Abstract
Influenza A virus infection is a major global health concern causing significant mortality, morbidity, and economic loss. Antiviral chemotherapeutics that target influenza A virus are available; however, rapid emergence of drug-resistant strains has been reported. Consequently, there is a burgeoning need to identify novel anti-influenza A drugs, particularly those that target host gene products required for virus replication, to reduce the likelihood of drug resistance. In this study, a small interfering RNA (siRNA) screen was performed to identify host druggable gene targets for anti-influenza A virus therapy. The host organic anion transporter-3 gene (OAT3), a member of the SLC22 family of transporters, was validated as being required to support influenza A virus replication. Probenecid, a prototypical uricosuric agent and chemical inhibitor of organic anion transporters known to target OAT3, was shown to be effective in limiting influenza A virus infection in vitro (50% inhibitory concentration [IC50] of 5.0 × 10−5 to 5.0 × 10−4 μM; P < 0.005) and in vivo (P < 0.05). Probenecid is widely used for treatment of gout and related hyperuricemic disorders, has been extensively studied for pharmacokinetics and safety, and represents an excellent candidate for drug repositioning as a novel anti-influenza A chemotherapeutic.
INTRODUCTION
Influenza A virus is a global public health concern, causing morbidity and substantial mortality (1), with increasing trends in disease severity in the members of the population who are age 65 or older (2). Influenza vaccines are available; however, due to high virus mutation rates and constant antigenic drift, vaccines need to be developed annually (3). Currently, several antiviral drugs, including the neuraminidase (NA) inhibitors zanamivir and oseltamivir and the M2 ion channel inhibitors (amantadine and rimantidine), are available to treat influenza A virus infection (3–6). These antiviral drugs target viral components, a feature which provides selective pressure for development of drug resistance. The rapid emergence of drug-resistant influenza A virus strains has been increasingly reported, with a limited number of new antiviral drugs in the pipeline (7), highlighting the imminent need to identify novel drug targets and for the subsequent development of a new class of antiviral drugs.
High cost and lengthy approval processes are associated with development of new drugs for clinical use. Due to these factors, along with the limited number of new antiviral drugs currently in the pipeline, there is an increasing need for repositioning available currently approved drugs for treatment of other diseases. Drug repositioning allows faster availability of new treatments for a fraction of the cost of developing new drugs. Furthermore, the safety and pharmacokinetics of these drugs have already been assessed. Examples of such repositioning include the use of anticancer drugs zidovudine (AZT), decitabine, and gemcitabine for HIV (human immunodeficiency virus) treatment (8) and of the erectile dysfunction drug sildenafil, which was originally developed for antihypertension treatment (9).
Recently, numerous studies have utilized an RNA interference (RNAi) screen to identify cellular factors involved in virus replication. This strategy takes advantage of the fact that viruses lack their own machinery to replicate independently; thus, most use or co-opt host-derived gene products to facilitate entry, replication, and release. Therefore, targeting host factors involved in virus replication may serve as a therapeutic and/or prophylactic disease intervention strategy. In this study, RNAi was used to identify novel host drug targets for influenza A antiviral therapies. A small interfering RNA (siRNA) screen targeting 4,795 druggable genes in human lung type II epithelial (A549) cells was performed with influenza A/WSN/33 virus (WSN). This gene library was chosen for the druggability properties of the host gene products previously shown to favor interaction with drug compounds, thereby increasing the likelihood of identification of pharmacological inhibitors (10). Various druggable genes were identified in the siRNA screen, but only a few host genes were validated to substantially affect influenza A virus replication. One of these host genes was the organic anion transporter-3 gene, OAT3, a member of the SLC22 gene family. Transfection of A549 cells with siRNA targeting the SLC22A8 gene, also known as OAT3, completely blocked influenza A/WSN/33(H1N1) virus replication. The solute carrier (SLC) superfamily comprises 298 members grouped into 43 families, including SLC22 (11). The SLC22 family is further subdivided into three subfamilies: organic cation transporters (OCT), zwitterion/cation transporters (OCTN), and organic anion transporters (OAT) (12). The role of OAT transporters in the lung is not well characterized, but their substrates in intestine, liver, and kidney are targeted by several drugs that include diuretics, nonsteroidal anti-inflammatory compounds, β-lactam antibiotics, several antiviral drugs, xenobiotics, and endogenous compounds, e.g., cyclic nucleotide endogenous metabolites (11–13). Elsewhere, OAT family members have been implicated in homeostasis and sensing in brain, heart, eye, muscle, and olfactory epithelium (13). OAT3 is mainly expressed in the basolateral membrane of the kidney proximal tubule; however, its expression also occurs elsewhere, including the luminal surface of choroid plexus in brain, skeletal muscle, developing bone, and adrenal glands (12). In this report, we show that OAT3 is also expressed in both human and mouse lung epithelial cells and that siRNA silencing of other closely related transporters, i.e., OAT1, OAT2, OAT4, OAT7, and URAT1, did not affect influenza A/WSN/33 virus replication, indicating a specific role of OAT3 to support influenza A virus replication.
Probenecid {4-[(dipropyl-amino)sulfonyl] benzoic acid} is a classical inhibitor of OAT and is widely prescribed for therapeutic treatment of gout and other hyperuricemic disorders (14, 15). Probenecid usage for treatment of other OAT-mediated disorders such as hypertension has also been explored, along with its use to extend the plasma level of drugs identified as OAT substrates (such as β-lactam antibiotics and several antiviral drugs) (16–19). In this study, probenecid was shown to reduce OAT3 mRNA and protein levels in vitro and in vivo. Administration of probenecid alone reduced influenza A virus titer in agreement with the finding that OAT3 is important for influenza A virus replication. Additionally, probenecid has been previously reported to elevate plasma concentrations of an active oseltamivir metabolite, oseltamivir carboxylate; thus, its coadministration with oseltamivir has been suggested (19–21). This report shows that probenecid, the classical OAT3 inhibitor, can potentially be repositioned for a new anti-influenza A therapy.
MATERIALS AND METHODS
Cell cultures, influenza A virus stocks, and mice used.
