Abstract
Multidrug-resistant Escherichia coli strains belonging to a single lineage frequently account for a large proportion of extraintestinal E. coli infections in many parts of the world. However, limited information exists on the community prevalence and clonal composition of drug-susceptible E. coli strains. Between July 2007 and September 2010, we analyzed all consecutively collected Gram-negative bacterial isolates from patients with bloodstream infection (BSI) admitted to a public hospital in San Francisco for drug susceptibility and associated drug resistance genes. The E. coli isolates were genotyped for fimH single nucleotide polymorphisms (SNPs) and multilocus sequence types (MLSTs). Among 539 isolates, E. coli accounted for 249 (46%); 74 (30%) of them were susceptible to all tested drugs, and 129 (52%) were multidrug resistant (MDR). Only five MLST genotypes accounted for two-thirds of the E. coli isolates; the most common were ST131 (23%) and ST95 (18%). Forty-seven (92%) of 51 ST131 isolates, as opposed to only 8 (20%) of 40 ST95 isolates, were MDR (P < 0.0001). The Simpson's diversity index for drug-susceptible ST genotypes was 87%, while the index for MDR ST genotypes was 81%. ST95 strains were comprised of four fimH types, and one of these (f-6) accounted for 67% of the 21 susceptible isolates (P < 0.003). A large proportion (>70%) of both MDR and susceptible E. coli BSI isolates represented community-onset infections. These observations show that factors other than the selective pressures of antimicrobial agents used in hospitals contribute to community-onset extraintestinal infections caused by clonal groups of E. coli regardless of their drug resistance.
INTRODUCTION
Recently, much attention has been given to the worldwide dissemination of extraintestinal pathogenic Escherichia coli (ExPEC) strains belonging to a related lineage, such as the drug-resistant E. coli 025b:H4 multilocus sequence type (MLST) ST131 (1–7). Retrospective studies have shown that from as early as 2000, multidrug-resistant (MDR) ST131 strains were implicated in a large proportion of community-acquired urinary tract infections (UTI) and bloodstream infections (BSI) in different regions of the world (5, 6, 8–10). This global dispersion of ST131 has been suggested to be mediated by activities such as global food trade, international travel, and other modes of transmission (5, 6, 10, 11).
There are other ExPEC strains belonging to a limited number of lineages that circulate globally (2, 4–7, 12, 13). However, most published reports on these lineages have focused on drug-resistant strains. The epidemiologic observation that certain drug-resistant E. coli lineages predominate in community and institutional settings is unexplained. One explanation may be that most studies focus on drug-resistant infections and that such study results are more likely to be reported. Another explanation is that drug-resistant strains have selective advantage because of their drug resistance. Finally, it is possible that they become predominant because of epidemiologic or biologic factors unrelated to drug resistance. We reasoned that if drug-susceptible E. coli strains are also found to exhibit clonal distribution in community settings, this would indicate that factors other than drug resistance contribute to the clonal spread of ExPEC. This study was undertaken to compare the clonal compositions of drug-resistant and -susceptible E. coli clinical isolates from patients with BSI who were treated at a large public hospital in San Francisco.
MATERIALS AND METHODS
Strain collection.
The clinical microbiology laboratory of San Francisco General Hospital (SFGH) is one of the largest in the Bay Area, and the cultures originate from all the hospital wards and jail clinics and San Francisco's city outpatient clinics. All Gram-negative bacillus (GNB) isolates from BSI at the clinical microbiology laboratory of SFGH are stored on agar slants for up to 3 months. Every 3 months, these isolates were provided to us for analysis. We examined all consecutively collected GNB BSI isolates from inpatients admitted to SFGH between July 2007 and September 2010. This study was approved by the University of California, San Francisco, Committee on Human Research.
The information on the dates of admission and first blood culture that yielded the GNB pathogen was available for most of the isolates. In this study, we considered BSI to be community onset if the time period between the date of admission and the date of the first blood culture that grew a GNB pathogen was ≤48 h.
Strain identification and susceptibility testing.
At SFGH, the BSI isolates were identified to the species level biochemically with API 20E (bioMérieux, Durham, NC) for fermenters or API 20NE for nonenteric bacteria. Antimicrobial susceptibility tests were performed by a MicroScan WalkAway Gram-negative panel (Dade Behring/Siemens USA, Deerfield, IL). This panel included the following 11 classes of antimicrobial agents (the drugs tested within each): aminoglycosides (amikacin, gentamicin, tobramycin), aminopenicillins (ampicillin), β-lactamase inhibitor combinations (ampicillin-sulbactam, piperacillin-tazobactam, ticarcillin-clavulanic acid), broad-spectrum 1st-generation cephalosporins (cefazolin), broad-spectrum 2nd-generation cephalosporins (cefotetan, cefuroxime, cefoxitin), extended-spectrum 3rd-generation cephalosporins (cefotaxime, ceftriaxone, ceftazidime), extended-spectrum 4th-generation cephalosporins (cefepime), fluoroquinolones (ciprofloxacin, levofloxacin, moxifloxacin), folate path inhibitors (trimethoprim-sulfamethoxazole), monobactams (aztreonam), and carbapenems (ertapenem, imipenem, meropenem). E. coli multidrug resistance (MDR) was defined as resistance to one or more agents in three or more classes of tested drugs (8, 14). Intermediate susceptibility was classified as resistant. Etest (bioMérieux, Marcy l'Etoile, France) was used to confirm susceptibility to carbapenems for select isolates.
DNA extraction and PCR amplification for gene detection.
The bacterial DNA was extracted by a freeze-boil method, and PCR amplification was carried out as previously described (15). Primer sequences for PCR analysis of fimH genotyping, β-lactamase genes, class 1 integron, and associated gene cassettes were used as previously described (Table 1).
Table 1.
