Abstract
A Klebsiella pneumoniae clinical isolate recovered in Tunisia showed resistance to all β-lactams and decreased susceptibility to carbapenems. K. pneumoniae 204 expressed the carbapenem-hydrolyzing β-lactamase OXA-204, differing from OXA-48 by two amino acid substitutions (Gln98His and Thr99Arg) (class D β-lactamase [DBL] numbering). OXA-48 and OXA-204 shared similar resistance profiles, hydrolyzing carbapenems but sparing broad-spectrum cephalosporins. The blaOXA-204 gene was located on a ca. 150-kb IncA/C-type plasmid, which also carried the blaCMY-4 gene. The blaOXA-204 gene was associated with an ISEcp1 element, whereas the blaOXA-48 genes are usually associated with IS1999.
TEXT
Among the different types of carbapenemases identified in Enterobacteriaceae, OXA-48 is one of the most commonly identified in many countries. OXA-48 is a class D carbapenem-hydrolyzing β-lactamase (CHDL) that was first identified in Turkey (1) and has subsequently been commonly reported, in particular, in the Middle East, North Africa, and Europe (2, 3). In addition, there have been recent reports of OXA-48-related enzymes, such as OXA-181, which has been identified in India (4) or in other countries with a link to India (5), as well as OXA-163 from Argentina (6). Those variants differ by few amino acid substitutions or deletions. OXA-48 and OXA-181 hydrolyze penicillins at a high level and carbapenems at a low level and spare broad-spectrum cephalosporins, whereas OXA-163 hydrolyzes broad-spectrum cephalosporins at a high level and carbapenems at a very low level. The blaOXA-48-like genes are plasmid-borne and have been identified with insertion sequences involved in their acquisition and expression (7). The spread of the blaOXA-48 gene is mostly linked to the dissemination of a single IncL/M-type self-transferable plasmid (8).
Our study was initiated by the isolation of a Klebsiella pneumoniae strain showing reduced susceptibility to carbapenems and recovered from urine specimens of a single patient who had been hospitalized in Tunis, Tunisia. Identification of K. pneumoniae isolate 204 was performed by using the API 20E system (bioMérieux, La Balme-les-Grottes, France), and susceptibility testing was done by disc diffusion assays and interpreted according to the CLSI guidelines (9). K. pneumoniae 204 was resistant to all penicillins, to β-lactamase inhibitor–penicillin combinations, to broad-spectrum cephalosporins, and to ertapenem and was susceptible to imipenem and meropenem (Table 1). Double-disc synergy testing revealed that it expressed an extended-spectrum β-lactamase (ESBL). In addition, K. pneumoniae 204 was resistant to all aminoglycosides (netilmicin, amikacin, kanamycin, tobramycin, and gentamicin), fluoroquinolones, sulfonamides, and tetracycline (data not shown). The MICs of fosfomycin, colistin, and tigecycline, determined using E-tests, were at 32, 0.094, and 2 μg/ml, respectively. Whole-cell DNA of K. pneumoniae 204 was extracted using a QIAamp DNA Minikit and following the recommendations of the manufacturer (Qiagen, Courtaboeuf, France) and then used as the template under standard PCR conditions (10) with a series of primers designed for the detection of class A, B, and D β-lactamase genes (5, 11, 12). K. pneumoniae 204 expressed the ESBL CTX-M-14 that belongs to the CTX-M-9 cluster (13), together with ß-lactamases OXA-1, CMY-4, and SHV-1. In addition, K. pneumoniae 204 possessed a novel blaOXA-48-like gene termed blaOXA-204 (www.lahey.org/studies/).
Table 1.
