromolecular transport mechanisms, called type IV secretion systems (24).
In this study our aim was to understand VRSA occurrence by identifying factors that contribute to conjugative transfer of vanA plasmids from Enterococcus donor isolates to S. aureus recipient isolates from VRSA case patients.
MATERIALS AND METHODS
Bacterial isolates.
The strains used in the present study are listed in Table 1. VRSA isolates 1 to 9 all were obtained in the United States. Either vancomycin-resistant E. faecalis or E. avium were collected from the associated VRSA cases (1). Vancomycin-susceptible S. aureus (MRSA) isolates are MRSA isolates that were either coisolated from the same body site as the VRSA (MRSA5, -7, and -8) or from other body sites (MRSA1 and -3) (see Table 1).
Table 1.
Strains from VRSA cases and other strains used in this study
| Case | U.S. state | Strain | Phenotypea |
Comment(s) | PCR result |
Reference(s) | |||
|---|---|---|---|---|---|---|---|---|---|
| OXA | VAN | GEN | pSK41 traE/traK | Inc18 traA/repR | |||||
| VR strains | |||||||||
| 1 | MI | VRSA1 | R | R | R | Isolated from plantar ulcer | +/+ | −/− | 2, 8 |
| MI | MRSA1 | R | S | R | Isolated from the nares swab | +/+ | −/− | 2, 8 | |
| MI | VR-E. faecalis1 | S | R | R | Coisolated with VRSA from plantar ulcer | −/− | +/+ | 2, 8 | |
| 2 | PA | VRSA2 | R | R | S | Isolated from plantar ulcer | −/− | −/− | 2 |
| 3 | NY | VRSA3 | R | R | R | Isolated from urine specimen | −/− | −/− | 2 |
| NY | MRSA3 | R | S | R | Isolated from a rectal swab | +/+ | −/− | 2, 24 | |
| 4 | MI | VRSA4 | R | R | S | Isolated from a toe wound | −/− | +/+ | 2 |
| MI | VR-E. faecalis4 | S | R | R | Isolated from a rectal swab | −/− | +/+ | 2 | |
| 5 | MI | VRSA5 | R | R | S | Isolated from a surgical wound site | −/− | +/+ | 2 |
| MI | MRSA5 | R | S | S | Coisolated with VRSA from wound | −/− | −/− | 2 | |
| MI | VR-E. faecalis5 | S | R | R | Coisolated with VRSA from wound | −/− | +/+ | 2 | |
| 6 | MI | VRSA6 | R | R | S | Isolated from a plantar ulcer | −/− | −/− | 2 |
| MI | VR-E. avium6 | S | R | R | Isolated from a rectal swab | −/− | +/+ | 2 | |
| 7 | MI | VRSA7 | R | R | S | Isolated from a triceps wound | −/− | +/+ | 2 |
| MI | MRSA7 | R | S | S | Coisolated with VRSA from wound | −/− | −/− | 2 | |
| 8 | MI | VRSA8 | R | R | R | Isolated from a foot wound | +/+ | −/− | 2 |
| MI | MRSA8 | R | S | R | Coisolated with VRSA from wound | +/+ | −/− | 2 | |
| 9 | MI | VRSA9 | R | R | R | Isolated from a surgical site wound | +/+ | −/− | 2 |
| Other strains | |||||||||
| JH2-2/pVREfs1 | R | S | R | E. faecalis JH2-2 transconjugant with vanA plasmid from VR-E. faecalis1 | −/− | +/+ | This study | ||
| MRSA5/pMRSA1 | R | S | R | MRSA5 transformed with the pSK41-like plasmid from MRSA1 | +/+ | −/− | This study | ||
| MRSA7/pMRSA1 | R | S | R | MRSA7 transformed with the pSK41-like plasmid from MRSA1 | +/+ | −/− | This study | ||
| S. aureus RN8325-4 | R | S | S | Derived from 8325, plasmid-free strain | −/− | −/− | 25 | ||
| S. aureus RN4220 | S | S | S | Lab strain derived from S. aureus 8325-4 | −/− | −/− | 26 | ||
| S. aureus RN4220/pSK41 | S | S | S | Strain carrying the prototype pSK41 plasmid | +/+ | −/− | 27 | ||
OXA, oxacillin; VAN, vancomycin; GEN, gentamicin.
PCR detection of pSK41-like and Inc18-like plasmids.
Two multiplex PCRs were performed for detection of pSK41-like and Inc18-like vanA plasmids. PCRs were carried out according to the manufacturer's instruction (multiplex PCR kit; Qiagen, Valencia, CA). Briefly, PCRs were performed in a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA) with the following reaction cycles: an initial denaturation step of 2 min at 94°C, followed by 30 cycles of 15 s at 95°C, 90 s at 55°C, and 90 s at 72°C, with a final elongation for 7 min at 72°C. PCR products were visualized on a 1% agarose gel. The primer sequences (2, 28–30) targeting Inc18-like plasmids were as follows: vanA F (CATGAATAGAATAAAAGTTGCTGCAATA), vanA R (CCCCTTTAACGCTAATACGATCAA), traA F (TAATCGCAATGGCTTCTTATC), traA R (TCTGCCCAATCTTTACGAAT), repR F (GCTTCATGACGGCTTGTTA), and repR R (TTGGCTGCTTTGACAGATTTA) and those targeting pSK41-like plasmids were traE F (ACAAATGCGTACTACAGACCCTAAACGA), traE R (GCCCTGCTGTTGCTGTATCCATATT), traK F (TTGCCGAAGATAGCGAATTG), and traK R (CTGCAATACCTTCGGTCAGTTC).
DNA transformation.