PCR primers used in this study
| Primer target | Primer namea | Sequence (5′–3′) | Expected product size (bp) | Reference |
|---|---|---|---|---|
| Class 1 integrase | 201F | CCTCCCGCACGATGATC | 280 | 1 |
| 202R | TCCACGCATCGTCAGGC | |||
| Class 1 gene cassette, variable region | 5′ CS | GGCATCCAAGCAGCAAG | Variable | 25 |
| 3′ CS | AAGCAGACTTGACCTGA | |||
| fimH SNP | fimH F | TCGAGAACGGATAAGCCGTGG | 506 | 49 |
| fimH R | GCAGTCACCTGCCCTCCGGTA | |||
| 16S ribosomal DNA, positive control | 16S8F | AGAGTTTGATCCTGGCTCAG | 800 | 1 |
| 16S8R | GGACTACCAGGGTATCTAATCC | |||
| ESBL multiplex | ||||
| TEM variants, including TEM-1 and -2 | MultiTSO-T_for | CATTTCCGTGTCGCCCTTATTC | 800 | 7 |
| MultiTSO-T_rev | CGTTCATCCATAGTTGCCTGAC | |||
| SHV variants, including SHV-1 | MultiTSO-S_for | AGCCGCTTGAGCAAATTAAAC | 713 | 7 |
| MultiTSO-S_rev | ATCCCGCAGATAAATCACCAC | |||
| OXA-1, -4, and -30 | MultiTSO-O_for | GGCACCAGATTCAACTTTCAAG | 564 | 7 |
| MultiTSO-O_rev | GACCCCAAGTTTCCTGTAAGTG | |||
| CTX-M universal | CTX-M Univ F | TTTGCGATGTGCAGTACCAGTAA | 500 | 45 |
| CTX-M Univ R | CTCCGCTGCCGGTTTTATC | |||
| CTX-M families | CTX-M-1-all F | ATGGTTAAAAAATCACTGCG | 876 | 45 |
| CTX-M-1-all R | TTACAAACCGTCGGTGACGAT | |||
| CTX-M-9-F | GCAGATAATACGCAGGTG | 392 | 45 | |
| CTX-M-9-R | CGGCGTGGTGGTGTCTCT | |||
| CTX-M multiplex | ||||
| Variants including CTX-M-1, -3, and -15 | MultiCTXMGp1_for | TTAGGAARTGTGCCGCTGYA | 688 | 7 |
| MultiCTXMGp1_rev | CGATATCGTTGGTGGTRCCAT | |||
| Variants including CTX-M-2 | MultiCTXMGp2_for | CGTTAACGGCACGATGAC | 404 | 7 |
| MultiCTXMGp2_rev | CGATATCGTTGGTGGTRCCAT | |||
| Variants including CTX-M-9 and -14 | MultiCTXMGp9_for | TCAAGCCTGCCGATCTGGT | 561 | 7 |
| MultiCTXMGp9_rev | TGATTCTCGCCGCTGAAG | |||
| Variants including CTX-M-8, -25, -26, -39, and -41 | MultiCTXMGp8_for | AACRCRCAGACGCTCTAC | 326 | 7 |
| MultiCTXMGp8_rev | TCGAGCCGGAASGTGTYAT | |||
| pabB | pabB F | TCCAGCAGGTGCTGGATCGT | 347 | 3 |
| pabB R | GCGAAATTTTTCGCCGTACTGT |
F, forward primer; R, reverse primer.
Sequence analysis.
A 5-μl aliquot of each PCR product that appeared as a unique band when visualized by gel electrophoresis was cleaned with 2 μl of a 1:10 dilution of ExoSAP-IT (Affymetrix, Inc., Santa Clara, CA) and subjected to direct sequencing. Sequencing was done on an Applied Biosystems 3730 DNA analyzer (Applied Biosystems, Foster City, CA) at the University of California, Berkeley, DNA Sequencing Facility. The DNA sequences were visually inspected, edited, and assembled with BioEdit version 7.0.1. ClustalW was used to perform multiple alignment analyses of the sequences. Sequences were compared with those deposited in the National Center for Biotechnology Information (NCBI) database by an updated version of the BLAST program.
Genotyping of E. coli strains.
We genotyped E. coli isolates by two methods. One was a single nucleotide polymorphism (SNP) analysis of the fimH locus that encodes a mannose-binding subunit protein located at the tip of type 1 fimbriae, performed according to a previously published report (17). Designation of fimH allele types followed the classification system established by Dias et al. (21); the SNP analysis of fimH can further discriminate strains genotyped by multilocus sequence typing (MLST). The other method was MLST based on a protocol published on the University College Cork website (http://mlst.ucc.ie/mlst/). The MLST analysis is based on sequence comparison of a portion of the following seven housekeeping genes: adk, fumC, recA, mdh, purA, gyrB, and icd.
Allele sequences of these genes were submitted to the MLST database curator for sequence type (ST) and sequence complex assignment. Sequence complexes include STs that differ from each other by no more than two of the seven loci, as defined by Wirth et al. (22). Strains that were genotyped as ST131 were based on all seven MLST alleles if all of the targets yielded interpretable sequences or a combination of at least three MLST alleles and ST131 allele-specific PCR for the pabB gene as established by Clermont et al. (20).
Statistics.
Categorical variables were compared by a chi-square or Fisher exact test (2-tailed). Genotype diversity was analyzed by Simpson's diversity index (23).
Nucleotide sequence accession numbers.
The fimH gene sequences from this study that were found to be unique (partial coding sequence [CDS] corresponding to the bp positions 360 to 781 of the E. coli strain K-12 sequence [GenBank accession number GQ487190]) were deposited in GenBank under accession numbers KC020174 to KC020181.
RESULTS
Bacterial species identified from patients with BSI.
Of 539 Gram-negative bacterial BSI isolates collected between July 2007 and September 2010, five species comprised 76% of the population. E. coli was the most frequent species with 249 (46%) isolates, followed by 70 (13.0%) Klebsiella pneumoniae isolates, 35 (7%) Proteus mirabilis isolates, 31 (6%) Pseudomonas aeruginosa isolates, and 26 (5%) Enterobacter cloacae isolates. This study focuses on the E. coli BSI isolates.
Antimicrobial drug resistance.
Susceptibility results for 11 classes of antimicrobial agents were available for 246 (99%) E. coli isolates. The following results are based on these 246 isolates. Among these isolates, 74 (30%) were susceptible to all tested drugs, 43 (17%) were resistant to one or two classes, and 129 (52%) were MDR. Based on the number of discharges from the medical and surgical services of the study hospital in the study period of 2007 to 2010 (average of 12,200/year), the incidence (per 1,000 discharges/year) was estimated to be 6.1 for E. coli BSI, 1.9 for drug-susceptible E. coli BSI, and 3.2 for MDR E. coli BSI. The susceptibility results for isolates in the study period were reported prior to the 2010 Clinical and Laboratory Standards Institute (CLSI) antimicrobial susceptibility breakpoint changes for carbapenems, cephalosporins, and aztreonam. All isolates were susceptible to ertapenem, imipenem, and meropenem, 29 (12%) were resistant to aztreonam, 108 (44%) were resistant to trimethoprim-sulfamethoxazole, 66 (27%) were resistant to fluoroquinolones, 29 (12%) were resistant to cefotaxime, ceftazidime, and ceftriaxone, and 24 (10%) were resistant to cefepime (Table 2). All 25 CTX-M gene-positive isolates were MDR, comprising 19% of the 129 MDR E. coli isolates, and were significantly more likely to be MDR than other drug-resistant strains (P < 0.0001).
Table 2.
