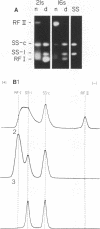
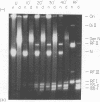
Abstract
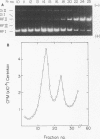
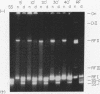
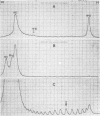
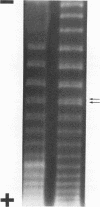
The replication cycle of bacteriophage phi X174 DNA has been analyzed by agarose gel electrophoresis. The electrophoretic behavior of the predominant species of parental and progeny DNA molecules formed between 5 and40 min after infection was deduced and quantitated. Migration through 1.4% agarose at 5 and 10 V/cm resolved all known viral DNA species as well as fragments of host chromosomal DNA. Among parental replicative form(RF) molecules synthesized, 1 to 3% were full length linear duplexes (RFIII) and approximately 65% were closed circular duplexes (RFI). Most of the input viral strands remained in a duplex structure throughout the period of infection studied here. Among progeny molecules, RFIII was not readily detected unless viral DNA synthesis was inhibited by chloramphenicol. Late in infection, 20% of the progeny RF were found to exist as form I dna. in addition, approximately 1% of the viral DNA was found as unit length linear single strands. Electrophoretic analysis of RF DNA after controlled denaturation suggests the existence of four populations of closed circular RF: (i) molecules of native superhelix density (RFI); (ii) a population of molecules of altered topological linking number, alpha, differing in increments of one superhelical turn (tau) between tau values of 0 and approximately -31; (iii) a superimposed population of topological isomers which under electrophoresis conditions have mean tau value (tau) equal to +5; and (iv) a population of "complexed" molecules with a reduced number of superhelical turns due to their association with single-stranded DNA and RNA. Complexed parental molecules isolated from cells infected at high multiplicity released FRI and homologous single-stranded DNA upon denaturation and are postulated to be intermediates in genetic recombination. Complexed RF DNA isolated from cells infected at low multiplicity release native supercoils upon reaction with RNase H and are observed by electron microscopy to contain displacement loops. Such molecules are likely intermediates in transcription. Our results are consistent with a structure of complexed RFI involving a partially triple-stranded helix in which a covalently closed circular duplex molecule contains a reduced number of superhelical turns due to the unwinding produced by base pairing between one strand of the supercoil and an associated homologous single strand of DNA or RNA.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baas P. D., Keegstra W., Teertstra W. R., Jansz H. S. R-loops in bacteriophage phiX174 RF DNA. J Mol Biol. 1978 Oct 25;125(2):187–205. doi: 10.1016/0022-2836(78)90344-3. [DOI] [PubMed] [Google Scholar]
- Baas P. D., Teertstra W. R., Jansz H. S. Bacteriophage phiX174 RF DNA replication in vivo: a biochemical study. J Mol Biol. 1978 Oct 25;125(2):167–185. doi: 10.1016/0022-2836(78)90343-1. [DOI] [PubMed] [Google Scholar]
- Bauer W. R. Structure and reactions of closed duplex DNA. Annu Rev Biophys Bioeng. 1978;7:287–313. doi: 10.1146/annurev.bb.07.060178.001443. [DOI] [PubMed] [Google Scholar]
- Beattie K. L., Wiegand R. C., Radding C. M. Uptake of homologous single-stranded fragments by superhelical DNA. II. Characterization of the reaction. J Mol Biol. 1977 Nov;116(4):783–803. doi: 10.1016/0022-2836(77)90271-6. [DOI] [PubMed] [Google Scholar]
- Benbow R. M., Zuccarelli A. J., Davis G. C., Sinsheimer R. L. Genetic recombination in bacteriophage phi chi 174. J Virol. 1974 Apr;13(4):898–907. doi: 10.1128/jvi.13.4.898-907.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan P., Wang J. C., Echols H. Effect of circularity and superhelicity on transcription from bacteriophagelambda DNA. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3077–3081. doi: 10.1073/pnas.70.11.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. L., Smith M. The mapping and sequence determination of the single site in phiX174am3 replicative form DNA cleaved by restriction endonuclease Pst I. FEBS Lett. 1976 Jun 15;65(3):284–287. doi: 10.1016/0014-5793(76)80130-5. [DOI] [PubMed] [Google Scholar]
