Abstract
Chlorhexidine and mupirocin are used in health care facilities to eradicate methicillin-resistant Staphylococcus aureus (MRSA) carriage. The objective of this study was to assess the frequency of chlorhexidine and mupirocin resistance in isolates from nares carriers in multiple nursing homes and to examine characteristics associated with resistance. Nasal swab samples were collected from approximately 100 new admissions and 100 current residents in 26 nursing homes in Orange County, CA, from October 2008 to May 2011. MRSA isolates were tested for susceptibility by using broth microdilution, disk diffusion, and Etest; for genetic relatedness using pulsed-field gel electrophoresis; and for qac gene carriage by PCR. Characteristics of the nursing homes and their residents were collected from the Medicare Minimum Data Set and Long-Term Care Focus. A total of 829 MRSA isolates were obtained from swabbing 3,806 residents in 26 nursing homes. All isolates had a chlorhexidine MIC of ≤4 μg/ml. Five (0.6%) isolates harbored the qacA and/or qacB gene loci. Mupirocin resistance was identified in 101 (12%) isolates, with 78 (9%) isolates exhibiting high-level mupirocin resistance (HLMR). HLMR rates per facility ranged from 0 to 31%. None of the isolates with HLMR displayed qacA or qacB, while two isolates carried qacA and exhibited low-level mupirocin resistance. Detection of HLMR was associated with having a multidrug-resistant MRSA isolate (odds ratio [OR], 2.69; P = 0.004), a history of MRSA (OR, 2.34; P < 0.001), and dependency in activities of daily living (OR, 1.25; P = 0.004). In some facilities, HLMR was found in nearly one-third of MRSA isolates. These findings may have implications for the increasingly widespread practice of MRSA decolonization using intranasal mupirocin.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) prevalence in nursing homes often exceeds that in acute care settings, including intensive care units (2–4, 39). The MRSA burden may be high because nursing home residents have multiple comorbidities, chronic wounds, frequent exposure to antibiotics, and require health care worker contact to perform activities of daily living (ADL) (2, 5, 6). Moreover, opportunities for MRSA transmission may be enhanced in nursing homes, where residents are encouraged to interact during social activities and group dining (6).
Some nursing homes and other health care facilities use antimicrobials and/or antiseptics to suppress MRSA carriage among patients in order to prevent health care-associated infections. Chlorhexidine has become a mainstay of health care-associated infection prevention, as it is used in oral care, patient bathing, skin antisepsis prior to line placement and surgical procedures, and as a component of some antimicrobial-impregnated catheters and dressings (7–13). Mupirocin is commonly used as a topical antibiotic for skin infections and is sometimes given to patients to eliminate MRSA nasal carriage (14).
As with any antimicrobial or antiseptic agent, increased use raises concerns about emerging resistance (15). The presence of the qac gene was associated with higher chlorhexidine MICs and with unsuccessful decolonization during an intensive care unit-based topical chlorhexidine intervention (16, 17). There are also recent data suggesting that failure of MRSA decolonization is more likely when qacA and/or qacB is present in conjunction with low-level mupirocin resistance (LLMR) (18). High-level mupirocin resistance (HLMR) is emerging in various hospitals around the world (19–22). In addition to mupA, mupB has been identified recently in isolates with HLMR (14, 23).
There is limited information regarding the prevalence of colonization with chlorhexidine- and mupirocin-resistant MRSA among residents of nursing homes. The objective of our study was to assess overall and facility-specific chlorhexidine and mupirocin resistance among a representative collection of MRSA isolates from colonized residents in nursing homes located in a large metropolitan county (Orange County, CA, with a population of 3.1 million) and to assess patient and isolate characteristics associated with high-level mupirocin-resistant MRSA carriage. This study will aid in understanding the scope of chlorhexidine and mupirocin resistance in nursing homes, which may impact treatment and decolonization practices used for nursing home residents.
(Presented in part at the 21st Annual Scientific Meeting of the Society for Healthcare Epidemiology of America, Dallas, TX, 1 to 4 April 2011.)
MATERIALS AND METHODS
Screening nursing home residents for MRSA.
As described previously, we measured MRSA carriage among residents in a convenience sample of 26 of the 72 nursing homes in Orange County, CA, from October 2008 to May 2011 (4). In each nursing home, a point prevalence screen was performed of the bilateral nares of 100 residents. Nasal samples were collected during 1 visit or over 2 to 3 visits if the nursing home had <100 beds. In addition, we performed an admission prevalence screening of up to 100 consecutive residents within 3 days of admission. For nursing homes with low bed turnover, a lesser number of 30 to 50 residents were screened, and for nursing homes with an average length of stay in years, admission screening was not performed. Bilateral nares swabs (BBL Culture Swabs, Sparks, MD) were transported to a central microbiology laboratory and plated within 12 h. Samples were cultured onto 5% sheep blood agar (BBL) and selective and differential chromogenic media for MRSA (Spectra MRSA; Remel, Lenexa, KS). The Institutional Review Board of the University of California Regents approved this study.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was performed to examine the resistance profiles of the isolates. All MRSA isolates were tested for antimicrobial susceptibility by the broth dilution method described by the Clinical and Laboratory Standards Institute (CLSI) (24). Isolates were tested for susceptibility to oxacillin, tetracycline, erythromycin, clindamycin, trimethoprim-sulfamethoxazole, quinupristin-dalfopristin, gentamicin, levofloxacin, linezolid, daptomycin, vancomycin, and rifampin. There is no CLSI method for testing of chlorhexidine, but this was done using the standard broth dilution approach described by CLSI, using a complete inhibition endpoint at 18 to 24 h of incubation (24). Chlorhexidine digluconate 20% aqueous solution (Sigma-Aldrich, St. Louis, MO) was used as the starting material for broth dilution testing. The S. aureus isolate ATCC 29213 was used for quality control. Mupirocin resistance (low and high level) was detected using disk diffusion and Etest methods (14, 25). Disks with 5 μg or 200 μg of mupirocin were placed on Mueller-Hinton agar plates with a 0.5 McFarland inoculum. Isolates with a zone of ≤13 mm around the 5-μg disk were also tested using 200 μg of mupirocin. Mupirocin resistance was defined as a zone of ≤13 mm when using the 5-μg disk (25). Among mupirocin-resistant isolates, HLMR was defined as a zone of ≤6 mm with the 200-μg disk, and LLMR was defined as a zone of ≥7 mm using the 200-μg disk (25). Additionally, correlations of resistance between various antibiotic classes were examined using Pearson's coefficient.