Antimicrobial resistance and selected β-lactamase genes identified among BSI E. coli isolates
| ST (total no. of isolates) | No. (%) of isolates resistant to each classa |
blaCTX-M type(s) (no. of isolates) | No. of isolates with: |
blaKPC type (no. of isolates) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| QUIN | SXT | CTX | FEP | ATM | CARB | blaTEM | blaOXA-1 | |||
| ST131 (51) | 46 (90) | 36 (71) | 17 (33) | 15 (29) | 16 (31) | 0 (0) | -15 (13), -1 (3), -14 (2) | 3 | 14 | |
| ST95 (40) | 1 (3) | 7 (18) | 1 (3) | 1 (3) | 2 (5) | 0 (0) | -14 (1) | 0 | 0 | -2 (1) |
| ST73 complex (20) | 0 (0) | 7 (35) | 1 (5) | 0 (0) | 1 (5) | 0 (0) | ||||
| ST69 (19) | 2 (11) | 12 (63) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||||
| ST12 complex (13) | 0 (0) | 9 (69) | 3 (23) | 3 (23) | 3 (23) | 0 (0) | -15 (1b), -14 (2b) | 3 | 0 | -2 or -3 (1) |
| ST10 complex (9) | 1 (11) | 6 (67) | 1 (11) | 1 (11) | 1 (11) | 0 (0) | -15 (1) | 1 | 0 | |
| ST1576 (2) | 2 (100) | 0 (0) | 2 (100) | 1 (50) | 2 (100) | 0 (0) | -15 (1), -14 (1) | 0 | 0 | |
| ST224 (1) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 0 | 0 | ||
| ST624 (1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | -14 (1) | 0 | 1 | |
| ST405 complex (4) | 1 (25) | 3 (75) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||||
| ST998 (1) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 0 | 0 | ||
| ST964 (1) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 | 0 | ||
| ST38 complex (4) | 1 (100) | 4 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 | 0 | ||
| Other cluster ST (39)c | 6 (15) | 10 (26) | 1 (3) | 1 (3) | 1 (3) | 0 (0) | ||||
| Unique ST (15)c | 0 (0) | 1 (7) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||||
| No ST (26) | 3 (12) | 10 (38) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||||
| All (246) | 66 (27) | 108 (44) | 29 (12) | 24 (10) | 29 (12) | 0 (0) | 26b | 8 | 15 | 2 |
QUIN, fluoroquinolones; SXT, trimethoprim-sulfamethoxazole; CTX, broad-spectrum cephalosporins (including cefotaxime, ceftazidime, and ceftriaxone); FEP, cefepime; ATM, aztreonam; CARB, carbapenem.
One strain had both blaCTX-M-14 and blaCTX-M-15.
Cluster ST, 2 or more isolates; unique ST, 1 isolate per group.
Identification of genotypes by MLST and fimH sequence analysis.
MLST was done to genotype 220 (89%) isolates, which fell into 41 distinct STs or ST complexes. MLST results were not able to be obtained for 26 (11%) isolates due to repeated failure of PCR amplification or interpretable sequence results of an insufficient number of the seven alleles to place them in a ST complex. Only five STs accounted for 143 (65%) of 220 typed E. coli BSI isolates (Table 3). ST131 comprised the largest ST cluster with 51 (23%) isolates, followed by ST95 identified in 40 (18%) isolates. Others included the ST73 complex with 20 (8%) isolates, ST69 with 19 (9%), and the ST12 complex with 13 (6%).
Table 3.
Distribution of E. coli genotypes by MLST
| MLST | No. (%) of isolates that are: |
P valuec | Total no. (%) of isolates typedd | fimH type(s) | Dominant f-type (no. in each type [% of total]) | ||
|---|---|---|---|---|---|---|---|
| Susceptible | Resistanta | MDRb | |||||
| ST131 | 0 (0) | 4 (8) | 47 (92) | <0.0005 | 51 (23) | f-1, -6, -9, -11, -17 | f-9 (44 [86]) |
| ST95 | 21 (53) | 11 (27) | 8 (20) | 0.001 | 40 (18) | f-1, -6, -9, -47 | f-6 (17[43]), f-1 (16[40]) |
| ST73 complex | 6 (30) | 7 (35) | 7 (35) | 0.806 | 20 (8) | f-3, -8, -9, -60 | f-8 (12[52]) |
| ST69 | 3 (16) | 4 (21) | 12 (63) | 0.198 | 19 (9) | f-1 | f-1 (19[100]) |
| ST12 complex | 1 (8) | 1 (8) | 11 (84) | 0.116 | 13 (6) | f-1, -6, -51, -52 | f-1 (6[46]) |
| ST10 complex | 2 (22) | 1 (11) | 6 (67) | 1.0 | 9 (4) | f-1, -4, -6, -20, -28 | f-6 (3[33]) |
| ST38 complex | 0 (0) | 0 (0) | 4 (100) | 0.319 | 4 (2) | f-12, -57 | f-12 (3[75]) |
| ST405 complex | 0 (0) | 0 (0) | 4 (100) | 0.319 | 4 (2) | f-1, -4 | f-1 (2[50]) |
| ST127 | 2 (67) | 1 (33) | 0 (0) | 0.216 | 3 (1) | f-2 | f-2 (3[100]) |
| ST1278 | 0 (0) | 3 (50) | 3 (50) | 0.182 | 6 (2) | f-9 | f-9 (6[100]) |
| ST14 complex | 0 (0) | 2 (67) | 1 (33) | 0.556 | 3 (1) | f-1, -9 | f-1 (2[67]) |
| ST144 | 2 (67) | 0 (0) | 1 (33) | 0.216 | 3 (1) | f-4 | f-4 (3[100]) |
| ST155 complex | 3 (100) | 0 (0) | 0 (0) | 0.026 | 3 (1) | f-7 | f-7 (3[100]) |
| ST23 complex | 1 (33) | 1 (33) | 1 (33) | 1.0 | 3 (1) | f-1, -6 | f-6 (2[67]) |
| ST31 complex | 0 (0) | 1 (25) | 3 (75) | 0.556 | 4 (2) | f-4 | f-4 (4[100]) |
| ST372 | 3 (100) | 0 (0) | 0 (0) | 0.026 | 3 (1) | f-3 | f-3 (3[100]) |
| ST1841 | 0 (0) | 0 (0) | 2 (100) | 1.0 | 2 (1) | f-7 | f-7 (2[100]) |
| ST1576 | 0 (0) | 0 (0) | 2 (100) | 1.0 | 2 (1) | f-1 | f-1 (2[100]) |
| ST420 | 2 (100) | 0 (0) | 0 (0) | 0.09 | 2 (1) | f-29 | f-29 (2[100]) |
| ST568 complex | 2 (100) | 0 (0) | 0 (0) | 0.09 | 2 (1) | f-2 | f-2 (2[100]) |
| ST62 | 0 (0) | 1 (50) | 1 (50) | 1.0 | 2 (1) | f-5 | f-5 (2[100]) |
| ST59 complex | 2 (67) | 0 (0) | 1 (33) | 0.512 | 3 (1) | f-47, 2 NTe | f-47 (1[50]) |
| Unique ST | 12 (63) | 2 (11) | 5 (26) | 0.001 | 19 (9) | f-1, -2, -3, -5, -7, -13, -27, -39, -49, -50, -59 | f-7 (4[21]) |
| Total isolates typed | 62 (28) | 39 (18) | 119 (54) | 220 | |||
| Not typed | 12 (46) | 4 (15) | 10 (39) | 26 | |||
| Total | 74 (30) | 43 (17) | 129 (52) | 246 | |||
Resistant to one or two classes of drugs.