- Brown W. M., Vinograd J. Restriction endonuclease cleavage maps of animal mitochondrial DNAs. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4617–4621. doi: 10.1073/pnas.71.11.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J. Renaturation of complementary single-stranded DNA circles: complete rewinding facilitated by the DNA untwisting enzyme. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5328–5332. doi: 10.1073/pnas.74.12.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- Eisenberg S., Griffith J., Kornberg A. phiX174 cistron A protein is a multifunctional enzyme in DNA replication. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3198–3202. doi: 10.1073/pnas.74.8.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa H., Hayashi M. Gene A product of phi X174 is required for site-specific endonucleolytic cleavage during single-stranded DNA synthesis in vivo. J Virol. 1976 Aug;19(2):416–424. doi: 10.1128/jvi.19.2.416-424.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Itoh T., Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni A. M., Hutton J. R., Smotkin D., Weinberg R. A. Proviral DNA of Moloney leukemia virus: purification and visualization. Science. 1976 Feb 13;191(4227):569–571. doi: 10.1126/science.1251192. [DOI] [PubMed] [Google Scholar]
- Hayashi Y., Hayashi M. Template activities of the phi X-174 replicative allomorphic deoxyribonucleic acids. Biochemistry. 1971 Nov;10(23):4212–4218. doi: 10.1021/bi00799a009. [DOI] [PubMed] [Google Scholar]
- Henry T. J., Knippers R. Isolation and function of the gene A initiator of bacteriophage phi-chi 174, a highly specific DNA endonuclease. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1549–1553. doi: 10.1073/pnas.71.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Holloman W. K., Radding C. M. Recombination promoted by superhelical DNA and the recA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3910–3914. doi: 10.1073/pnas.73.11.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloman W. K., Wiegand R., Hoessli C., Radding C. M. Uptake of homologous single-stranded fragments by superhelical DNA: a possible mechanism for initiation of genetic recombination. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2394–2398. doi: 10.1073/pnas.72.6.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Sinsheimer R. L. The process of infection with bacteriophage phi-X174. X. Mutations in a phi-X Lysis gene. J Mol Biol. 1966 Jul;18(3):429–447. doi: 10.1016/s0022-2836(66)80035-9. [DOI] [PubMed] [Google Scholar]
- Ikeda J. E., Yudelevich A., Hurwitz J. Isolation and characterization of the protein coded by gene A of bacteriophage phiX174 DNA. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2669–2673. doi: 10.1073/pnas.73.8.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. H., Grossman L. I. Electrophoresis of DNA in agarose gels. Optimizing separations of conformational isomers of double- and single-stranded DNAs. Biochemistry. 1977 Sep 20;16(19):4217–4225. doi: 10.1021/bi00638a014. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano T., Sinsheimer R. L. Preparation and purification of phi X-RF component I. Biochim Biophys Acta. 1968 Jan 29;155(1):295–298. doi: 10.1016/0005-2787(68)90360-2. [DOI] [PubMed] [Google Scholar]
- Newbold J. E., Sinsheimer R. L. The process of infection with bacteriophage phiX174. XXXII. Early steps in the infection process: attachment, eclipse and DNA penetration. J Mol Biol. 1970 Apr 14;49(1):49–66. doi: 10.1016/0022-2836(70)90375-x. [DOI] [PubMed] [Google Scholar]
- Puga A., Tessman I. Mechanism of transcription of bacteriophage S13. I. Dependence of messengerRNA synthesis on amount and configuration of DNA. J Mol Biol. 1973 Mar 25;75(1):83–97. doi: 10.1016/0022-2836(73)90530-5. [DOI] [PubMed] [Google Scholar]
- Richardson J. P. Attachment of nascent RNA molecules to superhelical DNA. J Mol Biol. 1975 Nov 5;98(3):565–579. doi: 10.1016/s0022-2836(75)80087-8. [DOI] [PubMed] [Google Scholar]
- Richardson J. P. Initiation of transcription by Escherichia coli RNA polymerase from supercoiled and non-supercoiled bacteriophage PM2 DNA. J Mol Biol. 1975 Feb 5;91(4):477–487. doi: 10.1016/0022-2836(75)90274-0. [DOI] [PubMed] [Google Scholar]
- SINSHEIMER R. L., STARMAN B., NAGLER C., GUTHRIE S. The process of infection with bacteriophage phi-XI74. I. Evidence for a "replicative form". J Mol Biol. 1962 Mar;4:142–160. doi: 10.1016/s0022-2836(62)80047-3. [DOI] [PubMed] [Google Scholar]
- Shure M., Vinograd J. The number of superhelical turns in native virion SV40 DNA and minicol DNA determined by the band counting method. Cell. 1976 Jun;8(2):215–226. doi: 10.1016/0092-8674(76)90005-2. [DOI] [PubMed] [Google Scholar]
- Stavrianopoulos J. G., Chargaff E. Purification and properties of ribonuclease H of calf thymus. Proc Natl Acad Sci U S A. 1973 Jul;70(7):1959–1963. doi: 10.1073/pnas.70.7.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truffaut N., Sinsheimer R. L. Use of bacteriophage phi chi 174 replicative from progeny DNA as templates for transcription. J Virol. 1974 Apr;13(4):818–827. doi: 10.1128/jvi.13.4.818-827.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosberg H. P., Grossman L. I., Vinograd J. Isolation and partial characterisation of the relaxation protein from nuclei of cultured mouse and human cells. Eur J Biochem. 1975 Jun 16;55(1):79–93. doi: 10.1111/j.1432-1033.1975.tb02140.x. [DOI] [PubMed] [Google Scholar]