qac PCR.
Testing for the presence of the qacA and/or qacB gene was performed to examine the correlation between the genes and the chlorhexidine MICs. For all isolates, single primer pair sequences were used to detect qacA and qacB. For isolates found to harbor qacA and/or qacB, real-time PCR was performed to further discriminate between qacA and qacB, as previously described (26).
Pulsed-field gel electrophoresis.
Pulsed-field gel electrophoresis (PFGE) was performed on the high-level mupirocin-resistant isolates according to previously published methods by using a CHEF-DR II unit (Bio-Rad Laboratories, Hercules, CA) (27). Chromosomal DNA was digested using the restriction enzyme SmaI (Sigma-Aldrich, St. Louis, MO). Gels were stained with ethidium bromide, and banding patterns were analyzed using Bionumerics software (Applied Maths, Kortrijk, Belgium). PFGE types were defined using a similarity coefficient of 0.75. The isolates were compared to CDC type strains USA100 to USA1200.
Nursing home variables.
Nursing home characteristics were obtained for the year 2009, the midpoint of swab collection from the Minimum Data Set (MDS), version 2.0 (28). The MDS is collected by nursing homes and transmitted to the Center for Medicare and Medicaid Services; it is an individual resident-level data set with assessments of physical, psychological, and psycho-social functioning for all residents of Medicare- and Medicaid-licensed nursing homes in the United States. Select characteristics were also obtained from Long-Term Care FocUS, a website created by the Brown University Center for Gerontology and Healthcare Research and supported by the National Institute on Aging (29). These characteristics included nursing home specific staffing levels, average resource utilization group (RUG) scores, and average ADL scores for 2007, the most recent data year available. RUG is a facility-level score that reflects the average level of care required by residents, based upon residents' comorbidities, ability to perform activities of daily living, and their required amount of physical and occupational therapy. Nursing home characteristics were grouped into several domains: demographics, comorbidities, measures of resident functional status (ADL and RUG scores), facility information (such as length of stay), and partner hospital characteristics. Partner hospitals were defined as the hospital that transferred the most patients to the given nursing home, based upon previously performed Orange County health care facility surveys in 2007 and 2008 (1). Partner hospital characteristics were also obtained from these surveys and included whether the hospital routinely decolonized MRSA-positive patients and whether mupirocin was used for decolonization (1). In addition, per swabbed resident, we recorded the nursing home's day of swab collection, whether the resident shared a room, and whether there was a known history of MRSA. History of MRSA was obtained by chart review from the resident's records at the nursing home, which also contained patient information from the transferring hospital.
Bivariate and multivariate analyses.
We tested the association of facility, resident, and isolate-level characteristics with individual carriage of MRSA with HLMR by using generalized estimating equation models that were clustered by nursing home. We compared residents that were colonized with MRSA that exhibited HLMR with residents that were not colonized with a high-level mupirocin-resistant MRSA strain. Variables with P values of <0.1 from bivariate testing were included in the multivariate model and were retained at an α of 0.05. Facility-level variables included MRSA point prevalence, annual admissions, average RUG score, average ADL score, average length of stay, and the percentages of residents with diabetes, skin lesions (e.g., pressure ulcers, significant rashes), or indwelling devices (e.g., ventilator, dialysis). Individual-level variables included whether the resident had a history of MRSA (infection or colonization), lived in a shared room, and whether the isolate was multidrug resistant (30). A multidrug-resistant isolate was defined using the criterion of Cadilla et al., which is an isolate resistant to ≥3 classes of non-β-lactam antimicrobials (30). Partner hospital variables included the percentage of physicians who routinely decolonized patients with multidrug-resistant organisms, the percentage of MRSA-positive patients who were routinely decolonized, and whether the hospital used mupirocin to decolonize MRSA-positive patients. All facility variables were continuous, except for RUG scores, which was dichotomized into high versus low groups based upon the median value. We excluded four nursing homes from the bivariate and multivariate analyses; these facilities were devoted to psychiatric care and reflected a substantially different patient population (younger, fewer exposures to health care, and fewer comorbidities).
RESULTS
Nasal samples were collected from 3,806 residents in 26 nursing homes. The facilities varied in size, with a range of 24 to 255 beds (Table 1). Facilities had a median of 262 admissions per year (range, 18 to 1,526) and a median length of stay of 101 days (range, 17 to 753). The majority of the residents were admitted from an acute care hospital (median, 82%; range, 15 to 98%), and the percentage of residents previously known to harbor MRSA varied by facility (range, 0 to 69%). Also, colonization with a multidrug-resistant MRSA isolate varied extensively between residents of nursing homes, with rates of 0 to 100%.