Human type II respiratory epithelial (A549) cells (ATCC; CCL-185) and Madin-Darby canine kidney (MDCK) cells (ATCC; CCL-34) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% heat-inactivated fetal bovine serum (FBS) (HyClone, Logan, UT) in a 37°C incubator with 5% CO2. Influenza virus strains A/WSN/33(H1N1), A/New Caledonia/20/99(H1N1) (New Caledonia), A/California/04/09(H1N1), A/California/07/09(H1N1) (CA), and A/Philippines/2/82/X-79(H3N2) (X-79) were propagated in 9-day-old embryonic chicken eggs, and titers in MDCK cells were determined as previously described (22, 23).
BALB/c female mice (6 to 8 weeks old) were obtained from the NCI (National Cancer Institute). All experiments and procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Georgia. All experiments were performed with 10 mice per group and repeated independently at least twice.
Human drug target library screen.
A primary RNAi screen using four pooled siRNAs to target each of the 4,795 genes in the human drug target library (SMARTpool; Dharmacon ThermoFisher, Lafayette, CO) was performed using A549 cells infected with influenza A/WSN/33(H1N1) virus at a multiplicity of infection (MOI) of 0.001 as previously described (24, 25). A siRNA pool targeting MEK (also known as mitogen-activated protein kinase kinase [MAPKK]) was used as a positive control due to the known role of MEK in influenza A virus replication (26). A nontargeting siRNA was also used as negative control. A549 cells were reverse transfected with siRNA using DharmaFECT-1 reagent (Dharmacon, Lafayette, CO) as previously described (25). Briefly, siRNAs resuspended in Hanks' balanced salt solution (HBSS) were incubated with DharmaFECT 1 to form transfection complexes in 96-well tissue culture dishes. Following formation of complexes, 1.5 × 104 A549 cells suspended in DMEM with serum were added into each well containing a siRNA transfection complex. The final concentration of pooled siRNA oligonucleotides was 50 nM per well, with a final DharmaFECT amount of 0.4 μl per well in a 100-μl culture volume. Transfections were carried out for 48 h to allow maximal expression knockdown before cells were infected with influenza A/WSN/33 virus at an MOI of 0.001. The level of infectious virus was measured 48 h postinfection by titration of A549 cell supernatants on MDCK cells (22), and the results were normalized to nontargeting siRNA-transfected cell levels. In addition, adherent A549 cells on culture plates were fixed and analyzed for the presence of influenza virus nucleoprotein (NP) by immunofluorescence staining as described below. All assays were run in duplicate, and the entire screening assay was repeated twice.
In vitro and in vivo inhibition assays.
Probenecid (Invitrogen, Carlsbad, CA), the OAT3 pharmacological inhibitor, was resuspended in phosphate-buffered saline (PBS), and its cellular toxicity on A549 cells was determined by a ToxiLight BioAssay (Lonza, Walkerville, MD). For in vitro analyses, A549 cells were pretreated with increasing concentrations of probenecid for 24 h. Cells were subsequently infected with influenza virus A/WSN/33(H1N1), A/New Caledonia/20/99(H1N1), A/California/07/09(H1N1), or A/Philippines/2/82/X-79(H3N2) at the indicated MOI. At 24 or 48 h postinfection, cells were fixed with cold methanol:acetone for subsequent anti-NP immunostaining as described below or collected for total RNA isolation using a Qiagen RNeasy kit (Qiagen) to assess the OAT3 expression level and influenza A virus copy number using quantitative reverse transcription-PCR (qRT-PCR).
For in vivo studies to evaluate lung virus burden, probenecid was administered intraperitoneally (i.p.) at doses and time points pre- or post-influenza A virus infection as indicated in Results. At each indicated time point, mice were intranasally inoculated with a mouse-adapted strain of influenza A/WSN/33(H1N1) virus, nonadapted influenza A/New Caledonia/20/99(H1N1) virus, or influenza A/California/04/09(H1N1) virus at their respective 50% lethal dose (LD50) of 70 PFU, 22 PFU, or 35 PFU. Lungs were collected 48 h postinfection, which corresponds to peak lung virus replication, and longitudinally sectioned for qRT-PCR and 50% tissue culture infective dose (TCID50) analyses. Left lobes of lungs were homogenized in serum-free DMEM to assess virus titer, and right lobes were homogenized in TRizol solution (Invitrogen, Carlsbad, CA) for total RNA isolation. For virus titration analyses, lung homogenates were serially diluted and the titer was determined on MDCK cells for 72 h. Hemagglutination (HA) assays were performed using turkey red blood cells (RBCs) and virus-infected MDCK cell supernatant as described previously (22, 27). The HA titer was determined from the highest dilution factor that produced a positive HA reading, and virus titers were calculated as the TCID50 using the Spearman-Karber formula (22, 27). For the in vivo survival study, mice were i.p. treated with PBS only or probenecid, as indicated in Results, pre- or postinfection with 10× LD50 of influenza A/WSN/33(H1N1) virus (2.2 × 103 PFU per mouse). Mice were monitored for 14 days postinfection for survival and weight loss under the University of Georgia IACUC guideline.
Gene expression analyses.
For measurement of the influenza A viral copy number, total RNA collected from infected A549 cells, or lungs of infected mice, was used for a quantitative RT-PCR assay using a OneStep RT-PCR kit (Qiagen). A 5-ng volume of total RNA was used per reaction. A universal influenza A virus primer-probe set was used for amplification and detection of influenza A virus RNA (InfA forward [SS118272-45], InfA reverse [SS118272-46], and InfA probe [SS118273-01]; Bioresearch Technologies, Inc., Novato, CA; provided by the CDC). RT-PCR was performed using an MX3005P thermocycler (Strategene; Agilent Technologies, Santa Clara, CA) under the following conditions: 30 min at 50°C (reverse transcription); 15 min at 95°C (Taq inhibitor activation); and 45 cycles of PCR amplifications (95°C for 15 s, 55°C for 30 s).
To assess OAT3 gene expression, cDNAs were synthesized from total RNA using a SuperScript Vilo cDNA synthesis kit (Invitrogen, Carlsbad, CA). cDNAs were subsequently used for quantitative PCR amplifications using OAT3 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene-specific primers and RT2 SYBR Green qPCR Master Mix (SABioscience) in an MX3005P thermocycler. The PCR conditions used for amplification were as follows: 10 min at 95°C followed by 40 cycles of 95°C for 30 s, 60°C for 1 min, and 72°C for 30 s. OAT3 expression was normalized to GAPDH expression, and its expression relative to mock-treated samples was calculated using the 2(−ΔΔCt) formula.