MICs of β-lactams for K. pneumoniae 204, E. coli pTOPO-OXA-204, and E. coli pTOPO-OXA-48 in E. coli TOP10 or E. coli HB4 reference strains and E. coli TOP10(p204-B), A. baumannii CIP70.10(p204-B), P. aeruginosa PU21(p204-B), and reference strains A. baumannii CIP70.10 and P. aeruginosa PU21c
| β-lactama | MIC (μg/ml) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K. pneumoniae 204 | E. coli TOP10 (pTOPO-OXA-204) | E. coli TOP10 (pTOPO-OXA-48) | E. coli TOP10 (p204-B) | E. coli TOP10 | E. coli HB4 (pTOPO-OXA-204) | E. coli HB4 (pTOPO-OXA-48) | E. coli HB4 | A. baumannii CIP70.10 (p204-B) | A. baumannii CIP70.10 | P. aeruginosa PU21 (p204-B) | P. aeruginosa PU21 | |
| Amoxicillin | >256 | >256 | >256 | >256 | 2 | >256 | >256 | 8 | >256 | 32 | >256 | >256 |
| Amoxicillin + CLA | >256 | >256 | >256 | >256 | 2 | >256 | >256 | 8 | >256 | 32 | >256 | >256 |
| Ticarcillin | >256 | >256 | >256 | >256 | 2 | >256 | >256 | 4 | >256 | 8 | >256 | 8 |
| Ticarcillin + CLA | >256 | >256 | >256 | >256 | 2 | >256 | >256 | 4 | >256 | 8 | >256 | 8 |
| Piperacillin | >256 | 64 | 64 | 64 | 1 | 128 | 128 | 4 | 256 | 16 | 128 | 4 |
| Piperacillin + TZB | 128 | 64 | 64 | 64 | 1 | 128 | 128 | 4 | 256 | 8 | 128 | 4 |
| Cephalothin | >256 | 8 | 8 | >256 | 4 | 128 | 128 | 64 | >256 | 32 | >256 | >256 |
| Cefoxitin | 128 | 2 | 2 | 32 | 2 | 256 | 256 | 256 | >256 | 32 | >256 | >256 |
| Ceftazidime | 64 | 0.12 | 0.12 | 12 | 0.12 | 0.75 | 0.75 | 0.75 | >256 | 4 | 24 | 1.5 |
| Cefotaxime | 128 | 0.25 | 0.25 | 12 | 0.06 | 1 | 1 | 0.38 | >256 | 8 | >256 | >256 |
| Aztreonam | 32 | 0.06 | 0.06 | 4 | 0.06 | 0.38 | 0.38 | 0.38 | 64 | 32 | 8 | 1.5 |
| Cefepime | 16 | 0.25 | 0.25 | 0.25 | 0.06 | 1 | 1 | 0.5 | 16 | 2 | 2 | 1 |
| Imipenem | 0.5 | 0.5 | 0.5 | 0.5 | 0.06 | 32 | 32 | 0.25 | 8 | 0.25 | 8 | 1 |
| Ertapenem | 2 | 0.25 | 0.25 | 0.25 | 0.06 | >32 | >32 | 1 | NDb | ND | ND | ND |
| Meropenem | 0.5 | 0.06 | 0.06 | 0.06 | 0.01 | 32 | 32 | 0.25 | 4 | 0.25 | 32 | 0.75 |
CLA, clavulanic acid at a fixed concentration of 4 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
ND, data not determined because of the intrinsic resistance.
Natural plasmid p204-B and recombinant plasmid pTOPO-OXA-204 expressed the OXA-204 β-lactamase.
Multilocus sequence typing, performed as described previously (14), identified K. pneumoniae 204 as a ST383 strain. This sequence type had been previously reported, corresponding to one isolate coproducing VIM-4, KPC-2, and CMY-4 from Greece (15) and to one isolate coproducing VIM-19 and a CTX-M-like β-lactamase from Sweden but with a Greek origin (16).