Resistant to three or more classes of drugs.
P value compares the susceptible column and any resistance.
% of those typed by MLST (n = 220).
NT, not typed.
Of 246 E. coli isolates, 236 (96%) had the fimH gene, comprised of 34 different fimH SNP types (17, 21). Only three fimH types (f-1, -6, and -9) accounted for 138 (58%) of 236 typed E. coli isolates; 115 (83%) of these belonged to the five most frequently detected STs (Table 4). The three most frequent fimH types comprised 49 (96%) of the ST131 isolates, 36 (90%) of the ST95 isolates, 19 (100%) of the ST69 isolates, 4 (20%) of the ST73 complex isolates, and 7 (54%) of the ST12 complex isolates. The eight most prevalent STs contained from one to five fimH types, with one or two types found to be dominant per ST (Table 3). Strains containing the f-9 type were more likely to be MDR (P = 0.001), and 44 (86%) of them were ST131, while strains containing the f-6 type were more likely to be susceptible (P = 0.02) and 17 (43%) of them were ST95.
Table 4.
Distribution of E. coli genotypes by fimH SNP analysis
| fimH typea | No. (%) of isolates that are: |
P valued | No. (%) of isolates among 5 most prevalent STs | Total no. of fimH type isolates | ||
|---|---|---|---|---|---|---|
| Susceptible | Resistantb | MDRc | ||||
| f-1 | 10 (19) | 13 (24) | 31 (57) | 0.024 | 43 (80) | 54 |
| f-2 | 7 (88) | 1 (13) | 0 (0) | 0.001 | 8 | |
| f-3 | 6 (75) | 0 (0) | 2 (25) | 0.01 | 3 (38) | 8 |
| f-4 | 3 (27) | 1 (9) | 7 (64) | 1.0 | 11 | |
| f-5 | 0 (0) | 1 (25) | 3 (75) | 0.319 | 4 | |
| f-6 | 14 (54) | 4 (15) | 8 (31) | 0.005 | 21 (81) | 26 |
| f-7 | 7 (78) | 0 (0) | 2 (22) | 0.004 | 9 | |
| f-8 | 2 (17) | 6 (50) | 4 (33) | 0.519 | 12 (100) | 12 |
| f-9 | 4 (7) | 9 (16) | 45 (77) | <0.0005 | 51 (88) | 58 |
| f-10 | 1 (33) | 0 (0) | 2 (67) | 0.512 | 3 | |
| f-11 | 1 (50) | 0 (0) | 1 (50) | 0.512 | 1 (50) | 2 |
| f-12 | 0 (0) | 0 (0) | 3 (100) | 0.556 | 3 | |
| f-13 | 1 (25) | 1 (25) | 2 (50) | 0.556 | 4 | |
| f-27 | 2 (100) | 0 (0) | 0 (0) | 0.09 | 2 | |
| f-29 | 2 (100) | 0 (0) | 0 (0) | 0.09 | 2 | |
| f-47 | 2 (40) | 2 (40) | 1 (20) | 0.638 | 4 (80) | 5 |
| f-51 | 2 (33) | 1 (17) | 3 (50) | 1.0 | 4 (67) | 6 |
| f-52 | 0 (0) | 0 (0) | 2 (100) | 1.0 | 2 (100) | 2 |
| f-53 | 2 (100) | 0 (0) | 0 (0) | 0.09 | 2 | |
| Unique f-type | 4 (27) | 3 (20) | 8 (53) | 1.0 | 2 (13) | 15 |
| Total typed | 70 (30) | 42 (18) | 124 (52) | 236 | ||
| Total untyped | 4 (40) | 1 (10) | 5 (50) | 0 (0) | 10 | |
| Total | 74 (30) | 43 (17) | 129 (52) | 143 (58) | 246 | |
fimH types (f-types) in bold are the three most prevalent fimH SNP types.
Resistant to one or two classes of drugs.
Resistant to three or more classes of drugs.
P value compares the susceptible column and any resistance.
fimH SNP types were used to further differentiate ST95 strains. ST95 strains were comprised of 4 fimH SNP types (f-1, -6, -9, and -47). Fourteen (67%) of the 21 drug-susceptible ST95 strains belonged to one fimH SNP type (f-6), whereas 16 (84%) of 19 resistant ST95 strains belonged to three other fimH types (P = 0.003) (Fig. 1). The f-6 type was the most prevalent SNP type, accounting for 17 (43%) of the ST95 strains. None of these were MDR (P < 0.05). The f-1 type comprised 16 (40%) of the ST95 strains, and 12 (75%) of these were resistant to at least one class of antimicrobial agents while 6 (38%) were MDR (P > 0.05). The f-9 and f-47 types comprised 3 and 4 ST95 strains, respectively, with one MDR strain in each.
Fig 1.
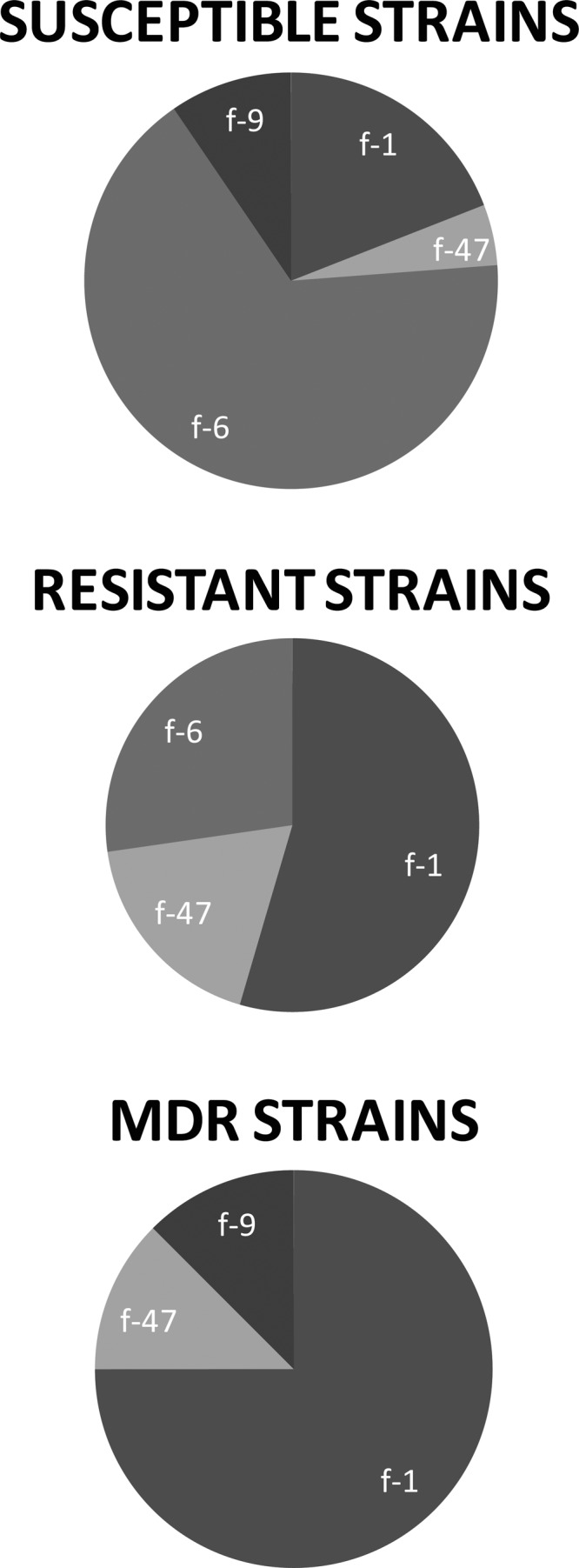
Distribution of fimH SNP types among ST95 strains by drug susceptibility.