Plasmid DNA was isolated from 50-ml overnight cultures of MRSA1 or S. aureus RN4220, which carried the prototype pSK41 plasmid using a plasmid kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. The pSK41 plasmid DNA was introduced by electroporation into isolates MRSA5 and -7 to generate strains MRSA5/pMRSA1, MRSA5/pSK41, MRSA7/pMRSA1, and MRSA7/pSK41, respectively. The electroporation was performed by preparing a bacterial suspension in 10% glycerol electroporation buffer with 1 μg of DNA in a 1-mm cuvette which was then subjected to a single pulse using the Bio-Rad Gene Pulser (Bio-Rad Laboratories, Hercules, CA) set at 2.5 kV and 25 μF, with the pulse controller set at infinity. The content of the cuvette was diluted into 2 ml of tryptic soy broth and incubated at 37°C for 2 h. After incubation, aliquots were plated onto Trypticase soy agar plates containing 15 μg of gentamicin/ml. The gentamicin-resistant transformants were tested for the presence of pSK41-like plasmids by PCR amplification of traE and traK.
To construct an E. faecalis isolate that carried only a vanA Inc18-like plasmid, the vanA plasmid from VR-E. faecalis1 was isolated and transferred to E. faecalis JH2-2 by electroporation. The methods were as described above except the transformants were selected on agar containing 6 μg of vancomycin/ml.
Conjugation assay.
Mating experiments between Enterococcus spp. with an Inc18-like vanA plasmid and MRSA isolates were carried out by combining mid-log-phase brain heart infusion (BHI) cultures at a donor to recipient ratio of 20:1 in a volume of 200 μl, which was applied to a 0.45-μm-pore-size filter. Filters were removed from the filter unit, placed on a BHI agar plate without antimicrobials, and incubated at 35°C for 24 h. In some experiments, 20 μl of cell-free filtrate from a bacterial culture was added to the conjugation mixture by applying the filtrate to the filter on the agar plate. After the 24-h incubation, filters were removed from the agar and placed in 5 ml of BHI broth and mixed by vortexing to remove the growth. Serial dilutions of the conjugation mixture were prepared, and 200 μl of each dilution was plated onto agar containing antimicrobial agents for selection. Each dilution was plated in triplicate. Transconjugants were selected on BHI agar with 5 μg of vancomycin/ml and 30 μg of ampicillin/ml and analyzed by PCR for vanA, Inc18 vanA plasmid genes (traA, repR), and pSK41 genes (traE, traK). In order to count the numbers of donors and recipients in each mating experiment, the dilutions were also plated onto BHI agar containing either vancomycin (5 μg/ml) or ampicillin (30 μg/ml) for selection of VRE (donor) and S. aureus (recipient) growth, respectively. Colony counts from these plates were used to calculate the transfer efficiency, which was expressed as transformants/recipient CFU. All mating experiments were performed in triplicate.
Cell-free filtrates were prepared by inoculating growth from an overnight culture of S. aureus on Trypticase soy agar with 5% sheep blood to 20 ml of BHI broth and incubated for 6 to 8 h at 35°C. The culture was filtered through a 0.45-μm-pore-size filter, and 20 μl was immediately added to the conjugation cell mixture and plated.
For evaluation of the ability of the VRSA-associated MRSA isolates to accept the other foreign DNA, conjugation assays were performed by using E. faecalis JH2-2 containing pAM378 (31), a pheromone-responsive conjugative plasmid that confers tetracycline resistance, as the donor and the respective MRSA strains as the recipients. The conjugation procedures were the same as those described above except the mating mix was composed of 50 μl of donor to 500 μl of recipient (1:10 ratio) and the selective plates contained BHI plus either 25 μg of ampicillin/ml (Amp), 10 μg of tetracycline/ml (Tet), or Amp-Tet. S. aureus 8325-4 and RN4220 were used as control recipient strains in the conjugation assays. Previous studies demonstrated that strain RN4220 can acquire this plasmid but that the parent strain of RN4220, S. aureus 8325-4, cannot. This difference is the result of a stop codon mutation in hsdR of RN4220 which encodes the restriction enzyme of a type I restriction system (32).
Stability assay.
The stability of vancomycin resistance in transconjugants was tested by subculturing one colony from nonselective BHI agar for 10 consecutive passages and then subculturing on BHI agar containing vancomycin at 6 μg/ml to select for cells which remained resistant to vancomycin.
DNA sequencing.
The DNA templates were prepared by a previously reported NaOH lysis procedure (8). The primers described by Waldron and Lindsay (32) were used to amplify and sequence hsdR from MRSA and VRSA isolates. The target was amplified using the AccuPrime Taq DNA polymerase kit (Invitrogen, Carlsbad, CA). The 50- μl reaction mix included 1× buffer II, 100 nM concentrations of each primer, 3 μl of template (1 to 200 ng), and 1 μl of AccuPrime Taq DNA polymerase. The reaction conditions included an initial denaturation step at 94°C for 5 min, 30 cycles of 94°C for 30 s, 54°C for 30 s, and 68°C for 3 min, an extension for 5 min at 68°C. A GeneAmp 9700 thermocycler (Applied Biosystems) was used for all of the PCR amplifications. PCR products were purified with QIA quick spin columns (Qiagen), and the DNA quantitated on an ND1000 spectrophotometer (NanoDrop, Inc., Rockland, DE) before sequencing. PCR products served as the template for sequencing reactions with the BigDye Terminator v.3.1 cycle sequencing kits (Applied Biosystems). Centri-Sep 8 strips and 96-well plates (Princeton Separation, Adelphia, NJ) were used to remove excess dye terminators from the sequencing reactions before the sequence was determined on an ABI 3130xl automated DNA genetic Analyzer (Applied Biosystems). DNA sequences were aligned and compared using Sequencher software (GeneCodes Corp., Ann Arbor, MI).
Susceptibility testing.
Susceptibility to vancomycin and other antimicrobial agents was determined by reference broth microdilution using in-house prepared panels with cation-adjusted Mueller-Hinton broth (BD Diagnostics Systems, Sparks, MD). The susceptibility methods and interpretation were performed according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (33, 34).