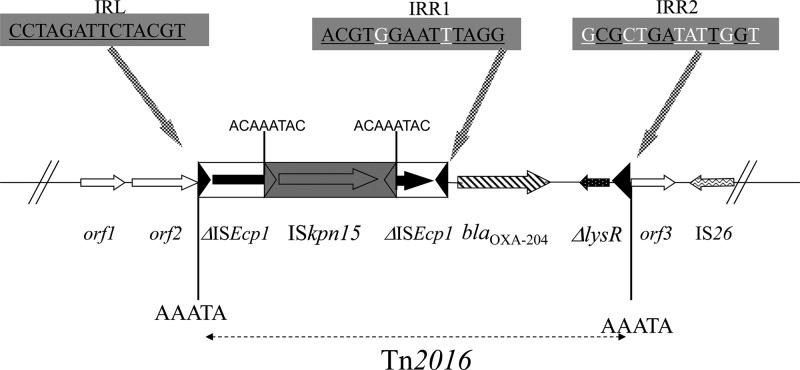
Shotgun cloning using HindIII-restricted genomic DNA and a HindIII-restricted pBK-CMV plasmid was performed as described previously (10). Recombinant plasmids were selected on Trypticase soy (TS) agar plates containing ticarcillin (100 μg/ml) and kanamycin (30 μg/ml). The resulting Escherichia coli (pBK-OXA-204) recombinant strain displayed a typical OXA-48-type phenotype, with high-level resistance to penicillins and penicillin-β-lactamase inhibitor combinations, reduced susceptibility to carbapenems, and susceptibility to expanded-spectrum cephalosporins (data not shown). Sequence analysis of the 7,688-bp cloned DNA fragment identified the blaOXA-204 gene. Compared to OXA-48, OXA-204 exhibited two amino acid substitutions, namely, Gln98His and Thr99Arg (class D β-lactamase [DBL] nomenclature) (17). Sequencing of the insertion of recombinant plasmid pBK-OXA-204 revealed that the insertion sequence (IS) ISEcp1 was located immediately upstream of the blaOXA-204 gene, as observed with the blaOXA-181 gene (5) but in contrast to what is observed for blaOXA-48, which is preceded by IS1999 (2). Interestingly, the segment separating the blaOXA-204 gene from the ISEcp1 element was 46 bp, whereas it was 128 bp for the blaOXA-181 gene, suggesting that the two genes were not derived from each other by mutations but had been mobilized independently through two distinct transposition processes from their natural progenitor, i.e., Shewanella xiamenensis. Noteworthy, detailed sequence analysis showed that ISEcp1 was truncated by another IS, namely, ISKpn15. That latter element possessed a transposase belonging to the IS66 family and showing 80% amino acid identity to that of ISUnCu16, an IS identified on plasmid pRSB113 isolated from a sewage treatment plant in Germany (18). ISKpn15 did not disrupt the −35 (TTGAAA) and −10 (TACAAT) promoter sequences located inside ISEcp1 and likely involved in the expression of the blaOXA-204 gene, as demonstrated for blaOXA-181 or blaCTX-M genes (19). The blaOXA-204 gene was part of a 4,420-bp potential transposon, named Tn2016, flanked by a 5-bp duplication of the target site (AAATA), being the signature of a transposition event, which occurred on the IncA/C scaffold (Fig. 1). This transposon was bracketed by two imperfect 14-bp inverted-repeat sequences, corresponding to the original inverted repeat left (IRL) of ISEcp1 and to a secondary inverted repeat right (IRR) IRR2 sequence showing identity of 6 of 14 bp with the IRL of ISEcp1, in accordance with the ISEcp1-mediated one-ended transposition process (Fig. 1). The identification of an 8-bp target site duplication (ACAAATAC) at both extremities of ISKpn15 was likely the signature of a transposition event, which occurred in the ISEcp1-blaOXA-204-made tranposon (Fig. 1). Consequently, this result suggests that this truncation may have stabilized the Tn2016 transposon on the IncA/C scaffold by disrupting the ISEcp1 transposase activity, therefore inactivating its one-ended transposition activity.
Fig 1.
Schematic map of the transposon Tn2016 structure and the surrounding sequences. Open reading frames are shown as arrows or as horizontal boxes with an arrow indicating the orientation of the coding sequence. The IRL, IRR1, and IRR2 motifs are indicated (blackened base pairs are identical, whereas whitened base pairs are different), and target site duplications (AAATA or ACAAATAC) are represented by black bars. orf1, orf2, and orf3 were similar to open reading frames encoding hypothetical proteins identified on IncA/C scaffolds.
In order to compare the hydrolytic activity of OXA-204 to that of OXA-48, the corresponding genes were amplified using primers preOXA-48A and preOXA-48B, with DNAs of K. pneumoniae 204 and 11978 used as the templates, respectively (1, 5). The amplicons were cloned in the pCR-Blunt II-TOPO vector (Invitrogen, Cergy-Pontoise, France) and expressed in the E. coli TOP10 reference strain by following the manufacturer recommendations. Two E. coli recombinant strains, harboring recombinant plasmids pTOPO-OXA-48 and pTOPO-OXA-204, respectively, were obtained and verified by sequencing. They expressed OXA-48 and OXA-204, respectively, and displayed exactly the same β-lactam resistance pattern (Table 1). The specific activities of β-lactamases OXA-204 and OXA-48 for carbapenems, measured as described previously (20) by using E. coli (pTOPO-OXA-204) and E. coli (pTOPO-OXA-48) as the templates, were very similar for benzylpenicillin, broad-spectrum cephalosporins, and carbapenems, suggesting that OXA-204 and OXA-48 possessed similar hydrolytic activities (Table 2). Then, these recombinant plasmids were electroporated into E. coli HB4, a porin-deficient strain (21), in order to evaluate the impact of their production in such an E. coli background. As expected, expression of both the blaOXA-204 and blaOXA-48 genes in E. coli HB4 conferred resistance to penicillins, β-lactamase inhibitor–penicillin combinations, and carbapenems at a quite high level (Table 1). The MICs of β-lactams for OXA-204 and OXA-48 were similar, confirming that OXA-204 and OXA-48 possessed very similar resistance patterns (Table 1).