Diversity of genotypes.
Among 220 E. coli isolates typed by MLST, 41 STs or ST complexes were identified. Of these, 201 (91%) belonged to one of 22 ST groups (Table 5). Nineteen isolates belonged to unique STs. Simpson's diversity index for all STs was 89% (95% confidence interval [CI], 86% to 91%). The index for drug-susceptible ST genotypes was 87% (95% CI, 80% to 94%), while the index for MDR ST genotypes was 81% (95% CI, 74% to 87%). The index for any drug-resistant (MDR and resistance to one or two classes of drug) ST genotypes was 85% (95% CI, 81% to 89%). A higher index represents greater diversity.
Table 5.
Simpson's diversity index by MLST and by antimicrobial resistance status
| Resistance level (no. of isolates) | No. (%) of isolates MLST typed | No. of MLST cluster types | No. (%) of isolates in MLST cluster typesa | No. of isolates in MLST unique typesa | Simpson's diversity index (%) (95% CI) |
|---|---|---|---|---|---|
| Susceptible (74) | 62 (84) | 13 | 50 (81) | 12 | 87 (80–94) |
| Resistant (43)b | 39 (91) | 12 | 37 (95) | 2 | 86 (80–93) |
| MDR (129)c | 119 (92) | 17 | 114 (96) | 5 | 81 (74–87) |
| Any resistance (172)d | 154 (90) | 18 | 147 (95) | 7 | 85 (81–89) |
| All E. coli isolates (246) | 220 (89) | 22 | 201 (91) | 19 | 89 (86–91) |
Cluster, 2 or more isolates; unique, 1 isolate per group.
Resistant to 1 or 2 antimicrobial agents.
Multidrug resistant (≥3 classes).
Resistance to ≥1 class.
Three of the five most prevalent E. coli clonal groups, ST131, ST69, and ST12 complex, were significantly more likely to be MDR than were others (P = 0.047). These three clonal groups comprised 70 (54%) of the MDR strains (Table 3). The other MDR strains were comprised of 19 distinct STs (10 strains were not typed). On the other hand, ST95 strains were more likely to be susceptible (P = 0.001). In fact, drug-susceptible strains belonging to this clonal group accounted for 28% of all 74 drug-susceptible isolates in the study. Of 119 typed MDR E. coli strains, 114 (96%) belonged to one of 17 STs represented by 2 or more members per ST. Of 62 typed susceptible strains, 50 (81%) belonged to one of 13 STs represented by 2 or more members per ST. A large proportion (63%) of the 19 unique STs were fully susceptible.
Distribution of β-lactamase genes.
The blaCTX-M gene was detected among 25 (10%) E. coli strains in five ST groups and only one nonclonal ST (Table 2). Fifteen had blaCTX-M-15 (group 1), 6 had blaCTX-M-14 (group 9), and one (an ST12 complex isolate) had both. For three strains with the blaCTX-M-1 group gene, CTX-M variants were not able to be further differentiated. ST131 accounted for 18 (72%) of the 25 isolates carrying blaCTX-M. Two carried blaCTX-M-14, and16 had blaCTX-M-1 group genes, of which 13 were blaCTX-M-15. Fifteen CTX-M gene-positive isolates also had blaOXA-1. PCR analysis of blaTEM and blaSHV was performed on 36 E. coli isolates. Three ST131 isolates were blaTEM gene positive by PCR, and two of these coharbored blaCTX-M. Isolates carrying blaTEM were also found among three ST12 complex isolates and one each of ST10 complex and ST38 complex isolates. Only one of 40 ST95 isolates was extended-spectrum β-lactamase (ESBL) gene positive. This strain was resistant to cefotaxime and carried blaCTX-M-14 and blaKPC. One ST12 complex isolate was also found to carry both blaCTX-M-14 and blaKPC. Both KPC gene-positive isolates were sensitive to all carbapenems tested. Susceptibility to imipenem and ertapenem was confirmed by Etest, with MICs for both strains of 0.25 and 0.023 for imipenem and ertapenem, respectively. No ESBL genes were found among the ST69 or ST73 complex strains. The blaSHV gene was not detected in any of the tested strains.
Strain distribution by time of first positive blood culture results.
Of 244 (99%) E. coli isolates with known time of first positive blood culture results, 198 (81%) were cultured from blood <48 h after patient admission (Table 6). Most of the cultures were done at the emergency department (ED) before admission. Among 73 fully susceptible and 128 MDR isolates with culture result data, 62 (85%) and 101 (79%), respectively, were isolated from blood <48 h after patient admission (P > 0.05). Large proportions (>70%) of each of the 5 most common ST strains were isolated from blood <48 h after patient admission. Based on the number of ED visits during the study period, the incidences of susceptible E. coli BSI and MDR E. coli BSI were 0.12 and 0.30 per 1,000 ED visits, respectively (P > 0.05).
Table 6.
Distribution of E. coli MLSTs by timing of the first collection of blood culture that grew E. coli
| ST or ST complex | No. (%) of isolates for each collection time |
|
|---|---|---|
| >48 h | <48 h | |
| ST131 | 14 (27) | 37 (73) |
| ST95a | 4 (10) | 36 (90) |
| ST73 complex | 4 (20) | 16 (80) |
| ST69a | 3 (17) | 15 (83) |
| ST12 complexa | 1 (8) | 11 (92) |
| Other STsa | 13 (18) | 61 (82) |
| Untyped | 6 (23) | 20 (77) |
| Totala | 46 (19) | 198 (81) |
Data on the timing of the first positive isolate collection are available for 244 of 249 E. coli isolates.