Table 1.
Characteristics of 26 nursing homes in Orange County, 2009
| Characteristic | Median value (range) |
|---|---|
| Nursing home characteristics | |
| No. of beds | 99 (24–255) |
| Annual no. of admissionsa | 262 (18–1,526) |
| Median length of stay (days) | 101 (17–753) |
| MRSA point prevalence (% MRSA+ residents) | 26 (0–52) |
| Demographics (as % of all facility residents) | |
| Age over 85 yrsa | 25 (2–72) |
| Male | 42 (21–67) |
| Non-Caucasian race | 16 (1–88) |
| Less than high school education | 24 (0–64) |
| Admitted from acute hospital | 82 (15–98) |
| Comorbidities (as % of all facility residents) | |
| Diabetes | 27 (11–59) |
| Skin lesions | 72 (4–100) |
| Indwelling devices | 2 (0–46) |
| History of MRSA | 11 (0–69) |
| Multidrug-resistant MRSA isolate | 68 (0–100) |
| Functional status (avg score among all facility residents) | |
| RUG scoreb | 0.92 (0.81–1.43) |
| ADL score | 19.82 (10.77–26.90) |
After excluding the 4 psychiatric facilities, the median annual admissions was 300 and the percentage of facility residents over 85 years old was 39%. The medians for the other variables did not differ substantially after excluding the 4 psychiatric nursing homes.
The RUG score is a facility-level score that reflects the average level of care required by residents, based upon residents' comorbidities, ability to perform activities of daily living, and the required amount of physical and occupational therapy.
There were 829 MRSA isolates collected from the nares of residents in 25 of the 26 nursing homes; 1 nursing home had no MRSA carriers. Each isolate was from a unique patient. The number of MRSA isolates collected from residents at a single nursing home ranged from 1 to 81, with a median of 34 isolates. Of the 829 isolates tested, all had chlorhexidine MICs that were ≤4 μg/ml (range, 0.5 to 4 μg/ml; MIC50 and MIC90, 2 and 4 μg/ml, respectively) (Fig. 1). Only 5 (0.6%) isolates were found to harbor the qacA and/or qacB gene loci, and all five isolates carried qacA. Four isolates with the qacA gene had a chlorhexidine MIC of 4 μg/ml while one isolate had a MIC of 2 μg/ml. The five isolates were also resistant to clindamycin, erythromycin, levofloxacin, tetracycline, trimethoprim-sulfamethoxazole, and gentamicin. The five isolates were collected from residents of three nursing homes.
Fig 1.
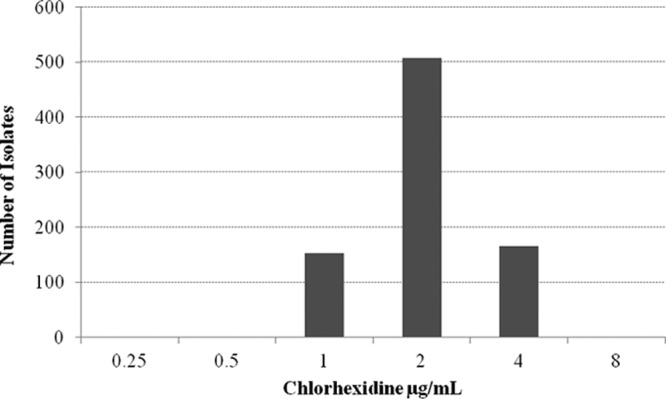
Distribution of chlorhexidine MICs of 829 MRSA isolates from residents of 25 nursing homes.
The majority of the MRSA isolates were resistant to clindamycin (55%), erythromycin (90%), and levofloxacin (98%) (Table 2). A few isolates were resistant to tetracycline (4%), trimethoprim-sulfamethoxazole (2%), gentamicin (12%), quinupristin-dalfopristin (<1%), linezolid (<1%), and rifampin (1%). All the isolates were susceptible to vancomycin and daptomycin. When examining correlations between antibiotics, clindamycin and erythromycin resistance were moderately correlated (0.43), as were gentamicin and HLMR (0.44) and tetracycline and trimethoprim-sulfamethoxazole (0.53).
Table 2.
Susceptibility of Orange County nursing home MRSA isolates (n = 829) to 13 antimicrobial agents
| Antimicrobial agent | MIC data (μg/ml) |
% resistant | ||
|---|---|---|---|---|
| Range | MIC50 | MIC90 | ||
| Chlorhexidine | 0.5–4 | 2 | 4 | NAa |
| Clindamycin | ≤0.12 to >16 | >16 | >16 | 55 |
| Erythromycin | ≤0.25 to >32 | >32 | >32 | 90 |
| Tetracycline | ≤0.5 to >64 | ≤0.5 | ≤0.5 | 4 |
| Trimethoprim-sulfamethoxazole | ≤0.25 to >32 | ≤0.25 | ≤0.25 | 2 |
| Gentamicin | ≤0.5 to >64 | ≤0.5 | 64 | 12 |
| Levofloxacin | ≤0.25 to >32 | 32 | >32 | 98 |
| Vancomycin | 0.5–2 | 1 | 1 | 0 |
| Daptomycin | ≤0.5–4 | ≤0.5 | ≤0.5 | NA |
| Quinupristin-dalfopristin | ≤0.5–4 | ≤0.5 | ≤0.5 | <1 |
| Linezolid | ≤1–8 | 2 | 2 | <1 |
| Rifampin | ≤0.5 to >4 | ≤0.5 | ≤0.5 | 1 |
| Mupirocinb | ||||
| Low-level resistance isolates | 3 | |||
| High-level resistance isolates | 9 | |||
NA, not available, because the Clinical and Laboratory Standards Institute has not yet established breakpoints for resistance to this agent.