Immunoblot analyses.
To evaluate OAT protein expression following siRNA transfection or probenecid treatment, A549 cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris HCl [pH 7.5], 150 mM NaCl, 0.5% sodium deoxycholate, 1% Nonidet P-40, 1 mM EDTA, and 0.1% sodium dodecyl sulfate) supplemented with a protease inhibitor cocktail tablet (Roche, Germany), followed by 4°C centrifugation at 16,000 × g for 10 min to clarify lysates. Equivalent protein amounts were analyzed by SDS-polyacrylamide gel electrophoresis followed by immunoblotting (Bio-Rad, Hercules, CA). Rabbit anti-OAT3 (Santa Cruz Biotechnology, Santa Cruz, CA) and rabbit anti-GAPDH polyclonal antibodies were used as primary antibodies for immunoblot analyses. Allophycocyanin (AP)-conjugated goat anti-rabbit IgG was used as a secondary antibody. Protein bands were visualized following addition of ECF substrate (GE Healthcare, Piscataway, NJ) and scanned using an Amersham Typhoon 9210 fluorescence scanner (GE Healthcare, Piscataway, NJ). Protein bands were quantified using ImageQuant software, and expression of OAT3 was normalized to expression of GAPDH.
Immunofluorescence staining.
A549 cells were fixed with cold methanol:acetone (80:20) for 15 min and incubated with primary antibodies (mouse anti-NP monoclonal antibody [ATCC; H16-L10-4R5] [5 μg/ml] and/or goat anti-OAT3 polyclonal antibody [Santa Cruz Biotechnology, Santa Cruz, CA] [4 μg/ml]) followed by incubation with appropriate secondary antibodies (Alexa 488-conjugated goat anti-mouse, Alexa 488-conjugated donkey anti-goat, and/or Alexa 546-conjugated goat anti-rabbit [Invitrogen, Carlsbad, CA] [1 μg/ml]) and 4′,6-diamidino-2-phenylindole (DAPI) counterstain (Invitrogen, Carlsbad, CA) (2 μg/ml). Cells were visualized and counted using a Cellomics ArrayScan system (Thermo Fisher Scientific), an automated fluorescence microscope coupled with image and analytical software. Where indicated, percentages of infected cells (positive for NP staining) were calculated for probenecid 50% inhibitory concentration (IC50) analysis.
Statistical analyses.
Statistical analyses were done using Student's t test or one-way analysis of variance (ANOVA), as indicated. Results were calculated as means ± standard errors. Values of P < 0.05 were considered significant.
RESULTS
siRNA screen of host drug target genes.
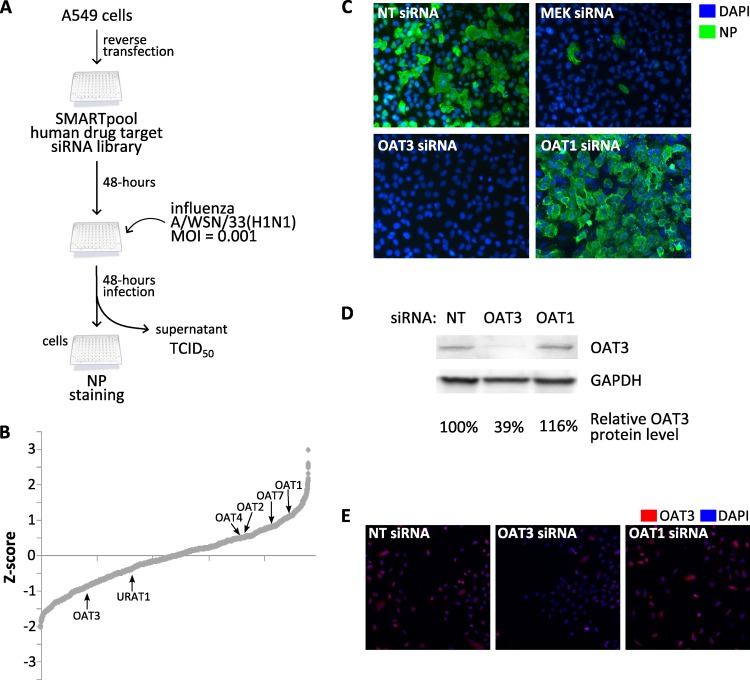
To identify novel host druggable targets, a screen was performed using a drug target siRNA library comprised of siRNA pools targeting 4,795 genes. The screen was performed in A549 cells infected with influenza A/WSN/33(H1N1) (WSN) virus at an MOI = 0.001 (Fig. 1A). As assay endpoints, infected cells were fixed and immunostained for influenza virus NP and culture supernatant was collected to measure virus titer by a hemagglutination (HA) assay 48 h following infection. Z-scores were calculated from HA assay results and plotted to display their distributions (Fig. 1B). Genes with negative Z-score values reduced influenza A virus titers when silenced with specific siRNA pools, indicating their importance for influenza A virus replication. Conversely, genes with positive Z-score values increased virus titer upon siRNA silencing, indicating their role in limiting influenza A virus replication. Transfection with MEK siRNA was used as a positive control due to its known importance in cellular signaling during influenza A virus infection. Genes with known pharmacological inhibitors available were chosen for further examinations in which the effects of these inhibitors on influenza A virus infection were assessed.
Fig 1.
siRNA screen of host drug target genes important for influenza A virus replication. (A) Schematic outline of siRNA screen performed. (B) Z-score values were calculated based on virus titers for siRNAs targeting host drug target genes. Their distributions were plotted based on Z-score rank order, from siRNA with the lowest to that with the highest virus titer. Labels denote OAT gene members within the drug target gene library. (C) A549 cells were transfected with nontargeting (NT) siRNA and pooled siRNA to MEK, OAT3, or OAT1 for 48 h prior to infection with influenza A/WSN/33(H1N1) virus at an MOI = 0.001. Cells were fixed 48 h postinfection, stained for viral NP (green) and nuclei (DAPI; blue), and visualized using fluorescence microscopy. (D and E) OAT3 protein expression was evaluated by immunoblot (D) and immunofluorescence microscopy (E) following transfection of nontargeting (NT) siRNA and pooled siRNA to OAT3 or OAT1. For immunoblot analysis, OAT3 protein bands were quantified and normalized to GAPDH, and percent normalized OAT3 protein levels relative to nontargeting siRNA control were calculated. For immunofluorescence analysis, cells were stained for OAT3 (red) and nuclei (DAPI; blue) and visualized using fluorescence microscopy.