RESULTS
Conjugation frequency of VRSA parent strains.
To determine whether VRSA parent strains were more likely to acquire foreign DNA than a control S. aureus strain, five MRSA strains, which were isolated from five different VRSA cases (1, 3, 5, 7, and 8), were chosen as recipient strains for conjugation experiments, since in each case the MRSA isolate demonstrated a SmaI PFGE pattern that was the same as the PFGE pattern from the VRSA isolate of the same case except for a band corresponding to the vanA plasmid (Table 1) (1; CDC, unpublished data). Therefore, these MRSA isolates likely represent the vancomycin-susceptible parent of the VRSA strain. The donor strain was E. faecalis JH2-2 carrying the conjugative plasmid pAM378, which confers a tetracycline resistance.
The MRSA isolates from VRSA cases 1, 3, 5, and 7 acquired pAM378 from E. faecalis at a frequency of 10−7 to 10−8/donor (Table 2). No transconjugants were isolated when the MRSA isolates from VRSA case 8 was the recipient strain. Conjugation frequencies were compared to those for control strains S. aureus RN4220 and S. aureus 8325-4, which were previously characterized for their ability to acquire pAM378 via conjugation (32). Our results are consistent with the previous results demonstrating that strain RN4220 can acquire this plasmid but the parent strain of RN4220, S. aureus 8325-4, cannot. None of the VRSA case-associated MRSA isolates acquired the plasmid at the same frequency of RN4220 (10−6/donor) and, unlike RN4220, no stop codon mutations were identified in the hsdR genes of the MRSA isolates (data not shown).
Table 2.
Conjugation assays for the ability of S. aureus to accept pAM378 from E faecalis JH2-2
| Recipient | Transfer frequency (recipient/donor) |
Avg transfer frequency | ||
|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | ||
| MRSA1 | 9.9 × 10−7 | 9.0 × 10−8 | 6.3 × 10−8 | 3.8 × 10−7 |
| MRSA3 | 4.3 × 10−8 | 1.8 × 10−7 | 5.2 × 10−8 | 9.1 × 10−8 |
| MRSA5 | 3.3 × 10−8 | 2.6 × 10−7 | 2.1 × 10−8 | 1.4 × 10−7 |
| MRSA7 | 1.4 × 10−7 | 4.2 × 10−8 | — | 9.1 × 10−8 |
| MRSA8 | —a | — | — | NAb |
| S. aureus 8325-4 | — | — | — | NA |
| S. aureus RN4220 | 2.5 × 10−6 | 3.5 × 10−7 | 5.6 × 10−7 | 1.1 × 10−6 |
–, No transformants.
NA, not applicable.
Transfer of Inc18-like vanA plasmids from different VRE donor isolates to a single MRSA recipient.
To determine whether vanA plasmids could be transferred in vitro from VRE to S. aureus, we attempted in vitro conjugations using a single S. aureus recipient isolate, MRSA1, in conjugation experiments with one of four different VRE isolates from VRSA case patients as donors: VR-E. faecalis1, VR-E. faecalis4, VR-E. faecalis5, and E. avium6 (Table 3). All of these VRE isolates carried vanA on Inc18-like plasmids (1, 2). Conjugations were successful in all four experiments with a frequency of transfer of ∼10−7 per donor cell in each experiment.
Table 3.
Characteristics of the donors (VRE), recipient (MRSA1), and transconjugants
| Expt | Isolate | Purpose of isolate | PCR result |
Vancomycin MIC (μg/ml) | |||
|---|---|---|---|---|---|---|---|
| vanA | Inc 18 traA/repR | pSK41 traE/traK | ddl (VR-E. faecalis) | ||||
| MRSA1, recipient | Recipient in expts 1 to 5 | − | −/− | +/+ | − | 1 | |
| 1 | VR-E. faecalis1 | Donor | + | +/+ | −/− | + | 512 |
| MRSA1/pVREfs1 | Transconjugant | + | +/+ | +/+ | − | 1,024 | |
| 2 | VR-E. faecalis4 | Donor | + | +/+ | −/− | + | 512 |
| MRSA1/pVREfs4 | Transconjugant | + | +/+ | +/+ | − | >512 | |
| 3 | VR-E. faecalis5 | Donor | + | +/+ | −/− | + | 1,024 |
| MRSA1/pVREfs5 | Transconjugant | + | +/+ | +/+ | − | >512 | |
| 4 | VR-E. avium6 | Donor | + | +/+ | −/− | − | 512 |
| MRSA1/pVREav6 | Transconjugant | + | +/+ | +/+ | − | >512 | |
| 5 | E. faecalis JH2-2/pVREfs1 | Donor | + | +/+ | −/− | + | |
| MRSA1/pJH2-2, VREfs1 | Transconjugant | + | +/+ | +/+ | − | >512 | |
Since it is possible that the VRE donor isolates in these experiments carry plasmids other than the vanA Inc18-like plasmid that facilitate conjugal transfer of the Inc18-like plasmid from the Enterococcus isolate to the S. aureus isolate, we constructed a strain that only carried a Inc18-like vanA plasmid, VR-E. faecalisJH2-2/pVREfs1. This strain was also used as a donor strain in a conjugation experiment with MRSA1 as the recipient. Transconjugants were isolated at the same frequency described above.
Transconjugants were characterized by PCR for vanA, Inc18-like plasmid markers traA and repR, pSK41-like plasmid markers traE and traK, and the E. faecalis-specific ddl gene (35) and by vancomycin susceptibility testing. The results of this analysis indicate that all transconjugants were positive for vanA and resistant to vancomycin, were positive for both an Inc18-like plasmid and a pSK41-like plasmid, and were not contaminated with VRE donor cells (i.e., negative by PCR for E. faecalis ddl) (Table 3). Plasmids from the transconjugants were analyzed by restriction analysis (Fig. 1). These results confirm that the Inc18-like plasmids were maintained, along with the naturally occurring pSK41-like plasmid from MRSA1. This is in contrast to the first VRSA isolates in which a VRE vanA plasmid was not detected, but the Tn1546-like vanA element was inserted in the pSK41-like plasmid (1, 8).