Disk diffusion testing was performed, and so no MICs are reported here.
Mupirocin resistance, at either a high or low level, was observed in 101 isolates (12%). Resistance rates among nursing homes varied from 0% to 40% (Fig. 2). The facility with a 40% resistance rate had only 10 MRSA isolates collected; nevertheless, resistance rates greater than 20% were observed in six facilities (24%). In contrast, mupirocin resistance was not identified in 10 nursing homes (38%). LLMR was identified in 23 isolates, with 11 (42%) facilities having at least one resident colonized with a low-level mupirocin-resistant isolate. HLMR was observed in 78 (9%) isolates, with HLMR rates varying considerably by facility, ranging from 0 to 31%. Fifteen of 25 (60%) nursing homes had at least one resident who carried a high-level mupirocin-resistant isolate, and 5 (20%) nursing homes had HLMR levels exceeding 20% of all MRSA isolates from the corresponding facility. There were no MRSA isolates that exhibited both HLMR and harbored the qacA gene. However, two isolates from the same nursing home harbored the qacA gene and exhibited LLMR.
Fig 2.
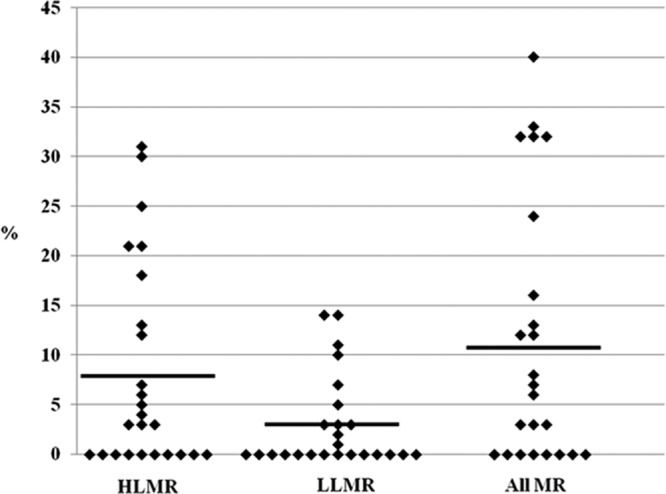
MRSA resistant to mupirocin (HLMR, LLMR, and all resistant [MR]) at each of 25 nursing homes in Orange County.
The 78 isolates with HLMR could be grouped into four PFGE types, and these types included 31 subtypes (Table 3). The majority of the isolates (60%) were closely related to USA100. These isolates were very widespread, being represented in almost half (48%) of the nursing homes included in this study. Twenty-five (32%) isolates from nine nursing homes were related to USA300. Furthermore, application of spa typing results from a previous study (31) along with our susceptibility data enabled us to determine that 11% (24/221) of the isolates that were spa type t008 (the type commonly associated with USA300) exhibited HLMR. Five (6%) isolates from a single facility were related to USA500, and one isolate was not related to any USA type strain or to any of the other isolates with HLMR in this study.
Table 3.
PFGE types of high-level mupirocin-resistant MRSA (n = 78)
| PFGE type | No. of isolates (%) | No. of subtypes | Related USA type | Nursing home(s) representeda |
|---|---|---|---|---|
| A | 47 (60) | 17 | 100 | 1–6, 8–10, 12, 14, 15 |
| B | 25 (32) | 11 | 300 | 3, 4, 6–11, 13 |
| C | 5 (6) | 2 | 500 | 5 |
| D | 1 (1) | 1 | NAb | 1 |
PFGE types A and B were both identified in nursing homes 3, 4, 6, 8, 9, and 10. PFGE types A and C were identified in nursing home 5. PFGE types A and D were identified in nursing home 1.
This isolate is not related to any of the current USA types (USA100 to USA1200).
In the bivariate analysis, carriage of MRSA with HLMR in the nares was associated with residing in a facility with a high RUG score, a high ADL score (indicating more dependence upon caregivers), or a higher percentage of residents with indwelling devices (P < 0.1) (Table 4). Also, residents harboring high-level mupirocin-resistant MRSA were 3-fold more likely to have a history of MRSA. When analyzing practices performed at partner hospitals, carriage of high-level mupirocin-resistant MRSA was not associated with the percentage of physicians who routinely decolonized patients, the percentage of MRSA-positive patients routinely decolonized, or whether mupirocin was used for decolonization. Additionally, none of the nursing homes routinely decolonized residents that were colonized with MRSA.
Table 4.
Bivariate analysis of nursing home characteristics and partner hospital characteristics associated with high-level mupirocin-resistant MRSAa
| Variable | Odds ratio | P value |
|---|---|---|
| Nursing home characteristics | ||
| Annual no. of admissions | 0.99 | 0.58 |
| Length of stay (median no. of days) | 1.00 | 0.83 |
| MRSA point prevalence | 1.02 | 0.43 |
| Demographics (as % of all facility residents) | ||
| Less than high school education | 1.03 | 0.88 |
| Admitted from acute hospital | 1.01 | 0.42 |
| Comorbidities (as % of all facility residents) | ||
| Diabetes | 1.01 | 0.59 |
| Skin lesions | 1.15 | 0.40 |
| Indwelling device | 1.30 | 0.006 |
| History of MRSAb | 2.99 | <0.001 |
| Multidrug-resistant MRSA isolateb | 2.55 | 0.004 |
| Functional status (avg score among all facility residents) | ||
| High RUG scorec | 2.97 | 0.04 |
| Avg ADL score | 1.28 | 0.001 |
| Partner hospital characteristicd | ||
| % physicians who routinely decolonize patients who have MDROe | 1.04 | 0.78 |
| % MRSA+ patients who are decolonized | 0.79 | 0.13 |
| % that use mupirocin to decolonize MRSA+ patients | 0.54 | 0.26 |
Note that four psychiatric facilities were removed from the bivariate analyses.