In the siRNA screen, the OAT3 (SLC22A8) gene was identified and subsequently validated as a druggable target gene for influenza A virus infection. Transfection of A549 cells with four pooled siRNAs targeting different seed regions on the OAT3 gene reduced virus titer in culture supernatant (Z-score = −0.854) compared to transfection of cells with nontargeting siRNA (Fig. 1B). Additionally, no influenza virus NP-positive cells were detectable following OAT3 siRNA transfection (Fig. 1C), supporting the idea of OAT3 as important for influenza A virus replication. siRNA transfection of other OAT family members in the library displayed weakly moderate or no reduction in virus titers or in the number of detectable NP-positive cells (Fig. 1B and C; see also Fig. S1 in the supplemental material). Curiously, transfection of siRNA targeting a closely related molecule, OAT1 (SLC22A6), which was previously reported to display tissue distributions and functions similar to those of OAT3 (12), resulted in increased virus titer (Z-score = 1.043) and numbers of NP-positive cells compared to nontargeting siRNA-transfected cell results (Fig. 1B and C), suggesting opposing roles of OAT1 and OAT3 during influenza A virus replication. Immunoblot analyses were performed to verify OAT3 protein expression following nontargeting (NT), OAT3, or OAT1 siRNA transfections (Fig. 1D and E). Transfection of OAT3 siRNA, but not nontargeting or OAT1 siRNA, resulted in a 61% reduction of OAT3 protein expression, indicating efficiency and specificity of OAT3 protein expression silencing by siRNA transfection. Together, these results demonstrate the importance of OAT3 during influenza A virus replication. Using gene pathway analysis, it was hypothesized that targeting OAT3 using its classical pharmacological inhibitor, probenecid, may limit influenza A virus replication.
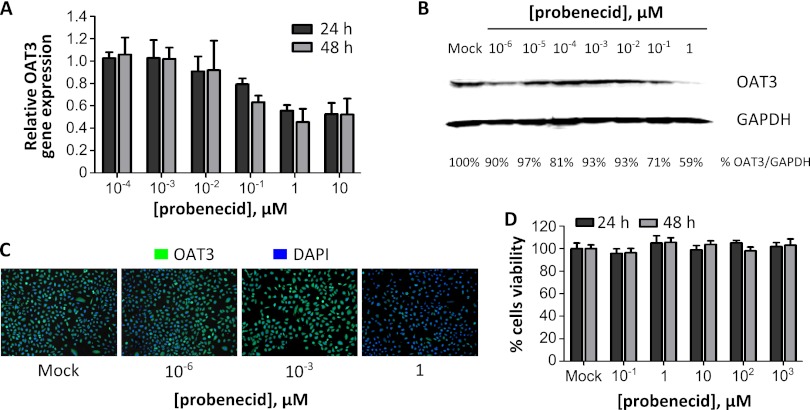
Probenecid reduces OAT3 mRNA and protein levels in a dose-dependent manner.
To examine the mechanism by which probenecid may affect OAT3 function, A549 cells were treated with increasing doses of probenecid, and OAT3 mRNA and protein levels were determined at 24 and 48 h following treatment. Twenty-four hours of probenecid treatment at a concentration of 1 μM or higher resulted in a >40% reduction in the OAT3 mRNA level as measured by qRT-PCR (Fig. 2A). A similar reduction was observed in cells treated with 0.1 μM probenecid for 48 h. Likewise, a 41% reduction of OAT3 protein expression was observed in cells treated with 1 μM probenecid for 24 h as assessed by immunoblotting (Fig. 2B) and a reduction of OAT3 staining was observed by immunofluorescence microscopy (Fig. 2C). Cytotoxicity assays were performed using A549 cell culture supernatant treated with increasing doses of probenecid for 24 or 48 h. Probenecid had a low level of cytotoxicity even when used at higher (1 mM) concentrations (Fig. 2D). These results show that probenecid acts to reduce OAT3 mRNA and protein expression and has minimal cytotoxicity and show for the first time that human lung epithelial (A549) cells express both OAT3 mRNA and protein.
Fig 2.
The OAT inhibitor probenecid reduces OAT3 mRNA and protein levels in vitro in a dose-dependent manner. (A to C) A549 cells were treated with increasing doses of probenecid. At 24 and 48 h posttreatment, cells were fixed for immunostaining or harvested for total RNA and protein. (A) OAT3 mRNA levels were assessed using qRT-PCR and normalized to GAPDH. Fold changes of OAT3 expression were calculated relative to mock-treated cell results. (B) OAT3 and GAPDH protein levels following 24 h of probenecid treatment were evaluated by immunoblot analysis, and protein bands were quantified. OAT3 protein levels were normalized to GAPDH, and percent OAT3 protein levels were calculated relative to mock-treated cell results. (C) Cells were fixed at 24 h following probenecid treatment, stained for OAT3 (green) and nuclei (DAPI; blue), and visualized using fluorescence microscopy. (D) Probenecid demonstrated low cellular cytotoxicity as evaluated by a ToxiLight BioAssay kit.