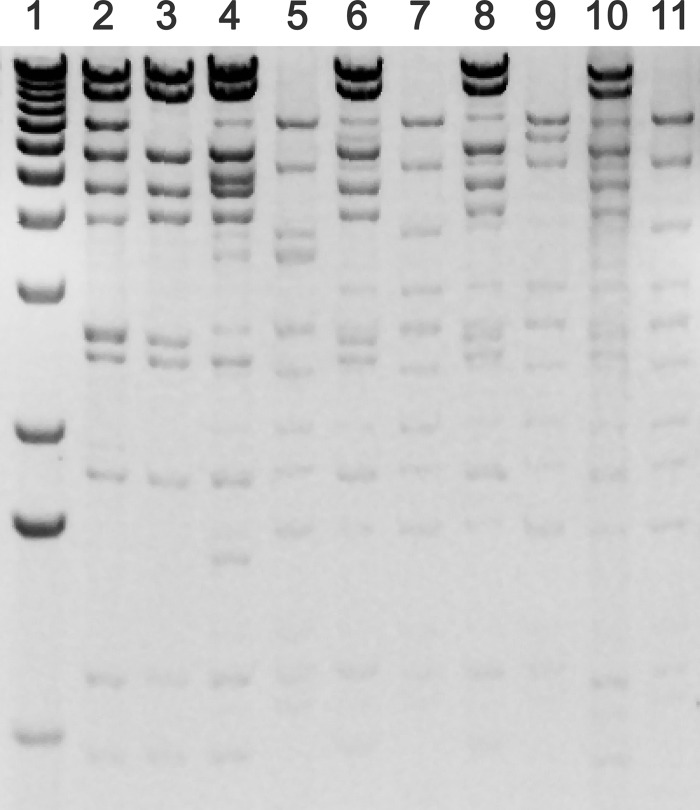
Fig 1.
Comparison of the HindIII restriction profiles of plasmids from donors, recipients, and their transconjugants. Lane 1, 1-kb DNA marker; lane 2, VRSA1; lane 3, recipient MRSA1; lane 4, transconjugant MRSA1/pVR-Efs1; lane 5, donor VR-E. faecalis1; lane 6, transconjugant MRSA1/pVR-Efs 4; lane 7, donor VR-E. faecalis4; lane 8, transconjugant MRSA1/pVR-Efs5; lane 9, donor VR-E. faecalis5; lane 10, transconjugant MRSA1/pVR-Ev6; lane 11, donor VR-E. avium6.
Stability of the Inc18-like vanA plasmid in transconjugants was tested by consecutive subculture of the transconjugants on nonselective BHI agar for 10 passages. Two transconjugants from each conjugation experiments 1 to 4 (Table 3) were analyzed for the stability of the vancomycin resistance. The results indicated that 6 of 8 tested transconjugants lost vancomycin resistance after 10 passages, and only two (MRSA1/pVREfs4 and MRSA1/pVREav6) were still resistant to vancomycin. Plasmid analysis indicated that the vancomycin-susceptible transconjugant derivatives lost the Inc18-like vanA plasmid, while the two resistant transconjugants still retained the plasmid (data not shown).
Conjugative transfer of an Inc18-like vanA plasmid from E. faecalis to different S. aureus recipients.
To determine whether all VRSA-associated MRSA isolates were equally functional as recipients of an Inc18-like vanA plasmid, we conducted conjugation experiments with a single donor, VR-E. faecalis4, which carries an Inc18-like vanA plasmid, with five different VRSA-associated MRSA isolates from cases 1, 3, 5, 7, and 8. The Inc18-vanA plasmid transferred from E. faecalis to only three MRSA isolates (MRSA1, -3, and -8) at a frequency of ∼10−7/recipient CFU. Despite repeated experiments, there was no detectable vanA transfer from the VRE donor to two original MRSA isolates, MRSA5 and -7. In all of the successful conjugation experiments, the MRSA recipient strain naturally carried a pSK41-like plasmid and in all cases where the conjugation failed, the MRSA isolate was negative for a pSK41-like plasmid (Table 4).
Table 4.
Conjugative transfer of an Inc18-like vanA plasmid from E. faecalis to different S. aureus recipientsa
| Recipient | pSK41-like plasmid in recipient | Avg transfer frequencyb (no. of successful conjugation expts/total no. of expts) |
|---|---|---|
| MRSA1 | + | 1.9 × 10−7 (3/3) |
| MRSA3 | + | 8.7 × 10−7 (3/3) |
| MRSA5 | − | NA (0/3) |
| MRSA7 | − | NA (0/3) |
| MRSA8 | + | 7.9 × 10−7 (3/3) |
The donor strain in all experiments was VR-E. faecalis4.
The transfer frequencies were expressed as the number of transconjugants per recipient. NA, not applicable because there were no successful conjugation experiments, the lower level of detection was 10−10.
We investigated whether the presence of a pSK41-like plasmid in the recipient strain facilitated conjugation of the vanA plasmid by introducing such a plasmid into MRSA5 and -7 and repeating the conjugation experiments. Two different pSK41-like plasmids were tested: either the pSK41-like plasmids isolated from MRSA1 or the prototype plasmid isolated from S. aureus strain RN4220. Conjugation of the vanA plasmid was successful when MRSA5 and -7 carried either pSK41-like plasmid (Table 5).
Table 5.