DISCUSSION
Of 246 E. coli isolates consecutively collected from patients with BSI in one San Francisco hospital over a 3-year period, we found that only five genotypes by MLST accounted for 146 (66%) of the typed E. coli isolates; these were ST131, ST95, ST73 complex, ST69, and ST12 complex. Interestingly, MDR strains belonging to ST131 and ST69 accounted for nearly half (46%) of all MDR isolates while drug-susceptible strains from ST95 and ST73 complex accounted for more than one-third (36%) of all drug-susceptible strains in this study. Thus, both MDR and drug-susceptible E. coli strains have clonal distribution and accounted for a large proportion of bloodstream isolates from patients admitted to a public hospital in San Francisco.
All of these STs, with the exception of ST69, which belongs to phylogenetic group D, are reported to belong to phylogenetic group B2. Both phylogenetic groups are typically associated with human ExPEC infections (13, 24, 25). Strains belonging to these clonal groups are also frequently isolated from food-producing and companion animals and from retail meats (10, 11, 26–35) and cause serious infections in otherwise healthy individuals (6, 13). Isolates of the enterobacterial repetitive intergenic consensus 2 (ERIC-2) PCR pattern CgA, later characterized as ST69 by MLST, were the most common uropathogenic E. coli genotype isolated from college students with UTI (36, 37). ST95 contains members of avian pathogenic E. coli (APEC) as well as human uropathogenic E. coli (UPEC) and neonatal meningitis E. coli (NMEC) (25). ST95 subgroups share common serotypes and virulence factors, including carriage of pap genes encoding P-type fimbriae which enhance colonization in avian and human epithelial cells. These characteristics may promote an opportunity for zoonotic transmission (38–41).
ST131, the most prevalent MLST genotype reported globally, was also the most common ST found in this study, accounting for 51 (23%) of typed E. coli isolates, 47 (36%) of all MDR isolates, and 18 (72%) of CTX-M gene-positive isolates. This clonal group was comprised of five fimH SNP types, for which 39 (76%) strains were f-9, and all but four of these were MDR. A large proportion of these ST131 infections appear to be community onset, since the blood cultures of 37 (73%) isolates grew E. coli within 48 h of hospital admission. This clonal group has worldwide distribution as a common cause of UTI and BSI (3–6, 42). ST131 is remarkable for its carriage of a wide array of virulence factors (8, 10, 43, 44) in addition to being a major producer of CTX-M-type ESBLs (1, 2, 7, 9, 10) and, more recently, for its acquisition of the carbapenemase-encoding genes blaKPC, blaVIM, and blaNDM-1 (45–48). These drug resistance determinants are often located on plasmids and associated with integrons and IS elements (3, 10, 49–51). A recent report found that by pulsed-field gel electrophoresis (PFGE), ST131 strains isolated from human clinical sources belonged to pulsotypes distinct from those obtained from foods or animal food sources (52).
ST95 was the second largest clonal group identified in this study. Unlike ST131, the majority (53%) were susceptible to all tested antimicrobial agents. Like ST131, a large proportion (90%) of ST95 isolates were classified as representing community-onset infection. ST95 was found in our study population with prevalence nearly equal to that of ST131 in spite of having significantly fewer drug-resistant strains (P = 0.001). In fact, among four fimH SNP types comprising ST95, one (f-6) was responsible for most (67%) of the drug-susceptible strains. Such an observation suggests that drug resistance is not a prerequisite for clonal dissemination.
The observation that drug-susceptible ST95 disseminates clonally is not new. Manges et al. found several ST95 isolates in their report of endemic and epidemic lineages of E. coli causing UTI in Montreal and California (13). All except one of the strains were susceptible to all drugs tested. Vincent et al. detected ST95 in human clinical isolates and in one case of ready-to-eat food product (11). All but two of these isolates were susceptible. ST95 was found to be the most prevalent cause of bacterial peritonitis and BSI in France (24). All ST95 strains isolated from UTI in a study in the northwest of England were susceptible to all tested antimicrobial agents (12). Our study, based on consecutively collected, population-based samples from one large hospital, confirms that drug-susceptible E. coli strains can disseminate clonally in community settings.
Near the end of the study period (April 2010), one ST95 isolate collected from a patient within 48 h of admission carried both blaCTX-M and blaKPC, which, to our knowledge, is the first example of acquisition of these genes by ST95. This ST95 isolate, an f-1 fimH SNP type, was distinct from the other ST95 isolates in that it was resistant to 12 drugs within nine classes of antimicrobial agents and harbored the dfrA17 and blaCTX-M-14 genes. No ESBL genes were found among any of 19 ST69 strains, while more than two-thirds of 25 CTX-M gene-positive strains belonged to ST131. These observations suggest intrinsic differences in the frequency of distinct E. coli lineages acquiring different types of mobile drug resistance genes or that these genes remain within the lineages after their “founder” strains acquire them.
Because we consecutively collected and analyzed all BSI isolates from one hospital in San Francisco over a period of 3 years, we were able to determine the relative proportion of infections caused by major clonal groups of both drug-resistant and -susceptible strains. Strain diversity was higher in drug-susceptible strains than in MDR strains, but some of the drug-susceptible strains were indeed clonal by MLST, by fimH SNP type, and by both combined. Furthermore, more than two-thirds of MDR as well as drug-susceptible E. coli infections were found to have community onset. We do not have additional information about prior health care exposures of these patients, so it is possible that some of these community-onset BSI cases had contact with health care settings prior to their infection. Our observation, of course, is limited to one geographic setting, and thus, a similar study should be performed at other sites to determine the generalizability of our findings. Nevertheless, our data suggest that the overrepresentation of both MDR and susceptible E. coli strains causing community-onset BSI may be explained by their enhanced capacity to cause extraintestinal infection rather than selection by antimicrobial agents used in hospitals.
ACKNOWLEDGMENTS
We thank Michael Jula, Li Basuino, and Patricia Denn of San Francisco General Hospital for assistance with data analyses and arranging to provide the study isolates. We thank Satowa Suzuki, Sara Tartof, Hillary Berman, and Julian Schmidt for assistance with protocols. We thank Shantell Nolen, Pooja Jaeel, and Roxane Raphael for technical support.
This study was supported in part by NIH grant AI059523.
All authors declare no conflict of interest.