Analyzed as a resident-level or isolate-level variable, not as a facility-level variable.
RUG is a facility-level score that reflects the average level of care required by residents, based upon residents' comorbidities, ability to perform activities of daily living, and the required amount of physical and occupational therapy. The RUG score was dichotomized into high and low values around the median.
A nursing home's partner hospital was the hospital that transferred the most patients to that nursing home in a year.
MDRO, multidrug-resistant organisms.
In the multivariate model, residents colonized with a high-level mupirocin-resistant strain were more likely to have a history of MRSA (OR, 2.43; P < 0.001), to have a multidrug-resistant strain (OR, 2.69; P = 0.004), and to reside in a nursing home with an elevated average ADL score (OR, 1.2; P = 0.004) (Table 5). In our model, average ADL score was interchangeable with the percentage of residents with an indwelling device.
Table 5.
Multivariate analysisa of factors associated with high-level mupirocin-resistant MRSA
| Variable | Odds ratio (95% CI) | P value |
|---|---|---|
| Multidrug-resistant MRSA isolateb | 2.69 (1.37, 5.28) | 0.004 |
| History of MRSAb | 2.34 (1.75, 3.12) | <0.001 |
| Avg ADL scorec | 1.25 (1.07, 1.45) | 0.004 |
Note that four psychiatric facilities were removed from the multivariate analysis.
Analyzed as a resident-level or isolate-level variable, not as a facility-level variable.
The ADL score ranges from 0 (completely independent) to 28 (completely dependent) and is expressed per 1 point increase. The reported average ADL score is the average score among all residents of one facility. The ADL score was colinear with the presence of indwelling devices.
When examining the four nursing homes that were also psychiatric facilities, none of the facilities had a resident harboring a MRSA strain that carried the qacA and/or qacB gene or a strain that exhibited mupirocin resistance. Furthermore, the medians for most characteristics did not substantially differ after excluding the four psychiatric nursing homes (Table 1). The two characteristics that changed slightly were the median annual admissions (262 to 300) and the percentage of facility residents older than 85 years (25 to 39%).
DISCUSSION
Nursing homes may be at high risk for harboring MRSA strains resistant to topical agents, since their residents have high rates of antimicrobial exposure. However, knowledge about the epidemiology of resistance to these topical agents is limited. The goal of this study was to examine chlorhexidine and mupirocin resistance among MRSA carriers residing in nursing homes in Orange County, CA, and to assess factors associated with mupirocin resistance.
We found that fewer than 1% of the MRSA isolates carried the putative chlorhexidine resistance genes qacA and/or qacB, and none had chlorhexidine MICs that were >4 μg/ml. Other health care facilities have reported a higher prevalence of qacA and/or qacB in MRSA isolates. Lee et al. identified qacA and/or qacB in 91% of the MRSA isolates from patients who had failed decolonization (18). The rarity of the qacA and/or qacB gene loci in our large collection of nursing home MRSA isolates is of interest, given the common use of chlorhexidine for preoperative bathing, as well as body surface antisepsis prior to placement of central lines or surgical incisions. At least one affiliated hospital was using it for daily bathing in the intensive care unit setting.
In contrast, mupirocin resistance was common among MRSA isolates from these nursing home patients. The majority of mupirocin-resistant isolates (over three-fourths) exhibited high-level mupirocin resistance, which has been associated with decolonization failure (32, 33). Although the overall mean rate was 9%, substantial variability across nursing homes was noted, and very high rates (>20%) were not uncommon.
These rates are substantially higher than previously reported from nationwide surveillance programs in the United States. Recent U.S. data from Richter and colleagues, who collected and tested 2,247 MRSA isolates from 43 centers nationwide, found HLMR in only 2.9%, while the Centers for Disease Control and Prevention recently reported national surveillance data from 823 USA300 MRSA strains that indicated 2.7% were HLMR (20, 34). Our PFGE results demonstrated heterogeneity among the strains exhibiting HLMR, as might be expected for a resistance gene, mupA, which is usually plasmid mediated (14). Strains demonstrating HLMR were found among USA100, USA300, and USA500 strains, which were the most common USA types found in this population. However, our rate of HLMR was obviously substantially higher than in prior surveys, especially when focused on strains associated with USA300. USA300 strains with HLMR accounted for >3% of all the MRSA in this study and approximately one-third of all the high-level mupirocin-resistant MRSA. Additionally, 11% of the isolates that were spa type t008 exhibited HLMR.
When examining chlorhexidine and mupirocin resistance together, none of the isolates in our study harbored qacA and/or qacB and displayed HLMR. However, two isolates from one nursing home harbored the qacA gene and had LLMR. Lee et al. reported the presence of qacA and/or qacB and resistance to mupirocin in approximately 63% of the MRSA isolates from patients who had failed decolonization (18). They also reported a higher decolonization failure rate in patients carrying a MRSA strain with qacA and/or qacB in conjunction with LLMR. None of the nursing homes was routinely using chlorhexidine for resident bathing or mupirocin for nasal decolonization. Affiliation with hospitals that were decolonizing patients with mupirocin was not significantly associated with high-level mupirocin-resistant MRSA colonization.