Conjugative transfer of different plasmids to MRSA5 and MRSA7a
| Recipient | Transfer frequencyb (no. of successful conjugation expts/no. of expts) |
|||||
|---|---|---|---|---|---|---|
| None | Culture of MRSA1 | MRSA5 | MRSA5/pMRSA1 | RN4220 | RN4220/pSK41 prototype | |
| Mating expts with VR-E. faecalis4 as donor | ||||||
| MRSA5 | NA (0/3) | <1.8 × 10−8 (2/3) | NA (0/3) | 1.5 × 10−8 (3/3) | NA (0/3) | <2.8 × 10−8 (2/3) |
| Inc18 traA/repR PCR | NA | +/+ | NA | +/+ | NA | +/+ |
| pSK41 traE/traK PCR | NA | −/− | NA | −/− | NA | −/− |
| MRSA5/pMRSA1 | 2.5 × 10−7 (3/3) | ND4 | ND | ND | ND | ND |
| Inc18 traA/repR PCR | +/+ | NA | NA | NA | NA | NA |
| pSK41 traE/traK PCR | +/+ | NA | NA | NA | NA | NA |
| MRSA5/pSK41 prototype | 1.7 × 10−7 (3/3) | ND | ND | ND | ND | ND |
| Inc18 traA/repR PCR | +/+ | NA | NA | NA | NA | NA |
| pSK41 traE/traK PCR | +/+ | NA | NA | NA | NA | NA |
| MRSA7 | NA (0/3) | <10−9 (2/3) | NA (0/3) | <1.6 × 10−9 (1/3) | NA (0/3) | <1.3 × 10−9 (1/3) |
| Inc18 traA/repR PCR | NA | +/+ | NA | +/+ | NA | +/+ |
| pSK41 traE/traK PCR | NA | −/− | NA | −/− | NA | −/− |
| MRSA7/pMRSA1 | 1.8 × 10−8 (3/3) | ND | ND | ND | ND | ND |
| Inc18 traA/repR PCR | +/+ | NA | NA | NA | NA | NA |
| pSK41 traE/traK PCR | +/+ | NA | NA | NA | NA | NA |
| MRSA7/pSK41 prototype | 10−8 (3/3) | ND | ND | ND | ND | ND |
| Inc18 traA/repR PCR | +/+ | NA | NA | NA | NA | NA |
| pSK41 traE/traK PCR | +/+ | NA | NA | NA | NA | NA |
| Mating expts with E. faecalis JH2-2/pAM373 as donor | ||||||
| MRSA5 | 1.4 × 10−7 (3/3)c | ND | ND | ND | ND | ND |
| MRSA5/pMRSA1 | <2.1 × 10−8 (1/3) | ND | ND | ND | ND | ND |
| MRSA7 | <9.1 × 10−8 (2/3) | ND | ND | ND | ND | ND |
| MRSA7/pMRSA1 | <1.4 × 10−8 (2/3)c | ND | ND | ND | ND | ND |
All transformants were positive for vanA and negative for E. faecalis ddl by PCR. PCR results for Inc18- and pSK41-specific genes in transconjugants are listed under each successful conjugation experiment.
The transfer frequencies were expressed as the number of transconjugants per recipient. Average transfer frequencies were reported when more than one conjugation experiment was successful. The cell-free filtrate added to the conjugation mixture is indicated in the subheadings. Some of these results are also in Table 4, but they are repeated here for reference purposes. NA, not applicable because there were no successful conjugation experiments (the lower level of detection was 10−10); ND, not done.
These results were also in Table 1, but they are repeated here as a reference.
The presence of a pSK41-like plasmid in the recipient S. aureus strain was necessary for in vitro conjugal transfer of an Inc18-like vanA plasmid. However, MRSA5 and -7, which were naturally negative for a pSK41-like plasmid, were also the likely recipient strains of an Inc18-like vanA plasmid in the fifth and seventh VRSA cases. It could be that conjugation of an Inc18-like vanA plasmid to a pSK41-negative S. aureus strain might be promoted by a helper cell, e.g., a third cell that carried pSK41. To test this hypothesis, we repeated the conjugation experiments with MRSA5 and -7 as recipients but added a cell-free filtrate from a bacterial culture to determine whether any of the cell-free filtrate facilitated conjugal transfer of the vanA plasmid. Transfer of vanA was evaluated in the presence of a cell-free filtrate from a culture of MRSA1 that carries a pSK41-like plasmid, from a culture of MRSA5, from a culture of MRSA5 transformed with the pSK41-like plasmid from MRSA1 (MRSA5/pMRSA1), from a culture of RN4220, and from a culture of RN4220 transformed with the pSK41 prototype plasmid. Successful conjugal transfer of vanA to MRSA5 and MRSA7 was observed in the presence of a cell-free culture filtrate when the bacterial strain carried a pSK41-like plasmid (MRSA1, MRSA5/pMRSA1, and RN4220/pSK41). Not all conjugation attempts were successful and the conjugation frequency was close to the limit of detection (e.g., a frequency of 10−9 transconjugants per recipient cell with a lower level of detection of 10−10). No transconjugants were detected when the cell extract was from a culture of MRSA5 or RN4220.
We investigated whether the presence of a pSK41-like plasmid could affect conjugation of the pheromone responsive, non-Inc18-like plasmid, pAM378. Mating experiments were conducted using MRSA5/pMRSA1 and MRSA7/pMRSA1 as recipients. The presence of a pSK1-like plasmid in MRSA5 and -7 did not appear to affect the conjugation frequencies (Table 5).
DISCUSSION
Previously, we reported the association of Inc18-like vanA Enterococcus plasmids with VRSA cases. A total of 13 VRSA cases have been identified in the United States and an Inc18-like vanA plasmid was identified in the VRE or VRSA isolates from 8 of 13 cases. The presence of this plasmid in Enterococcus appears to be important for the in vivo transfer of vanA from Enterococcus to S. aureus. In the present study we identified a characteristic of the recipient S. aureus that is necessary for in vitro transfer of this Enterococcus plasmid to S. aureus. Three VRSA-associated MRSA isolates (MRSA1, -3, and -8) which naturally carry the pSK41-like plasmids could accept the Inc18-like vanA plasmids from enterococci, while the other two isolates (MRSA5 and -7), which carry no pSK41-like plasmids had no detectable conjugative transfer. However, when these two isolates were transformed with pSK41-like plasmids or conjugation experiments were performed in the presence of the culture filtrate from S. aureus containing a pSK41-like plasmid, transfer was successful. Furthermore, our results (Table 2) showed that pSK41 did not promote the transfer frequency of pAM378, a sex pheromone-responsive conjugative plasmid.