Table 2.
Specific activities of β-lactamases OXA-204 and OXA-48 for benzylpenicillin, broad-spectrum cephalosporins, and carbapenemsa
| Antimicrobial agent | Specific activity (mU/mg of protein ± SEM) |
|
|---|---|---|
| OXA-204 | OXA-48 | |
| Benzylpenicillin | 4,700 ± 330 | 4,700 ± 800 |
| Imipenem | 75 ± 7 | 80 ± 9 |
| Ertapenem | 2.5 ± 0.3 | 3 ± 0.4 |
| Meropenem | 2.8 ± 0.4 | 4 ± 0.7 |
| Cefotaxime | 100 ± 8 | 100 ± 8 |
| Cefepime | 7 ± 0.5 | 7 ± 0.5 |
| Ceftazidime | NHb | NH |
In each case, three independent experiments were performed, and the mean and the standard error of the mean (SEM) were calculated.
NH, hydrolysis not detectable.
Plasmid DNA analysis, performed as described previously (1), showed that K. pneumoniae 204 possessed three plasmids, namely, p204-A, p204-B, and p204-C, that were ca. 70, 150, and 180 kb in size, respectively (data not shown). Mating-out assays performed as described previously and using ertapenem (0.5 μg/ml) or ticarcillin (50 μg/ml) and sodium azide (100 μg/ml) for selection gave two types of transconjugants, E. coli J53(p204-A) and E. coli J53(p204-B). Transconjugant E. coli J53(p204-B) was resistant to penicillins, β-lactamase inhibitor–penicillin combinations, and broad-spectrum cephalosporins and showed decreased susceptibility to carbapenems (Table 1). It harbored the blaCMY-4 and blaOXA-204 genes. In addition, E. coli J53(p204-B) was resistant to tetracycline and to sulfamethoxazole-trimethoprim. Transconjugant E. coli J53(p204-A) displayed an ESBL phenotype and expressed the blaCTX-M-14 gene. Using PCR-based replicon typing (PBRT) as described previously (22), plasmid p204-B was typed as an IncA/C and plasmid p204-A as an IncL/M.
This is the first identification of a blaOXA-48-like gene on an IncA/C-type scaffold. Those plasmids possess a broad host range and are responsible for the spread of many resistance genes, in particular, the blaCMY-like Ambler class C ß-lactamase genes (23). They have been detected worldwide, and carbapenemase genes such as blaNDM-1 and blaVIM-4 have recently been identified on that plasmid scaffold (24, 25). In our study, the broad host range of plasmid p204-B was confirmed by obtaining electrotransformants using Acinetobacter baumannii as the recipient strain and transconjugants using Pseudomonas aeruginosa as the recipient strain, as described previously (Table 1) (2, 5). PCR experiments confirmed the positivity for the blaOXA-204 gene in the A. baumannii transformant and in the P. aeruginosa transconjugant. Plasmid DNA analysis further confirmed that the two recombinant strains harbored the same ca. 150-kb plasmid.
This study characterized the OXA-204 β-lactamase possessing a β-lactam resistance profile similar to that of OXA-48. The acquisition of the blaOXA-204 gene was linked to ISEcp1, as described for the blaOXA-181 gene and many other acquired broad-spectrum β-lactamase genes, such as blaCMY and blaCTX-M genes. Overall, the results of this study indicate that at least two different genetic backgrounds are associated with OXA-48-like carbapenemase-encoding genes in North Africa. Interestingly, the blaOXA-204 gene was associated with the blaCMY-4 gene on a widely diffused plasmid scaffold, giving rise to a successful genetic structure compromising the efficacy of all available β-lactams. Considering that IncA/C-type plasmids are known to spread efficiently, we might speculate that the diffusion of the blaOXA-204 gene among Gram-negative bacteria will be significant.
The nucleotide sequence data reported in this work has been deposited in the GenBank nucleotide database under accession no JQ809466.