In our study, a history of MRSA was associated with colonization with a isolate exhibiting HLMR. This may represent the likelihood that MRSA carriers are increasingly being offered decolonization therapy by inpatient or outpatient providers. Although we did not collect individual-level receipt of chlorhexidine and mupirocin, others have reported that mupirocin use within 1 year prior to infection was a significant risk factor for both low- and high-level mupirocin resistance (35).
High-level mupirocin-resistant MRSA strains were frequently multidrug resistant. This finding may be explained by the fact that the mupA gene is carried on a large conjugative plasmid that may facilitate cotransfer of other resistance determinants (14, 36). The relationship between HLMR and multidrug resistance has been reported in other studies. Cadilla et al. reported HLMR in 6.8% of the multidrug-resistant MRSA in their study (30). We did not find any association between HLMR among a nursing home's isolates and decolonization practices at each facility's hospital partner. However, our assessment was ecological, and we restricted our analysis to include only one partner hospital per nursing home. Since nursing homes may exchange patients with multiple hospitals, a more inclusive analysis may be needed to definitively test this association at the population level.
We found that nursing homes with a higher average ADL score, indicating more dependence upon caregivers, were more likely to have residents colonized with high-level mupirocin-resistant MRSA. As far as we know, this is the first study to identify an association between higher average ADL scores and carriage of high-level mupirocin-resistant MRSA. This may reflect the fact that residents needing greater assistance may have higher health care worker contact and spend more time in health care facilities. Previous studies have reported an association between colonization with MRSA or S. aureus and greater reliance upon others to perform ADL (5, 37, 38). Daeschlein et al. found that nursing home residents who needed 3 to 4 h of assistance with daily activities were more likely to carry S. aureus in their nares (37). Also, Washio et al. reported a relationship between greater ADL dependence and MRSA colonization in elderly Japanese patients admitted to a geriatric hospital (38). Additionally, March et al. identified physical disability to be a risk factor for colonization with MRSA in nursing home residents in Italy (5). Our study also found a correlation between a more dependent ADL score and patients having an indwelling device. Additional studies are needed to determine if this reflects a higher likelihood of providers to offer decolonization to those with devices or those with a greater dependency on the health care system. Regardless, monitoring of rising resistance in this important patient population is needed.
In summary, chlorhexidine resistance was not commonly found in MRSA isolates from nursing homes, but mupirocin resistance rates were higher in nursing homes than previously found in the community and from acute care facilities and varied substantially across facilities. Importantly, in contrast to other studies which have found a predominance of LLMR, we found that nearly all mupirocin-resistant isolates exhibited HLMR. These elevated HLMR rates in nursing homes are concerning and suggest that emerging resistance will be a barrier to prevention programs that include widespread use of mupirocin. Additional information regarding mupirocin use practices will help to better understand the high rates and wide variability of mupirocin resistance across these nursing homes.
ACKNOWLEDGMENTS
We thank Fathollah Riahi for assisting with the antimicrobial susceptibility testing.
This project was funded under contract number HHSA290-2005-0033I from the Agency for Healthcare Research and Quality, U.S. Department of Health and Human Services, as part of the Developing Evidence to Inform Decisions about Effectiveness (DEcIDE) program.
The authors of this report are responsible for its content. Statements in the report should not be construed as endorsement by the Agency for Healthcare Research and Quality or the U.S. Department of Health and Human Services.
J.S.M., C.R.M., V.Q., D.S.K., E.M.P., K.D.E., G.L.T., and S.S.H. report no conflicts of interest relevant to this article. D.J.D. has received research funding from bioMérieux, Innovative Biosensors, PurThread Technologies, Cerexa, Pfizer, and Merck. M.K.H. has received products for research from Sage, Inc.
Footnotes
Published ahead of print 12 November 2012
REFERENCES
- 1. Elkins KR, Nguyen CM, Kim DS, Meyers H, Cheung M, Huang SS. 2011. Successful strategies for high participation in three regional healthcare surveys: an observational study. BMC Med. Res. Method. 11:176 doi:10.1186/1471-2288-11-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lederer SR, Riedelsdorf G, Schiffl H. 2007. Nasal carriage of methicillin-resistant Staphylococcus aureus: the prevalence, patients at risk and the effect of elimination on outcomes among outpatient haemodialysis patients. Eur. J. Med. Res. 12:284–288 [PubMed] [Google Scholar]