There is much to learn about the transfer of foreign DNA to S. aureus. In the present study, we used MRSA from VRSA case patients as the recipient strains of the Inc18-like vanA plasmid. We did this because our purpose in conducting these studies was to understand how VRSA arise in nature rather than a study of Gram-positive conjugation. For this reason, we limited conjugation experiments to naturally occurring donor and recipient strains. As a result, we have not ruled out other characteristics that may be specific to these strains which contribute to their ability to acquire the Inc18-like vanA plasmid. Others have shown that mutations in genes of the SauI type I restriction-modification system (32, 36) or mutations in the CRISPR (clustered, regularly interspaced, short palindromic repeat) loci can increase a strain's ability to accept foreign DNA (37, 38). In the present study, conjugation experiments with pAM378 demonstrated that most MRSA acquire foreign DNA at a rate that is somewhere between control strains RN4220 and its parent strain 8325-4. Unlike its parent strain, S. aureus RN4220 has a critical stop codon in the hsdR gene of the SauI restriction-modification system that allows the strain to accept foreign DNA at a rate that is higher than its parent strain and most clinical isolates of S. aureus. In fact, the conjugation rates reported here are consistent with the conjugation rates for clinical isolates of S. aureus reported by Waldron and Lindsay (32). Indeed, by these criteria, one MRSA strain, MRSA8, is below average in its ability to accept foreign plasmid DNA. However, this strain was able to acquire the Inc18-like vanA plasmid, suggesting that a feature other than a general ability to acquire foreign DNA was at play.
The observation that pSK41-like plasmids can promote conjugation of an Inc18-like plasmid from another cell to the pSK41-like plasmid host cell is novel. Similarly, the ability of pSK41-like plasmids to promote conjugation via diffusion of gene products in a cell-free culture filtrate is also novel. Our experiments suggest that the strain carrying a pSK41-like plasmid produces an extracellular substance that promotes conjugation. It should be noted that the effect of the cell-free culture filtrate on conjugation was a bit weaker than the effect of a pSK41-like plasmid in the recipient strain. All of these conjugations were infrequent events, but the conjugations that occurred in the presence of a cell-free filtrate were at the lower limit of detection. It could be that the transfer of Inc18-like vanA plasmids occurs just below our detection level and that by repeating the experiments enough times we were able to detect this low frequency event. However, our controls suggest some benefit of using a cell extract from a pSK41-like positive strain and having pSK41-like plasmid in the recipient clearly promoted conjugation.
Additional studies are needed to understand the role and mechanism of pSK41 in this process. The extracellular factor produced by strains carrying a pSK41-like plasmid may be the pheromone-like lipoprotein encoded by pSK41 (39). Pheromones promote conjugation of pheromone-responsive plasmids between enterococci by inducing aggregation of the donor and recipient strains and in vitro pheromones can induce aggregation of the donor strain alone. Inc18-like plasmids are not pheromone responsive, and it was previously shown that S. aureus with pSK41-like plasmids do not induce aggregation of VRE with Inc18-like vanA plasmids (1, 31). If this lipoprotein is the extracellular substance that promotes conjugation, the mechanism is unclear, but it may be involved in an intracellular signal pathway that ultimately promotes conjugation (40).
pSK41- and Inc18-like plasmids do have homologous conjugation systems which encode a type IV secretion system. In general, type IV secretion systems facilitate cell-to-cell transfer of intracellular material, such as a conjugative plasmid or a protein. There are three components to this system: a coupling protein that recognizes and binds the intracellular material, a transport channel that allows the material to pass through the cellular membrane, and a cell surface adhesion or pilus that facilitates cell-to-cell contact. Although both plasmids have homologous conjugation systems, pSK41-like plasmids are only found in staphylococci, and Inc18-like plasmids are found in streptococci and enterococci, which suggests that these plasmids lack some component that is necessary for transfer between these different groups of bacteria. We hypothesize that pSK41 may promote Inc18 plasmid conjugation to S. aureus by complementing the missing component of the Inc18 type IV secretion system (10, 24). We also propose that this missing component may be an extracellular protein that can be supplied via a nongenetic mechanism in a cell-free culture filtrate, perhaps an adhesion or pilus that is necessary for cell-to-cell contact. Little is known about the mechanics of Gram-positive conjugation, but the role of a pilus in the conjugation of pInc18-like plasmids has been hypothesized (41).
The positive effect of a cell-free culture filtrate on conjugation raises the possibility that the recipient strain does not need to carry a pSK41-like plasmid. Evidence for a pSK41-like plasmid in the recipient strain, (i.e., the VRSA is positive for the plasmid or both the VRSA and a related MRSA are positive for the plasmid) only exists for five VRSA cases—i.e., cases 1, 3, 8, 9, and 11 (the present study; CDC, unpublished). In four VRSA cases—cases 2, 4, 6, and 12—no pSK41-like plasmid was found in the VRSA isolates, and a related MRSA isolate was not identified from the case (the present study; CDC, unpublished). It could be that the recipient isolate was either pSK41 negative or lost the plasmid either in vivo or in vitro. For three VRSA cases—cases 5, 7, and 10—no pSK41-like plasmid was identified in the VRSA isolate or the related MRSA isolates. Again, these isolates could have lost the plasmid, but here the case for the recipient never carrying pSK41 is a bit stronger because both the MRSA isolates and the VRSA isolates are negative. All of the VRSA isolates described here occurred in polymicrobial infections, which provides an opportunity for products from another microbe to contribute to conjugative transfer. We do not know whether other isolates in these infections were positive for a pSK41-like plasmid and may have acted as “helper cells” for conjugation. It is also possible that the beneficial effect derived from pSK41 could be overcome by another mechanism. For example, if pSK41 does facilitate cell-cell adhesion, this limitation could potentially be overcome by the physical proximity of strains in a biofilm.