Footnotes
Published ahead of print 12 November 2012
REFERENCES
- 1. Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Canton R, Nordmann P. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg. Infect. Dis. 14:195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fam N, Leflon-Guibout V, Fouad S, Aboul-Fadl L, Marcon E, Desouky D, El-Defrawy I, Abou-Aitta A, Klena J, Nicolas-Chanoine MH. 2011. CTX-M-15-producing Escherichia coli clinical isolates in Cairo (Egypt), including isolates of clonal complex ST10 and clones ST131, ST73, and ST405 in both community and hospital settings. Microb. Drug Resist. 17:67–73 [DOI] [PubMed] [Google Scholar]
- 3. Naseer U, Haldorsen B, Tofteland S, Hegstad K, Scheutz F, Simonsen GS, Sundsfjord A. 2009. Molecular characterization of CTX-M-15-producing clinical isolates of Escherichia coli reveals the spread of multidrug-resistant ST131 (O25:H4) and ST964 (O102:H6) strains in Norway. APMIS 117:526–536 [DOI] [PubMed] [Google Scholar]
- 4. Pitout JD, Church DL, Gregson DB, Chow BL, McCracken M, Mulvey MR, Laupland KB. 2007. Molecular epidemiology of CTX-M-producing Escherichia coli in the Calgary Health Region: emergence of CTX-M-15-producing isolates. Antimicrob. Agents Chemother. 51:1281–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pitout JD, Gregson DB, Campbell L, Laupland KB. 2009. Molecular characteristics of extended-spectrum-β-lactamase-producing Escherichia coli isolates causing bacteremia in the Calgary Health Region from 2000 to 2007: emergence of clone ST131 as a cause of community-acquired infections. Antimicrob. Agents Chemother. 53:2846–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66:1–14 [DOI] [PubMed] [Google Scholar]
- 7. Suzuki S, Shibata N, Yamane K, Wachino J, Ito K, Arakawa Y. 2009. Change in the prevalence of extended-spectrum-beta-lactamase-producing Escherichia coli in Japan by clonal spread. J. Antimicrob. Chemother. 63:72–79 [DOI] [PubMed] [Google Scholar]
- 8. Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 51:286–294 [DOI] [PubMed] [Google Scholar]
- 9. Peirano G, Pitout JD. 2010. Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int. J. Antimicrob. Agents 35:316–321 [DOI] [PubMed] [Google Scholar]
- 10. Platell JL, Johnson JR, Cobbold RN, Trott DJ. 2011. Multidrug-resistant extraintestinal pathogenic Escherichia coli of sequence type ST131 in animals and foods. Vet. Microbiol. 153:99–108 [DOI] [PubMed] [Google Scholar]
- 11. Vincent C, Boerlin P, Daignault D, Dozois CM, Dutil L, Galanakis C, Reid-Smith RJ, Tellier PP, Tellis PA, Ziebell K, Manges AR. 2010. Food reservoir for Escherichia coli causing urinary tract infections. Emerg. Infect. Dis. 16:88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lau SH, Reddy S, Cheesbrough J, Bolton FJ, Willshaw G, Cheasty T, Fox AJ, Upton M. 2008. Major uropathogenic Escherichia coli strain isolated in the northwest of England identified by multilocus sequence typing. J. Clin. Microbiol. 46:1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Manges AR, Tabor H, Tellis P, Vincent C, Tellier PP. 2008. Endemic and epidemic lineages of Escherichia coli that cause urinary tract infections. Emerg. Infect. Dis. 14:1575–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marquez C, Labbate M, Raymondo C, Fernandez J, Gestal AM, Holley M, Borthagaray G, Stokes HW. 2008. Urinary tract infections in a South American population: dynamic spread of class 1 integrons and multidrug resistance by homologous and site-specific recombination. J. Clin. Microbiol. 46:3417–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ajiboye RM, Solberg OD, Lee BM, Raphael E, Debroy C, Riley LW. 2009. Global spread of mobile antimicrobial drug resistance determinants in human and animal Escherichia coli and Salmonella strains causing community-acquired infections. Clin. Infect. Dis. 49:365–371 [DOI] [PubMed] [Google Scholar]
- 16. Levesque C, Piche L, Larose C, Roy PH. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tartof SY, Solberg OD, Riley LW. 2007. Genotypic analyses of uropathogenic Escherichia coli based on fimH single nucleotide polymorphisms (SNPs). J. Med. Microbiol. 56:1363–1369 [DOI] [PubMed] [Google Scholar]
- 18. Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495 [DOI] [PubMed] [Google Scholar]
- 19. Shibata N, Kurokawa H, Doi Y, Yagi T, Yamane K, Wachino J, Suzuki S, Kimura K, Ishikawa S, Kato H, Ozawa Y, Shibayama K, Kai K, Konda T, Arakawa Y. 2006. PCR classification of CTX-M-type β-lactamase genes identified in clinically isolated gram-negative bacilli in Japan. Antimicrob. Agents Chemother. 50:791–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clermont O, Dhanji H, Upton M, Gibreel T, Fox A, Boyd D, Mulvey MR, Nordmann P, Ruppe E, Sarthou JL, Frank T, Vimont S, Arlet G, Branger C, Woodford N, Denamur E. 2009. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J. Antimicrob. Chemother. 64:274–277 [DOI] [PubMed] [Google Scholar]
- 21. Dias RC, Moreira BM, Riley LW. 2010. Use of fimH single-nucleotide polymorphisms for strain typing of clinical isolates of Escherichia coli for epidemiologic investigation. J. Clin. Microbiol. 48:483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simpson EH. 1949. Measurement of diversity. Nature 163:688 [Google Scholar]
- 24. Bert F, Johnson JR, Ouattara B, Leflon-Guibout V, Johnston B, Marcon E, Valla D, Moreau R, Nicolas-Chanoine MH. 2010. Genetic diversity and virulence profiles of Escherichia coli isolates causing spontaneous bacterial peritonitis and bacteremia in patients with cirrhosis. J. Clin. Microbiol. 48:2709–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson JR, Russo TA. 2005. Molecular epidemiology of extraintestinal pathogenic (uropathogenic) Escherichia coli. Int. J. Med. Microbiol. 295:383–404 [DOI] [PubMed] [Google Scholar]
- 26. Ewers C, Grobbel M, Stamm I, Kopp PA, Diehl I, Semmler T, Fruth A, Beutlich J, Guerra B, Wieler LH, Guenther S. 2010. Emergence of human pandemic O25:H4-ST131 CTX-M-15 extended-spectrum-beta-lactamase-producing Escherichia coli among companion animals. J. Antimicrob. Chemother. 65:651–660 [DOI] [PubMed] [Google Scholar]
- 27. Hannah EL, Johnson JR, Angulo F, Haddadin B, Williamson J, Samore MH. 2009. Molecular analysis of antimicrobial-susceptible and -resistant Escherichia coli from retail meats and human stool and clinical specimens in a rural community setting. Foodborne Pathog. Dis. 6:285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johnson JR, Delavari P, O'Bryan TT, Smith KE, Tatini S. 2005. Contamination of retail foods, particularly turkey, from community markets (Minnesota, 1999-2000) with antimicrobial-resistant and extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 2:38–49 [DOI] [PubMed] [Google Scholar]