- 3. Mody L, Kauffman CA, Donabedian S, Zervos M, Bradley SF. 2008. Epidemiology of Staphylococcus aureus colonization in nursing home residents. Clin. Infect. Dis. 46:1368–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reynolds C, Quan V, Kim D, Peterson E, Dunn J, Whealon M, Terpstra L, Meyers H, Cheung M, Lee B, Huang SS. 2011. Methicillin-resistant Staphylococcus aureus (MRSA) carriage in 10 nursing homes in Orange County, California. Infect. Control Hosp. Epidemiol. 32:91–93 [DOI] [PubMed] [Google Scholar]
- 5. March A, Aschbacher R, Dhanji H, Livermore DM, Böttcher A, Sleghel F, Maggi S, Noale M, Larcher C, Woodford N. 2010. Colonization of residents and staff of a long-term-care Facility and adjacent acute-care hospital geriatric unit by multiresistant bacteria. Clin. Microbiol. Infect. 16:934–944 [DOI] [PubMed] [Google Scholar]
- 6. Smith PW, Bennett G, Bradley S, Drinka P, Lautenbach E, Marx J, Mody L, Nicolle L, Stevenson K, Society for Healthcare Epidemiology of America, Association for Professionals in Infection Control and Epidemiology 2008. SHEA/APIC guideline: infection prevention and control in the long-term care facility. Infect. Control Hosp. Epidemiol. 29:785–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chaiyakunapruk N, Veenstra DL, Lipdsky BA, Saint S. 2002. Chlorhexidine compared with povidone-iodine solution for vascular catheter-site care: a meta-analysis. Ann. Intern. Med. 136:792–801 [DOI] [PubMed] [Google Scholar]
- 8. Darouiche RO, Wall MJ, Itani KMF, Otterson MF, Webb AL, Carrick MM, Miller HJ, Awad SS, Crosby CT, Mosier MC, Alsharif A, Berger DH. 2010. Chlorhexidine-alcohol versus povidone-iodine for surgical-site antisepsis. N. Engl. J. Med. 362:18–26 [DOI] [PubMed] [Google Scholar]
- 9. Milstone AM, Passaretti CL, Perl TM. 2008. Chlorhexidine: expanding the armamentarium for infection control and prevention. Clin. Infect. Dis. 46:274–281 [DOI] [PubMed] [Google Scholar]
- 10. Popovich KJ, Hota B, Hayes R, Weinstein RA, Hayden MK. 2009. Effectiveness of routine patient cleansing with chlorhexidine gluconate for infection prevention in the medical intensive care unit. Infect. Control Hosp. Epidemiol. 30:959–963 [DOI] [PubMed] [Google Scholar]
- 11. Rupp ME, Lisco SJ, Lipsett PA, Perl TM, Keating K, Civetta JM, Mermel LA, Lee D, Dellinger EP, Donahoe M, Giles D, Pfaller MA, Maki DG, Sherertz R. 2005. Effect of a second-generation venous catheter impregnated with chlorhexidine and silver sulfadiazine on central catheter-related infections: A randomized controlled trial. Ann. Intern. Med. 143:570–580 [DOI] [PubMed] [Google Scholar]
- 12. Tantipong H, Morkchareonpong C, Jaiyindee S, Thamlikitkul V. 2008. Randomized controlled trial and meta-analysis of oral decontamination with 2% chlorhexidine solution for the prevention of ventilator-associated pneumonia. Infect. Control Hosp. Epidemiol. 29:131–136 [DOI] [PubMed] [Google Scholar]
- 13. Timsit JF, Schwebel C, Bouadma L, Geffroy A, Garrouste-Orgeas M, Pease S, Herault MC, Haouache H, Calvino-Gunther S, Gestin B, Armand-Lefevre L, Leflon V, Chaplain C, Benali A, Francais A, Adrie C, Zahar JR, Thuong M, Arrault X, Croize J, Lucet JC, Dressing Study Group 2009. Chlorhexidine impregnated sponges and less frequent dressing changes for prevention of catheter-related infections in critically-ill patients: a randomized controlled trial. JAMA 301:1231–1241 [DOI] [PubMed] [Google Scholar]
- 14. Patel JB, Gorwitz RJ, Jerigan JA. 2009. Mupirocin resistance. Clin. Infect. Dis. 49:935–941 [DOI] [PubMed] [Google Scholar]
- 15. Meyer B, Cookson B. 2010. Does microbial resistance or adaptation to biocides create a hazard in infection prevention and control? J. Hosp. Infect. 76:200–205 [DOI] [PubMed] [Google Scholar]
- 16. Batra R, Cooper BS, Whitely C, Patel AK, Wyncoll D, Edgeworth JD. 2010. Efficacy and limitation of a chlorhexidine-based decolonization strategy in preventing transmission of methicillin-resistant Staphylococcus aureus in an intensive care unit. Clin. Infect. Dis. 50:210–217 [DOI] [PubMed] [Google Scholar]
- 17. Wang JT, Sheng WH, Wang JL, Chen D, Chen ML, Chen YC, Chang SC. 2008. Longitudinal analysis of chlorhexidine susceptibilities of nosocomial methicillin-resistant Staphylococcus aureus isolates at a teaching hospital in Taiwan. J. Antimicrob. Chemother. 62:514–517 [DOI] [PubMed] [Google Scholar]
- 18. Lee AS, Macedo-Vinas M, Francois P, Renzi G, Schrenzel J, Vernaz N, Pittet D, Harbarth S. 2011. Impact of combined low-level mupirocin and genotypic chlorhexidine resistance on persistent methicillin-resistant Staphylococcus aureus carriage after decolonization therapy: a case-control study. Clin. Infect. Dis. 52:1422–1430 [DOI] [PubMed] [Google Scholar]
- 19. Liu QZ, Wu Q, Zhang YB, Liu MN, Hu FP, Xu XG, Zhu DM, Ni YX. 2010. Prevalence of clinical methicillin-resistant Staphylococcus aureus (MRSA) with high-level mupirocin resistance in Shanghai and Wenzhou, China. Int. J. Antimicrob. Agents 35:114–118 [DOI] [PubMed] [Google Scholar]