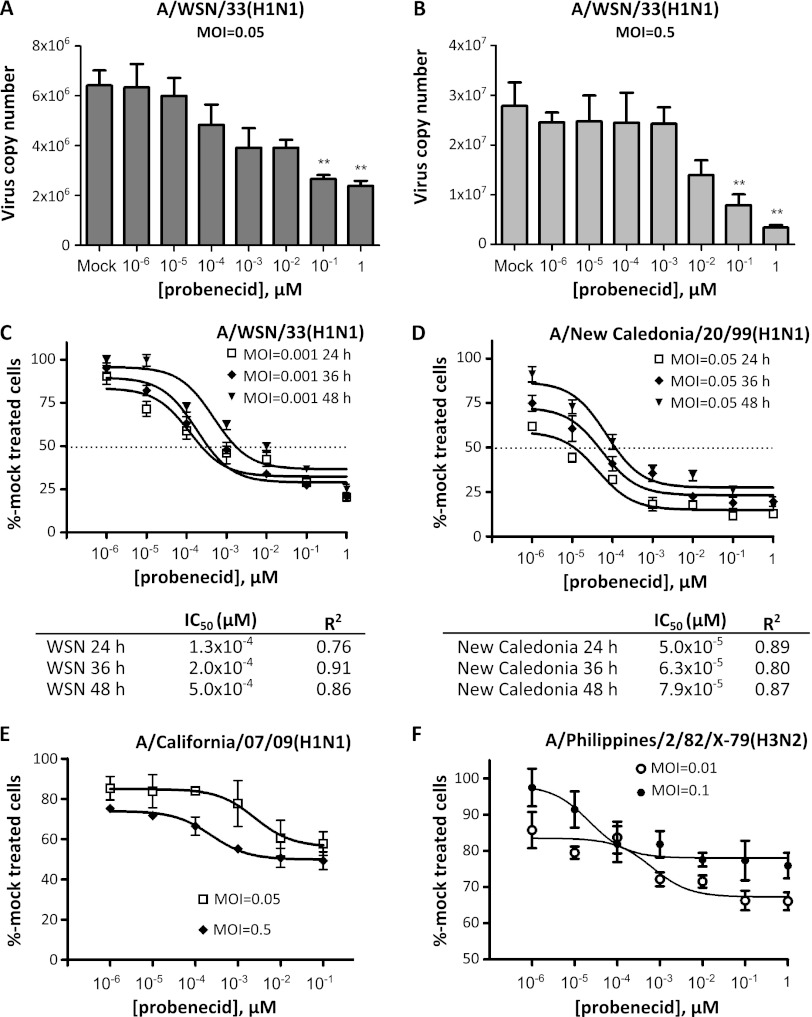
Probenecid reduces influenza A virus replication in vitro.
The effect of probenecid treatment on influenza A virus replication was evaluated in A549 cells pretreated with increasing doses of probenecid prior to infection with WSN at two different MOIs for 24 h. Pretreatment of cells with 10−1 μM probenecid significantly (P < 0.005) reduced virus copy numbers at both a low and a higher MOI of 0.05 and 0.5, respectively (Fig. 3A). A substantial reduction of virus copy numbers was also observed in cells infected with influenza A/New Caledonia/20/99 virus (H1N1; New Caledonia) (data not shown). As in Fig. 2A, probenecid treatment also reduced the OAT3 mRNA level in cells infected with WSN or New Caledonia virus (see Fig. S2 in the supplemental material). The percentage of NP-positive cells in probenecid-pretreated cells infected with WSN or New Caledonia virus for 24, 36, or 48 h was determined using a Cellomics ArrayScan immunofluorescence system and normalized to mock-treated cells to calculate the probenecid IC50 under each infection condition. It was determined that probenecid is effective in limiting influenza A virus replication in vitro at a IC50 ranging between 5.0 × 10−5 and 5.0 × 10−4 μM, depending on the influenza A virus strains and length of infection (Fig. 3C). To evaluate the effectiveness of probenecid in limiting replication of other influenza A virus strains, A549 cells were pretreated with increasing doses of probenecid and infected with influenza A/California/07/09(H1N1) (CA) (Fig. 3D) or A/Philippines/2/82/X-79(H3N2) (X-79) at a low and a higher MOI. In both cases, probenecid treatment reduced the percentage of NP-positive cells, although these reductions plateaued at 50% or higher versus mock-treated cells.
Fig 3.
The OAT inhibitor probenecid reduces influenza A virus replication in vitro in a dose-dependent manner. A549 cells were treated with increasing doses of probenecid for 24 h prior to infection with various strains of influenza A virus. (A and B) At 24 h following infection with influenza A/WSN/33(H1N1) virus at an MOI of 0.05 (A) or 0.5 (B), total RNA was collected and influenza A virus copy numbers were assessed using qRT-PCR. Graphs represent results from triplicate experiments, and error bars denote standard errors of the means. Statistical analyses were performed using one-way ANOVA. **, P < 0.005. (C to F) At 24, 36, or 48 h postinfection with A/WSN/33(H1N1) (C) or A/New Caledonia/20/99(H1N1) (D) or 24 h postinfection with A/California/07/09(H1N1) (E) or A/Philippines/2/82/X-79(H3N2) (F) at the indicated MOI, cells were fixed and stained for viral NP (green) and nuclei (DAPI; blue). Percent values of NP-positive cells were quantified using the Cellomics array scan system, normalized to mock treated cells, and plotted. Dose curves and IC50s (when indicated) were determined using nonlinear regression. Graphs represent results from six replicate experiments, and error bars denote standard errors of the means.
Probenecid reduces lung OAT3 expression and limits influenza A virus replication in vivo.
As probenecid was effective in limiting replication of several strains of influenza A virus in vitro, its effects were evaluated in mice. Two probenecid doses were tested in BALB/c mice prior to infection with WSN, New Caledonia, or CA virus at their respective LD50s (Fig. 4A). The levels of OAT3 mRNA were assessed in the lungs of mock- or influenza A virus-infected mice. Mice treated with 200 mg probenecid/kg of body weight at 24 h prior to infection with WSN (Fig. 4B), New Caledonia, or CA (data not shown) had a reduced lung OAT3 mRNA level as measured by qRT-PCR, with moderate decreases associated with lower doses of probenecid treatment. Importantly, there was a statistically significant reduction of lung virus load in mice pretreated with probenecid and infected with WSN, New Caledonia, or CA compared to PBS-treated mice as measured by qRT-PCR (Fig. 4C) and by evaluation of lung virus titers (TCID50) (Fig. 4D; P < 0.0001). Maximum reductions of lung virus load were evident when mice were treated with 200 mg probenecid/kg at 24 h prior to infection (P < 0.0001), although significant (P < 0.05) reductions were also observed with a lower dose of probenecid (10 mg/kg).
Fig 4.