We found that the presence of vanA in our transconjugants was relatively unstable, and yet most of the naturally occurring VRSA isolates stably maintained vanA (data not shown). For the first VRSA, it was hypothesized that the Inc18-like vanA plasmid moved by conjugation into the S. aureus isolate and then Tn1546 transposed from the Inc18-like plasmid to the pSK41-like plasmid. The Inc18-like plasmid was not maintained in the S. aureus recipient, but the Tn1546-carrying pSK41-like plasmid was maintained. Thus, the pSK41-like plasmid appears to have served a role in rescuing the resistance marker. This may also be the case for three of the first nine VRSA isolates, where vanA was found on a pSK41-like plasmid (1; the present study). Characterization of subsequent VRSA isolates is ongoing.
If our hypothesis for how pSK41-like plasmids contribute to conjugation of Inc18-like plasmids from E. faecalis to S. aureus is correct, then conjugative transfer of other Inc18-like plasmids to S. aureus should also be possible when other pSK41-like plasmids are present. It should also be possible to identify the conjugation-promoting factor of pSK41-like plasmids, synthesize it in vitro and promote conjugation transfer by supplying the purified product in trans. The implication of understanding Gram-positive conjugation in greater detail can have very real public health impact. Right now, VRSA isolates are occurring at a slow rate. This could be because the number of potential donor VRE with Inc18-like vanA plasmids is low, <2% among vanA-positive VRE strains isolated from intensive care unit patients (2). Likewise, in a survey of 2,455 methicillin-resistant S. aureus isolates from invasive infections, <4% of the isolates were potential recipients positive for a pSK41-like plasmid (29). In addition to the relatively low numbers, even when these strains do happen to meet, the conjugation transfer is infrequent. These factors may explain the low frequency at which VRSA isolates are identified. However, bacteria are very good adapters, and if this plasmid transfer between Enterococcus and Staphylococcus becomes more efficient, then we could encounter a resistance problem in S. aureus similar to that we are currently encountering with Enterobacteriaceae where multidrug-resistant plasmids are circulating and intergenus plasmid transfer is common. Understanding conjugation at the molecular level may reveal opportunities to disrupt plasmid transmission and in turn slow the dissemination of resistance.
ACKNOWLEDGMENTS
We thank Brandi Limbago for her scientific review of this work. We also thank David Lonsway and Betty Wong for their expert technical assistance. We are grateful to Richard P. Novick for the generous gift of strain S. aureus 4220 and Ronald A. Skurray for the prototype pSK41.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control.
Footnotes
Published ahead of print 22 October 2012
REFERENCES
- 1. Zhu W, Clark NC, McDougal LK, Hageman J, McDonald LC, Patel JB. 2008. Vancomycin-resistant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob. Agents Chemother. 52:452–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu W, Murray PR, Huskins WC, Jernigan JA, McDonald LC, Clark NC, Anderson KF, McDougal LK, Hageman JC, Olsen-Rasmussen M, Frace M, Alangaden GJ, Chenoweth C, Zervos MJ, Robinson-Dunn B, Schreckenberger PC, Reller LB, Rudrik JT, Patel JB. 2010. Dissemination of an Enterococcus Inc18-like vanA plasmid associated with vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 54:4314–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Banerjee T, Anupurba S. 2012. Colonization with vancomycin-intermediate Staphylococcus aureus strains containing the vanA resistance gene in a tertiary-care center in north India. J. Clin. Microbiol. 50:1730–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tiwari HK, Sen MR. 2006. Emergence of vancomycin resistant Staphylococcus aureus (VRSA) from a tertiary care hospital from northern part of India. BMC Infect. Dis. 6:156 doi:10.1186/1471-2334-6-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aligholi M, Emaneini M, Jabalameli F, Shahsavan S, Dabiri H, Sedaght H. 2008. Emergence of high-level vancomycin-resistant Staphylococcus aureus in the Imam Khomeini Hospital in Tehran. Med. Principles Pract. 17:432–434 [DOI] [PubMed] [Google Scholar]
- 6. Sievert DM, Rudrik JT, Patel JB, McDonald LC, Wilkins MJ, Hageman JC. 2008. Vancomycin-resistant Staphylococcus aureus in the United States, 2002–2006. Clin. Infect. Dis. 46:668–674 [DOI] [PubMed] [Google Scholar]
- 7. Gould IM. 2010. VRSA-doomsday superbug or damp squib? Lancet Infect. Dis. 10:816–818 [DOI] [PubMed] [Google Scholar]
- 8. Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569–1571 [DOI] [PubMed] [Google Scholar]
- 9. Weigel LM, Donlan RM, Shin DH, Jensen B, Clark NC, McDougal LK, Zhu W, Musser KA, Thompson J, Kohlerschmidt D, Dumas N, Limberger RJ, Patel JB. 2007. High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob. Agents Chemother. 51:231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grohmann E, Muth G, Espinosa M. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thompson JK, Collins MA. 1988. Evidence for the conjugal transfer of the broad host range plasmid pIP501 into strains of Lactobacillus helveticus. J. Appl. Bacteriol. 65:309–319 [DOI] [PubMed] [Google Scholar]
- 12. Langella P, Chopin A. 1989. Conjugal transfer of plasmid pIP501 from Lactococcus lactis to Lactobacillus delbrueckii subsp. bulgaricus and Lactobacillus helveticus. FEMS Microbiol. Lett. 51:149–152 [DOI] [PubMed] [Google Scholar]