- 20. Richter SS, Heilmann KP, Dohrn CL, Riahi F, Costello AJ, Kroeger JS, Biek D, Critchley IA, Diekema DJ, Doern GV. 2011. Activity of ceftaroline and epidemiologic trends in Staphylococcus aureus isolates collected from 43 medical centers in the United States in 2009. Antimicrob. Agents Chemother. 55:4154–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rossney A, O'Connell S. 2008. Emerging high-level mupirocin resistance among MRSA isolates in Ireland. Euro Surveill. 13:8084 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=8084 [PubMed] [Google Scholar]
- 22. Shittu AO, Udo EE, Lin J. 2009. Phenotypic and molecular characterization of Staphylococcus aureus isolates expressing low- and high-level mupirocin resistance in Nigeria and South Africa. BMC Infect. Dis. 9:10 doi:10.1186/1471-2334-9-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seah C, Alexander DC, Louie L, Simor A, Low DE, Longtin J, Melano RG. 2012. MupB, a new high-level mupirocin resistance mechanism in Staphylococcus aureus. Antimicrob. Agents Chemother. 56:1916–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clinical and Laboratory Standards Institute 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 25. Malaviolle X, Nonhoff C, Denis O, Rottiers S, Stuelens MJ. 2008. Evaluation of disc diffusion methods and Vitek 2 automated system for testing susceptibility to mupirocin in Staphylococcus aureus. J. Antimicrob. Chemother. 62:1018–1023 [DOI] [PubMed] [Google Scholar]
- 26. Kroeger J, Diekema D, Murphy C, Quan V, Kim DS, Peterson EM, Evans KD, Tan GL, Hayden MK, Huang SS. 2011. Chlorhexidine and mupirocin susceptibility of methicillin-resistant Staphylococcus aureus (MRSA) from long-term care facility residents, abstr 395. Progr. Abstr. 21st Annu. Sci. Meet. Soc. Healthcare Epidemio. Am., 1 to 4 April 2011, Dallas, TX Society of Healthcare Epidemiology of America, Arlington, VA [Google Scholar]
- 27. Pfaller MA, Hollis RJ, Sader HS. 1994. Chromosomal restriction fragment analysis by pulsed field gel electrophoresis, p 10.5.c.1–10.5c.12 In Isenberg HD. (ed), Clinical microbiology procedures handbook, suppl 1 American Society for Microbiology, Washington, DC [Google Scholar]
- 28. University of Minnesota School of Public Health Research Data Assistance Center 2010. MDS (Minimum Data Set) database. University of Minnesota School of Public Health, Minneapolis, MN: http://www.resdac.org/search/site/MDS Accessed 1 September 2011 [Google Scholar]
- 29. Brown University Alpert Medical School 2007. Long-term care: facts on care in the US. Brown University, Providence, RI: http://ltcfocus.org Accessed 1 September 2011 [Google Scholar]
- 30. Cadilla A, David M, Daum RS, Boyle-Vavra S. 2011. Association of high-level mupirocin resistance and multidrug-resistant methicillin-resistant Staphylococcus aureus at an academic center in the midwestern United States. J. Clin. Microbiol. 49:95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murphy CR, Hudson LO, Spratt BG, Quan V, Kim D, Peterson E, Tan G, Evans K, Meyers H, Cheung M, Lee BY, Mukamel DB, Enright MC, Whealon M, Huang SS. Predicting high prevalence of community MRSA strains in nursing homes. Infect. Control Hosp. Epidemiol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Simor AE, Phillips E, McGeer A, Konvalinka A, Loeb M, Devlin RH, Kiss A. 2007. Randomized controlled trial of chlorhexidine gluconate for washing, intranasal mupirocin, and rifampin and doxycycline versus no treatment for the eradication of methicillin-resistant Staphylococcus aureus colonization. Clin. Infect. Dis. 4:178–185 [DOI] [PubMed] [Google Scholar]
- 33. Walker ES, Vasquez JE, Dula R, Bullock H, Sarubbi FA. 2003. Mupirocin-resistant, methicillin-resistant Staphylococcus aureus: does mupirocin remain effective? Infect. Control Hosp. Epidemiol. 24:342–346 [DOI] [PubMed] [Google Scholar]
- 34. McDougal LK, Fosheim GE, Nicholson A, Bulens SN, Limbago BM, Shearer JE, Summers AO, Patel JB. 2010. Emergence of resistance among USA300 methicillin-resistant Staphylococcus aureus isolates causing invasive disease in the United States. Antmicrob. Agents Chemother. 54:3804–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Caffrey AR, Quilliam BJ, LaPlante KL. 2010. Risk factors associated with mupirocin resistance in methicillin-resistant Staphylococcus aureus. J. Hosp. Infect. 76:206–210 [DOI] [PubMed] [Google Scholar]
- 36. Pawa A, Noble WC, Howell SA. 2000. Co-transfer of plasmids in association with conjugative transfer of mupirocin or mupirocin and penicillin resistance in methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 49:1103–1107 [DOI] [PubMed] [Google Scholar]
- 37. Daeschlein G, Assadian O, Rangous I, Kramer A. 2006. Risk factors for Staphylococcus aureus nasal carriage in residents of three nursing homes in Germany. J. Hosp. Infect. 63:216–220 [DOI] [PubMed] [Google Scholar]
- 38. Washio M, Kiyohara C, Honjo N, Aoyagi K, Okada K, Arai Y, Fujishima M, Ito Y. 1997. Methicillin-resistant Staphylococcus aureus (MRSA) isolation from pharyngeal swab cultures of Japanese elderly at admission to a geriatric hospital. J. Epidemiol. 7:167–172 [DOI] [PubMed] [Google Scholar]
- 39. Furuno JP, Hebden JN, Standjford HC, Perencevich EN, Miller RR, Moore AC, Strauss SM, Harris AD. 2008. Prevalence of methicillin-resistant Staphylococcus aureus and Acinetobacter baumannii in a long-term acute care facility. Am. J. Infect. Control 36:468–471 [DOI] [PMC free article] [PubMed] [Google Scholar]