Probenecid treatment reduces OAT3 expression and influenza A virus load in lungs of infected mice. (A to D) BALB/c mice were intraperitoneally (i.p.) mock treated or treated with probenecid at 10 or 200 mg/kg of body weight 24 h prior to intranasal inoculation with 70 PFU of influenza A/WSN/33(H1N1) virus, 22 PFU of influenza A/New Caledonia/20/99(H1N1) virus, or 35 PFU of influenza A/California/04/09(H1N1) virus. At 48 h postinfection, animals were sacrificed and their lungs were collected. (A) Experimental outline; 0 h on the x axis denotes the start of infection. (B) OAT3 gene expression in lungs of PBS- or probenecid-treated mice infected with influenza A/WSN/33(H1N1) virus was assessed using qRT-PCR and normalized to GAPDH. Fold gene expression levels were calculated relative to PBS-treated mouse levels. (C) Lung influenza A virus copy numbers per 5 ng of total RNA were evaluated using qRT-PCR and graphed. (D) Lung influenza A virus titers are represented as TCID50. Graphs represent results from 10 animals per experimental group, and error bars denote standard errors of the means. Dotted lines represent the limit of assay detection. *, P < 0.05; **, P < 0.001; ***, P < 0.0001.
Therapeutic treatment with probenecid effectively reduces influenza A lung virus burden and provides partial protection against lethal influenza A virus infection in vivo.
To compare different probenecid treatment regimens, mice were prophylactically (24 h preinfection) or therapeutically (24 h postinfection) administered probenecid at 25 mg/kg and challenged with mouse-adapted WSN. Mice treated pre- or post-WSN infection had significantly (P < 0.05) reduced lung virus titers compared to mice subjected to mock treatment (Fig. 5A). The ability of probenecid to protect mice against infection with WSN at a lethal dose (i.e., 10× LD50, or 2.2 × 103 PFU/mouse) was evaluated. Mice were treated prophylactically with 200 mg/kg of probenecid (24 h preinfection) or therapeutically with 200 mg/kg probenecid (24 h postinfection) or were administered 25 mg/kg probenecid daily for 3 days following infection (days 1, 2, and 3) and monitored for mortality or morbidity for the duration of the 14-day study. Mice treated daily over 3 days with 25 mg/kg probenecid following lethal infection were partially protected (60% survival; P < 0.05), whereas single-dose probenecid treatment given either prophylactically or therapeutically resulted in less protection, although the data were statistically nonsignificant (Fig. 5B). The reduced efficacy may relate to pharmacokinetics and drug decay over that time. These results indicate that probenecid can be utilized as a chemoprophylactic or chemotherapeutic treatment; however, multiple administrations of probenecid are required for ideal protection.
Fig 5.
Probenecid administered either prophylactically or therapeutically reduces influenza A virus load in lung of infected animals. (A) BALB/c mice were i.p. treated with probenecid at 25 mg/kg body weight at 24 h prior to (−24 h; prophylactic) or following (24 h; therapeutic) infection with 70 PFU of influenza A/WSN/33 virus. In the experimental outline, 0 h on the x axis denotes the start of infection. At 72 h postinfection, animals were sacrificed and lungs were collected to evaluate viral load by TCID50. Graphs represent results from 10 animals per experimental group, and error bars denote standard errors of the means. (B) BALB/c mice were subjected to a single-dose treatment with probenecid (at 200 mg/kg) i.p. 24 h prior to infection (prophylactic) or 24 h following infection (therapeutic) or to a three-dose treatment regimen (at 25 mg/kg each dose) on days 1, 2, and 3 postinfection with 2.2 × 103 PFU of influenza A/WSN/33 virus. Mice were monitored daily for 14 days for morbidity and mortality, and their survival was recorded and graphed. *, P < 0.05.
Since probenecid coadministration with oseltamivir has been reported to result in elevated oseltamivir carboxylate serum levels due to perturbation of OAT (19), a combinational probenecid/oseltamivir treatment regimen was evaluated in vivo. Mice infected with WSN or New Caledonia virus were treated 24 h later with different single or combinational probenecid and/or oseltamivir regimens once or twice daily for 72 h (see Fig. S3 in the supplemental material). All treatment conditions evaluated resulted in a significant (P < 0.0001) reduction of lung virus load. Treatment with a probenecid/oseltamivir combination twice daily better reduced New Caledonia lung virus titers than the recommended oseltamivir treatment for humans, which is a twice-daily regimen (see Fig. S3C in the supplemental material; P = 0.0003) (6). This regimen also appeared to better to limit WSN infection, although the findings were not statistically significant (see Fig. S3B in the supplemental material; P = 0.115). Collectively, these data feature probenecid as a new candidate for drug positioning for a novel class of anti-influenza A therapeutics.
DISCUSSION
Despite the availability of two different classes of approved anti-influenza drugs and the availability of vaccines, influenza A virus infection remains a major worldwide concern due to significant morbidity, mortality, and pandemic and epidemic potential. The occurrence of influenza A virus strains resistant to approved anti-influenza chemotherapeutics is a significant concern. Furthermore, there are limited numbers of novel anti-influenza A drugs under development, highlighting the need for anti-influenza drug discovery.
In this study, a siRNA screen was performed using a host drug target siRNA library in A549 cells infected with influenza A/WSN/33 virus. The screen and subsequent validation confirmed that a member of the organic anion transporter (OAT) family, OAT3, was important for influenza A virus replication. While it remains unknown how silencing OAT3 expression specifically decreases influenza A virus replication, it is hypothesized that transport of a particular host or virus factor through OAT3 is required during the course of infection. As siRNA silencing of OAT3 results in a complete block of influenza virus NP expression, it is likely that that this event occurs early during infection, i.e., before viral protein synthesis takes place. Further studies are needed to identify the OAT3 substrate that is required for influenza A virus infection.
Interestingly, this study showed that OAT3 functions to specifically facilitate influenza A virus replication among members of the OAT family, as siRNA transfection targeting other OAT members such as OAT1, −2, -4, and -7 and URAT1 did not result in substantial reduction of influenza A virus replication. All genes, with the exception of URAT1, appear to have roles opposite that of OAT3 during influenza A virus replication. Transporters of the OAT family are polyspecific such that they can transport multiple substrates and can translocate anions in either direction (12). Therefore, it is possible that OAT3 specificity may be explained in part by the directionality of a particular substrate or protein involved during influenza A replication, particularly during transport across cellular membranes. A known example of this directionality of transport is the urate reabsorption carried out by OAT1, OAT3, OAT4, and URAT1. OAT4 and URAT1 located at the luminal side of human kidney proximal tubule mediate reabsorption of urate into the proximal tubule cells. Conversely, OAT1 and OAT3 which are located on the basolateral surface mediate urate transport from the proximal tubule cells into plasma (12, 15, 28). Modulation of urate transport by OATs has been utilized for treatment of gout, a condition that is caused by accumulation of uric acid. Probenecid, a chemical inhibitor for OAT transport, is currently used to promote uricosuria, or the release of urate through urinal excretion. Note, however, that probenecid is a nonspecific inhibitor of OATs, as it blocks transport by OAT1, OAT3, and URAT1, although it yields net excretion of urate. To understand the role of OAT3 during influenza A virus infection, future studies are planned to examine localization of OAT3 on the surface of polarized lung epithelial cells, in addition to identification of the substrate required during infection.