- 13. Schaberg DR, Clewell DB, Glatzer L. 1982. Conjugative transfer of R-plasmids from Streptococcus faecalis to Staphylococcus aureus. Antimicrob. Agents Chemother. 22:204–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gonzalez CF, Kunka BS. 1983. Plasmid transfer in Pediococcus spp.: intergeneric and intrageneric transfer of pIP501. Appl. Environ. Microbiol. 46:81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shearer JE, Wireman J, Hostetler J, Forberger H, Borman J, Gill J, Sanchez S, Mankin A, Lamarre J, Lindsay JA, Bayles K, Nicholson A, O'Brien F, Jensen SO, Firth N, Skurray RA, Summers AO. 2011. Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. G3 1:581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Berg T, Firth N, Apisiridej S, Hettiaratchi A, Leelaporn A, Skurray RA. 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J. Bacteriol. 180:4350–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clewell DB, Francia MV. 2004. Conjugation in gram-positive bacteria, p 227–256 In Funnell BE, Phillips GJ. (ed), Plasmid biology. ASM Press, Washington, DC [Google Scholar]
- 18. Archer GL, Dietrick DR, Johnston JL. 1985. Molecular epidemiology of transmissible gentamicin resistance among coagulase-negative staphylococci in a cardiac surgery unit. J. Infect. Dis. 151:243–251 [DOI] [PubMed] [Google Scholar]
- 19. Archer GL, Johnston JL. 1983. Self-transmissible plasmids in staphylococci that encode resistance to aminoglycosides. Antimicrob. Agents Chemother. 24:70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jaffe HW, Sweeney HM, Nathan C, Weinstein RA, Kabins SA, Cohen S. 1980. Identity and interspecific transfer of gentamicin-resistance plasmids in Staphylococcus aureus and Staphylococcus epidermidis. J. Infect. Dis. 141:738–747 [DOI] [PubMed] [Google Scholar]
- 21. Apisiridej S, Leelaporn A, Scaramuzzi CD, Skurray RA, Firth N. 1997. Molecular analysis of a mobilizable theta-mode trimethoprim resistance plasmid from coagulase-negative staphylococci. Plasmid 38:13–24 [DOI] [PubMed] [Google Scholar]
- 22. Projan SJ, Archer GL. 1989. Mobilization of the relaxable Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J. Bacteriol. 171:1841–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thomas WD, Jr, Archer GL. 1992. Mobilization of recombinant plasmids from Staphylococcus aureus into coagulase negative Staphylococcus species. Plasmid 27:164–168 [DOI] [PubMed] [Google Scholar]
- 24. Zechner EL, Lang S, Schildbach JF. 2012. Assembly and mechanisms of bacterial type IV secretion machines. Philos. Trans. R. Soc. London Ser. B Biol. Sci. 367:1073–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Novick R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155–166 [DOI] [PubMed] [Google Scholar]
- 26. Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712 [DOI] [PubMed] [Google Scholar]
- 27. Firth N, Apisiridej S, Berg T, O'Rourke BA, Curnock S, Dyke KG, Skurray RA. 2000. Replication of staphylococcal multiresistance plasmids. J. Bacteriol. 182:2170–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Evers S, Sahm DF, Courvalin P. 1993. The vanB gene of vancomycin-resistant Enterococcus faecalis V583 is structurally related to genes encoding d-Ala:d-Ala ligases and glycopeptide-resistance proteins vanA and vanC. Gene 124:143–144 [DOI] [PubMed] [Google Scholar]
- 29. Nicholson A, Schoonover MDV, McDougal L, Limbago B. 2005. Identification and molecular characterization of pSK41-bearing methicillin-resistant Staphylococcus aureus (MRSA) from ongoing, large-scale surveillance of invasive disease, United States–06. SHEA/IDSA 2010, Atlanta, GA [Google Scholar]
- 30. Zhu W, Clark N, Patel JB. 2008. Conjugal transfer of vancomycin resistance from Enterococcus faecalis to Staphylococcus aureus, abstr. C2-269. 48th Intersci. Conf. Antimicrob. Agents Chemother [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Flannagan SE, Clewell DB. 2002. Identification and characterization of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol. Microbiol. 44:803–817 [DOI] [PubMed] [Google Scholar]
- 32. Waldron DE, Lindsay JA. 2006. SauI: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 188:5578–5585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clinical and Laboratory Standards Institute 2010. Methods of antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline, 2nd ed M45-A2 CLSI, Wayne, PA [Google Scholar]
- 34. Clinical and Laboratory Standards Institute 2009. Performance standards for antimicrobial susceptibility testing. CLSI, Wayne, PA [Google Scholar]
- 35. Dutka-Malen S, Evers S, Courvalin P. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sung JM, Lindsay JA. 2007. Staphylococcus aureus strains that are hypersusceptible to resistance gene transfer from enterococci. Antimicrob. Agents Chemother. 51:2189–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barrangou R, Horvath P. 2009. The CRISPR system protects microbes against phages, plasmids: palindromic DNA repeat sequences immunize microorganisms against phages and plasmids, while also directing their evolution. Microbe 4:224–230 [Google Scholar]
- 38. Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Firth N, Fink PD, Johnson L, Skurray RA. 1994. A lipoprotein signal peptide encoded by the staphylococcal conjugative plasmid pSK41 exhibits an activity resembling that of Enterococcus faecalis pheromone cAD1. J. Bacteriol. 176:5871–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lazazzera BA. 2001. The intracellular function of extracellular signaling peptides. Peptides 22:1519–1527 [DOI] [PubMed] [Google Scholar]
- 41. Abajy MY, Kopec J, Schiwon K, Burzynski M, Doring M, Bohn C, Grohmann E. 2007. A type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in gram-positive bacteria. J. Bacteriol. 189:2487–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]