- 29. Johnson JR, McCabe JS, White DG, Johnston B, Kuskowski MA, McDermott P. 2009. Molecular analysis of Escherichia coli from retail meats (2002-2004) from the United States National Antimicrobial Resistance Monitoring System. Clin. Infect. Dis. 49:195–201 [DOI] [PubMed] [Google Scholar]
- 30. Johnson TJ, Logue CM, Wannemuehler Y, Kariyawasam S, Doetkott C, DebRoy C, White DG, Nolan LK. 2009. Examination of the source and extended virulence genotypes of Escherichia coli contaminating retail poultry meat. Foodborne Pathog. Dis. 6:657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, Voets GM, van den Munckhof MP, van Essen-Zandbergen A, Platteel T, Fluit AC, van de Sande-Bruinsma N, Scharinga J, Bonten MJ, Mevius DJ. 2011. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 17:873–880 [DOI] [PubMed] [Google Scholar]
- 32. Manges AR, Smith SP, Lau BJ, Nuval CJ, Eisenberg JN, Dietrich PS, Riley LW. 2007. Retail meat consumption and the acquisition of antimicrobial resistant Escherichia coli causing urinary tract infections: a case-control study. Foodborne Pathog. Dis. 4:419–431 [DOI] [PubMed] [Google Scholar]
- 33. Ramchandani M, Manges AR, DebRoy C, Smith SP, Johnson JR, Riley LW. 2005. Possible animal origin of human-associated, multidrug-resistant, uropathogenic Escherichia coli. Clin. Infect. Dis. 40:251–257 [DOI] [PubMed] [Google Scholar]
- 34. Randall LP, Clouting C, Horton RA, Coldham NG, Wu G, Clifton-Hadley FA, Davies RH, Teale CJ. 2011. Prevalence of Escherichia coli carrying extended-spectrum beta-lactamases (CTX-M and TEM-52) from broiler chickens and turkeys in Great Britain between 2006 and 2009. J. Antimicrob. Chemother. 66:86–95 [DOI] [PubMed] [Google Scholar]
- 35. Xia X, Meng J, McDermott PF, Zhao S. 2011. Escherichia coli from retail meats carry genes associated with uropathogenic Escherichia coli, but are weakly invasive in human bladder cell culture. J. Appl. Microbiol. 110:1166–1176 [DOI] [PubMed] [Google Scholar]
- 36. Manges AR, Johnson JR, Foxman B, O'Bryan TT, Fullerton KE, Riley LW. 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 345:1007–1013 [DOI] [PubMed] [Google Scholar]
- 37. Smith SP, Manges AR, Riley LW. 2008. Temporal changes in the prevalence of community-acquired antimicrobial-resistant urinary tract infection affected by Escherichia coli clonal group composition. Clin. Infect. Dis. 46:689–695 [DOI] [PubMed] [Google Scholar]
- 38. Ewers C, Antao EM, Diehl I, Philipp HC, Wieler LH. 2009. Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Appl. Environ. Microbiol. 75:184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ewers C, Li G, Wilking H, Kiessling S, Alt K, Antao EM, Laturnus C, Diehl I, Glodde S, Homeier T, Bohnke U, Steinruck H, Philipp HC, Wieler LH. 2007. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 297:163–176 [DOI] [PubMed] [Google Scholar]
- 40. Johnson TJ, Wannemuehler Y, Johnson SJ, Stell AL, Doetkott C, Johnson JR, Kim KS, Spanjaard L, Nolan LK. 2008. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl. Environ. Microbiol. 74:7043–7050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mora A, Lopez C, Dabhi G, Blanco M, Blanco JE, Alonso MP, Herrera A, Mamani R, Bonacorsi S, Moulin-Schouleur M, Blanco J. 2009. Extraintestinal pathogenic Escherichia coli O1:K1:H7/NM from human and avian origin: detection of clonal groups B2 ST95 and D ST59 with different host distribution. BMC Microbiol. 9:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Croxall G, Hale J, Weston V, Manning G, Cheetham P, Achtman M, McNally A. 2011. Molecular epidemiology of extraintestinal pathogenic Escherichia coli isolates from a regional cohort of elderly patients highlights the prevalence of ST131 strains with increased antimicrobial resistance in both community and hospital care settings. J. Antimicrob. Chemother. 66:2501–2508 [DOI] [PubMed] [Google Scholar]
- 43. Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, Zhanel GG. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob. Agents Chemother. 53:2733–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Canica MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273–281 [DOI] [PubMed] [Google Scholar]
- 45. Mantengoli E, Luzzaro F, Pecile P, Cecconi D, Cavallo A, Attala L, Bartoloni A, Rossolini GM. 2011. Escherichia coli ST131 producing extended-spectrum beta-lactamases plus VIM-1 carbapenemase: further narrowing of treatment options. Clin. Infect. Dis. 52:690–691 [DOI] [PubMed] [Google Scholar]
- 46. Morris D, Boyle F, Ludden C, Condon I, Hale J, O'Connell N, Power L, Boo TW, Dhanji H, Lavallee C, Woodford N, Cormican M. 2011. Production of KPC-2 carbapenemase by an Escherichia coli clinical isolate belonging to the international ST131 clone. Antimicrob. Agents Chemother. 55:4935–4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Naas T, Cuzon G, Gaillot O, Courcol R, Nordmann P. 2011. When carbapenem-hydrolyzing β-lactamase KPC meets Escherichia coli ST131 in France. Antimicrob. Agents Chemother. 55:4933–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peirano G, Schreckenberger PC, Pitout JD. 2011. Characteristics of NDM-1-producing Escherichia coli isolates that belong to the successful and virulent clone ST131. Antimicrob. Agents Chemother. 55:2986–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cullik A, Pfeifer Y, Prager R, von Baum H, Witte W. 2010. A novel IS26 structure surrounds blaCTX-M genes in different plasmids from German clinical Escherichia coli isolates. J. Med. Microbiol. 59:580–587 [DOI] [PubMed] [Google Scholar]
- 50. Eckert C, Gautier V, Arlet G. 2006. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 57:14–23 [DOI] [PubMed] [Google Scholar]
- 51. Poirel L, Naas T, Nordmann P. 2008. Genetic support of extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 14(Suppl 1):75–81 [DOI] [PubMed] [Google Scholar]
- 52. Johnson JR, Nicolas-Chanoine MH, Debroy C, Castanheira M, Robicsek A, Hansen G, Weissman S, Urban C, Platell J, Trott D, Zhanel G, Clabots C, Johnston BD, Kuskowski MA. 2012. Comparison of Escherichia coli ST131 pulsotypes, by epidemiologic traits, 1967-2009. Emerg. Infect. Dis. 18:598–607 [DOI] [PMC free article] [PubMed] [Google Scholar]