In this study, the anti-influenza A virus properties of probenecid were shown. In vitro, probenecid pretreatment of A549 cells resulted in a dose-dependent reduction of virus titer upon infections with several influenza A virus strains, A/WSN/33(H1N1), A/New Caledonia/20/99, and, to a lesser extent, A/California/07/09(H1N1) and A/Philippines/2/82/X-79 (Fig. 3). The IC50 of probenecid against influenza A infection in vitro for WSN and New Caledonia ranged from 5 × 10−5 to 5 × 10−4 μM. Additionally, it was shown that probenecid reduced mRNA and protein levels of OAT3, which suggests that probenecid blocks OAT-mediated transport by regulating the expression of transporter molecules itself (Fig. 2; see also Fig. S2 in the supplemental material). Probenecid administered prophylactically prior to influenza A virus infection resulted in reduced lung virus titers in vivo (Fig. 4 and 5). Likewise, probenecid given therapeutically at 24 h following influenza A virus infection also resulted in reduced lung virus titers, demonstrating the versatility of probenecid as an influenza A virus chemotherapeutic (Fig. 5; see also Fig. S3 in the supplemental material). Future studies should evaluate the efficacy of probenecid against other influenza A virus strains and determine optimal regimens for treatment of infections by a broad range of influenza A virus strains in vivo.
In addition to its direct anti-influenza A activity, probenecid has also been reported to maintain the plasma level of oseltamivir over a longer period of time (19, 21, 29, 30). Oseltamivir has been reported in several studies as a substrate for OAT1 and OAT3 (20, 31); therefore, blocking of OAT1/OAT3 transport with probenecid is thought to prevent oseltamivir renal excretion. For this reason, probenecid administration in conjunction with oseltamivir has previously been suggested (29, 32, 33). However, these studies were done in healthy individuals and are based on measurements of oseltamivir active as a metabolite in the plasma; thus, the efficacy of the oseltamivir-probenecid combinational therapy for treatment of influenza A virus infection has not been previously evaluated. In this study, combinational oseltamivir/probenecid therapy produced an anti-influenza A virus effect that was slightly higher than that seen with oseltamivir treatment alone using a mouse model.
Lastly, this study also identified host drug target genes, including many members of the OAT family such as OAT1, OAT2, OAT4, and OAT7, whose silencing led to increased influenza A virus replication. This suggests that they have a role in limiting influenza A virus replication; thus, silencing their expression or preventing their function with small-molecule chemical inhibitors could be useful in applications where amplification of virus replication is beneficial such as in the propagation of viruses during vaccine production. It also highlighted the breadth of new information obtained from siRNA screens performed during influenza A virus infection, both in the study reported here and in those done by others (24, 25, 34–39).
Supplementary Material
ACKNOWLEDGMENTS
We thank Jon Karpilow from the RNAi Global team for his technical expertise, Kate Oakley for critical reading of the manuscript, and the Georgia Research Alliance for supporting aspects of the study.
This study was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (HHSN266200700006C).
Footnotes
Published ahead of print 5 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01532-12.
REFERENCES
- 1. Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. 2003. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289:179–186 [DOI] [PubMed] [Google Scholar]
- 2. Simonsen L, Reichert TA, Viboud C, Blackwelder WC, Taylor RJ, Miller MA. 2005. Impact of influenza vaccination on seasonal mortality in the US elderly population. Arch. Intern. Med. 165:265–272 [DOI] [PubMed] [Google Scholar]
- 3. Kandel R, Hartshorn KL. 2005. Novel strategies for prevention and treatment of influenza. Expert Opin. Ther. Targets 9:1–22 [DOI] [PubMed] [Google Scholar]
- 4. Beigel J, Bray M. 2008. Current and future antiviral therapy of severe seasonal and avian influenza. Antiviral Res. 78:91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dreitlein WB, Maratos J, Brocavich J. 2001. Zanamivir and oseltamivir: two new options for the treatment and prevention of influenza. Clin. Ther. 23:327–355 [DOI] [PubMed] [Google Scholar]
- 6. Harper SA, Bradley JS, Englund JA, File TM, Gravenstein S, Hayden FG, McGeer AJ, Neuzil KM, Pavia AT, Tapper ML, Uyeki TM, Zimmerman RK. 2009. Seasonal influenza in adults and children—diagnosis, treatment, chemoprophylaxis, and institutional outbreak management: clinical practice guidelines of the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1003–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beigel JH. 2010. Antiviral compounds in the pipeline to tackle H1N1 influenza infection. Drugs Future 35:385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clouser CL, Patterson SE, Mansky LM. 2010. Exploiting drug repositioning for discovery of a novel HIV combination therapy. J. Virol. 84:9301–9309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goldenberg MM. 1998. Safety and efficacy of sildenafil citrate in the treatment of male erectile dysfunction. Clin. Ther. 20:1033–1048 [DOI] [PubMed] [Google Scholar]
- 10. Russ AP, Lampel S. 2005. The druggable genome: an update. Drug Discov. Today 10:1607–1610 [DOI] [PubMed] [Google Scholar]
- 11. Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, Bruford EA. 2004. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins. Pflugers Archiv. 447:465–468 [DOI] [PubMed] [Google Scholar]
- 12. Koepsell H, Endou H. 2004. The SLC22 drug transporter family. Pflugers Archiv. 447:666–676 [DOI] [PubMed] [Google Scholar]