ACKNOWLEDGMENTS
This work was partially funded by a grant from the INSERM (U914) and the Université Paris XI, Paris, France, and by a grant from the European Community (TEMPOtest-QC, HEALTH-2009-241742).
Footnotes
Published ahead of print 31 October 2012
REFERENCES
- 1. Poirel L, Héritier C, Tolün V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carrër A, Poirel L, Yilmaz M, Akan OA, Feriha C, Cuzon G, Matar G, Honderlick P, Nordmann P. 2010. Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob. Agents Chemother. 54:1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17:1791–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. 2011. Early dissemination of NDM-1 and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY antimicrobial surveillance program, 2006–2007. Antimicrob. Agents Chemother. 55:1274–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Potron A, Nordmann P, Lafeuille E, Al Maskari Z, Al Rashdi F, Poirel L. 2011. Characterization of OXA-181, a carbapenem-hydrolyzing class D β-lactamase from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 55:4896–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Poirel L, Castanheira M, Carrër A, Rodriguez CP, Jones RN, Smayevsky J, Nordmann P. 2011. OXA-163, an OXA-48-related class D β-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 55:2546–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases, the phantom menace. J. Antimicrob. Chemother. 67:1597–1606 [DOI] [PubMed] [Google Scholar]
- 8. Poirel L, Bonnin RA, Nordmann P. 2012. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 56:559–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing. CLSI M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 10. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 11. Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 55:5403–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70:119–123 [DOI] [PubMed] [Google Scholar]
- 13. Poirel L, Bonnin RA, Nordmann P. 2012. Genetic support and diversity of acquired extended-spectrum β-lactamases in Gram-negative rods. Infect. Genet. Evol. 12:883–893 [DOI] [PubMed] [Google Scholar]
- 14. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Papagiannitsis CC, Giakkoupi P, Vatopoulos AC, Tryfinopoulou K, Miriagou V, Tzouvelekis LS. 2010. Emergence of Klebsiella pneumoniae of a novel sequence type (ST383) producing VIM-4, KPC-2 and CMY-4 β-lactamases. Int. J. Antimicrob. Agents 36:573–574 [DOI] [PubMed] [Google Scholar]
- 16. Samuelsen O, Toleman MA, Hasseltvedt V, Fuursted K, Leegaard TM, Walsh TR, Sundsfjord A, Giske CG. 2011. Molecular characterization of VIM-producing Klebsiella pneumoniae from Scandinavia reveals genetic relatedness with international clonal complexes encoding transferable multidrug resistance. Clin. Microbiol. Infect. 17:1811–1816 [DOI] [PubMed] [Google Scholar]
- 17. Couture F, Lachapelle J, Levesque RC. 1992. Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol. Microbiol. 6:1693–1705 [DOI] [PubMed] [Google Scholar]
- 18. Girlich D, Poirel L, Szczepanowski R, Schlüter A, Nordmann P. 2012. Carbapenem-hydrolyzing GES-5-encoding gene on different plasmid types recovered from a bacterial community in a sewage treatment plant. Appl. Environ. Microbiol. 78:1292–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poirel L, Decousser JW, Nordmann P. 2003. Insertion sequence ISEcp1B is involved in the expression and mobilization of a blaCTX-M ß-lactamase gene. Antimicrob. Agents Chemother. 47:2938–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Poirel L, Héritier C, Nordmann P. 2004. Chromosome-encoded Ambler class D β-lactamase of Shewanella oneidensis as a progenitor of carbapenem-hydrolyzing oxacillinase. Antimicrob. Agents Chemother. 48:348–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mammeri H, Nordmann P, Berkani A, Eb F. 2008. Contribution of extended-spectrum AmpC (ESAC) ß-lactamases to carbapenem resistance in Escherichia coli. FEMS Microbiol. Lett. 282:238–240 [DOI] [PubMed] [Google Scholar]
- 22. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 23. Call DR, Singer RS, Meng D, Broschat SL, Orfe LH, Anderson JM, Herndon DR, Kappmeyer LS, Daniels JB, Besser TE. 2010. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonelle enterica are a distinct component of a larger lineage of plasmids. Antimicrob. Agents Chemother. 54:590–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53:2227–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carattoli A, Villa L, Poirel L, Bonnin RA, Nordmann P. 2012. Evolution of IncA/C blaCMY-2-carrying plasmids by acquisition of the blaNDM-1 carbapenemase gene. Antimicrob. Agents Chemother. 56:783–786 [DOI] [PMC free article] [PubMed] [Google Scholar]