- 13. Ahn SY, Nigam SK. 2009. Toward a systems level understanding of organic anion and other multispecific drug transporters: a remote sensing and signaling hypothesis. Mol. Pharmacol. 76:481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dantzler WH, Evans KK, Wright SH. 1995. Kinetics of interactions of para-aminohippurate, probenecid, cysteine conjugates and N-acetyl cysteine conjugates with basolateral organic anion transporter in isolated rabbit proximal renal tubules. J. Pharmacol. Exp. Ther. 272:663–672 [PubMed] [Google Scholar]
- 15. Stamp LK, O'Donnell JL, Chapman PT. 2007. Emerging therapies in the long-term management of hyperuricaemia and gout. Intern. Med. J. 37:258–266 [DOI] [PubMed] [Google Scholar]
- 16. Barza M, Brusch J, Bergeron MG, Kemmotsu O, Weinstein L. 1975. Extraction of antibiotics from the circulation by liver and kidney: effect of probenecid. J. Infect. Dis. 131:S86–S97 [DOI] [PubMed] [Google Scholar]
- 17. Cunningham RF, Israili ZH, Dayton PG. 1981. Clinical pharmacokinetics of probenecid. Clin. Pharmacokinet. 6:135–151 [DOI] [PubMed] [Google Scholar]
- 18. Laskin OL, de Miranda P, King DH, Page DA, Longstreth JA, Rocco L, Lietman PS. 1982. Effects of probenecid on the pharmacokinetics and elimination of acyclovir in humans. Antimicrob. Agents Chemother. 21:804–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rayner CR, Chanu P, Gieschke R, Boak LM, Jonsson EN. 2008. Population pharmacokinetics of oseltamivir when coadministered with probenecid. J. Clin. Pharmacol. 48:935–947 [DOI] [PubMed] [Google Scholar]
- 20. Hill G, Cihlar T, Oo C, Ho ES, Prior K, Wiltshire H, Barrett J, Liu B, Ward P. 2002. The anti-influenza drug oseltamivir exhibits low potential to induce pharmacokinetic drug interactions via renal secretion—correlation of in vivo and in vitro studies. Drug Metab. Dispos. 30:13–19 [DOI] [PubMed] [Google Scholar]
- 21. Holodniy M, Penzak SR, Straight TM, Davey RT, Lee KK, Goetz MB, Raisch DW, Cunningham F, Lin ET, Olivo N, Deyton LR. 2008. Pharmacokinetics and tolerability of oseltamivir combined with probenecid. Antimicrob. Agents Chemother. 52:3013–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 27:493–497 [Google Scholar]
- 23. Woolcock PR. 2008. Avian influenza virus isolation and propagation in chicken eggs. Methods Mol. Biol. 436:35–46 [DOI] [PubMed] [Google Scholar]
- 24. König R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, Garcia-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK. 2010. Human host factors required for influenza virus replication. Nature 463:813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meliopoulos VA, Andersen LE, Brooks P, Yan X, Bakre A, Coleman JK, Tompkins SM, Tripp RA. 2012. MicroRNA regulation of human protease genes essential for influenza virus replication. PLoS One 7:e37169 doi:10.1371/journal.pone.0037169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pleschka S, Wolff T, Ehrhardt C, Hobom G, Planz O, Rapp UR, Ludwig S. 2001. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat. Cell Biol. 3:301–305 [DOI] [PubMed] [Google Scholar]
- 27. Szretter KJ, Balish AL, Katz JM. 2006. Influenza: propagation, quantification, and storage. Curr. Protoc. Microbiol. 3:15G.1.1–15G.1.22 [DOI] [PubMed] [Google Scholar]
- 28. Burns CM, Wortmann RL. 2011. Gout therapeutics: new drugs for an old disease. Lancet 377:165–177 [DOI] [PubMed] [Google Scholar]
- 29. He G, Massarella J, Ward P. 1999. Clinical pharmacokinetics of the prodrug oseltamivir and its active metabolite Ro 64-0802. Clin. Pharmacokinet. 37:471–484 [DOI] [PubMed] [Google Scholar]
- 30. Wattanagoon Y, Stepniewska K, Lindegardh N, Pukrittayakamee S, Silachamroon U, Piyaphanee W, Singtoroj T, Hanpithakpong W, Davies G, Tarning J, Pongtavornpinyo W, Fukuda C, Singhasivanon P, Day NP, White NJ. 2009. Pharmacokinetics of high-dose oseltamivir in healthy volunteers. Antimicrob. Agents Chemother. 53:945–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ose A, Ito M, Kusuhara H, Yamatsugu K, Kanai M, Shibasaki M, Hosokawa M, Schuetz JD, Sugiyama Y. 2009. Limited brain distribution of [3R,4R,5S]-4-acetamido-5-amino-3-(1-ethylpropoxy)-1-cyclohexene-1-carboxylate phosphate (Ro 64-0802), a pharmacologically active form of oseltamivir, by active efflux across the blood-brain barrier mediated by organic anion transporter 3 (Oat3/Slc22a8) and multidrug resistance-associated protein 4 (Mrp4/Abcc4). Drug Metab. Dispos. 37:315–321 [DOI] [PubMed] [Google Scholar]
- 32. Butler D. 2005. Wartime tactic doubles power of scarce bird-flu drug. Nature 438:6. [DOI] [PubMed] [Google Scholar]
- 33. Howton JC. 2006. Probenecid with oseltamivir for human influenza A (H5N1) virus infection? N. Engl. J. Med. 354:879–880 [DOI] [PubMed] [Google Scholar]
- 34. Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Karlas A, Machuy N, Shin Y, Pleissner K-P, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822 [DOI] [PubMed] [Google Scholar]
- 37. Meliopoulos VA, Andersen LE, Birrer KF, Simpson KJ, Lowenthal JW, Bean AG, Stambas J, Stewart CR, Tompkins SM, van Beusechem VW, Fraser I, Mhlanga M, Barichievy S, Smith Q, Leake D, Karpilow J, Buck A, Jona G, Tripp RA. 2012. Host gene targets for novel influenza therapies elucidated by high-throughput RNA interference screens. FASEB J. 26:1372–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shapira SD, Gat-Viks I, Shum BOV, Dricot A, DE Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sui B, Bamba D, Weng K, Ung H, Chang S, Van Dyke J, Goldblatt M, Duan R, Kinch MS, Li W-B. 2009. The use of random homozygous gene perturbation to identify novel host-oriented targets for influenza. Virology 387:473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.