Abstract
Multidrug resistance in Pseudomonas aeruginosa is increasingly becoming a threat for human health. Indeed, some strains are resistant to almost all currently available antibiotics, leaving very limited choices for antimicrobial therapy. In many such cases, polymyxins are the only available option, although as their utilization increases so does the isolation of resistant strains. In this study, we screened a comprehensive PA14 mutant library to identify genes involved in changes of susceptibility to polymyxin B in P. aeruginosa. Surprisingly, our screening revealed that the polymyxin B resistome of this microorganism is fairly small. Thus, only one resistant mutant and 17 different susceptibility/intrinsic resistance determinants were identified. Among the susceptible mutants, a significant number carried transposon insertions in lipopolysaccharide (LPS)-related genes. LPS analysis revealed that four of these mutants (galU, lptC, wapR, and ssg) had an altered banding profile in SDS-polyacrylamide gels and Western blots, with three of them exhibiting LPS core truncation and lack of O-antigen decoration. Further characterization of these four mutants showed that their increased susceptibility to polymyxin B was partly due to increased basal outer membrane permeability. Additionally, these mutants also lacked the aminoarabinose-substituted lipid A species observed in the wild type upon growth in low magnesium. Overall, our results emphasize the importance of LPS integrity and lipid A modification in resistance to polymyxins in P. aeruginosa, highlighting the relevance of characterizing the genes that affect biosynthesis of cell surface structures in this pathogen to follow the evolution of peptide resistance in the clinic.
INTRODUCTION
Pseudomonas aeruginosa is a prominent nosocomial pathogen, as well as a major cause of chronic infections in cystic fibrosis patients (1–3). This microorganism is notably resistant to most antibiotics available in the market (hence its designation as a “superbug”). Indeed, the number of antimicrobials that can be used to treat Pseudomonas infections, particularly those caused by multiresistant strains, is quite limited (4). For this reason, there has been resurgence in the use of polymyxins (5), lipopeptides of bacterial origin, of which polymyxin B and colistin (polymyxin E) are the best-known representatives. The mechanism of action of polymyxins involves an initial stage of interaction with the lipid A of lipopolysaccharide (LPS) leading to self-promoted uptake across this membrane followed by cell death by mechanisms that have not been well defined (6). Even though polymyxins were discovered in the 1940s, their use since the mid-1980s has been restricted to topological applications due to concerns regarding their toxicity, although new manufacturing methods and the use of methane sulfonate derivatives as prodrugs seem to have alleviated some of these issues. Nevertheless, a legitimate concern about more intensive use of this class of drugs is the development of resistance by bacteria.
To date, the incidence of polymyxin-resistant microorganisms in the clinic is relatively low. However, the increased use of these antibiotics for the treatment of multiresistant strains has led to a greater frequency of resistant clinical isolates (7). In the case of Pseudomonas, a growing number of clinical strains display resistance to polymyxins due to mutations in the two-component systems PhoPQ, PmrAB, or ParRS (8–10). These mutations result in the constitutive expression of the LPS modification (arn) operon, which encodes the proteins necessary for the aminoarabinosylation of the lipid A moiety of LPS. This modification reduces the negative charge of the cell surface, thereby limiting its interaction with the positively charged polymyxins and consequent self-promoted uptake. This is the most common mechanism of adaptive resistance to antimicrobial peptides observed in many Gram-negative bacteria. In P. aeruginosa, this modification is triggered under low-Mg2+ conditions and upon exposure to subinhibitory concentrations of certain antimicrobial peptides in a process that involves several two-component systems, including those mentioned above (11–13). Despite recent advances in the understanding of peptide adaptive resistance in this pathogen, our knowledge of intrinsic and mutationally driven resistance of P. aeruginosa to polymyxins is somewhat limited.
Recently, screening of transposon mutant libraries has permitted the mapping of many of the genes that constitute the resistome of P. aeruginosa to different types of antimicrobials, including β-lactams, aminoglycosides, and fluoroquinolones (14–19). These comprehensive studies revealed that the genetic determinants of antibiotic resistance in this pathogen are far more complex than originally thought. Thus, in addition to traditional resistance mechanisms, such as porins and efflux pumps, genes encoding proteins with functions related to metabolism, transcriptional regulation, or motility were identified. Also of note, these studies reported the identification of numerous genes that conferred low-level resistance upon mutation. This is of particular interest as the accumulation of low-level-impact mutations is thought to eventually lead to breakthrough clinical resistance in a stepwise manner (20). In this study, we carried out a high-throughput screening of the Harvard transposon mutant library (21) of P. aeruginosa strain PA14, with the aim of identifying genes that participate in the polymyxin resistance of this microorganism. Surprisingly, a relatively modest resistome with respect to this cationic peptide antibiotic was identified.
MATERIALS AND METHODS
Growth conditions and antimicrobials.
Bacteria were routinely grown in Luria-Bertani (LB) broth or agar at 37°C. Mueller-Hinton medium and the defined medium BM2-glucose [62 mM potassium phosphate buffer (pH 7), 7 mM (NH4)2SO4, 10 μM FeSO4, 0.4% (wt/vol) glucose] containing a high (2 mM) or low (20 μM) MgSO4 concentration were used for all experiments concerning antibiotic susceptibility determinations. When required for plasmid or mutant maintenance in P. aeruginosa, gentamicin, chloramphenicol, or carbenicillin was supplemented to each growth medium at 15 μg/ml, 200 μg/ml, and 500 μg/ml, respectively. Chloramphenicol was supplemented at a concentration of 30 μg/ml for selection in Escherichia coli. All antibiotics were purchased from Sigma. The cationic peptides CP28, indolicidin, and LL37 were synthesized by 9-fluorenylmethoxy carbonyl (Fmoc) methods at either the Brain Research Center (University of British Columbia, Vancouver, Canada) or GenScript (Piscataway, NJ) and were 95% pure as determined by high-pressure liquid chromatography (HPLC) and mass spectrometry (MS).
Identification and confirmation of transposon mutants with altered susceptibility to polymyxin B.
The identification of genes that participate in the polymyxin B resistance of P. aeruginosa was performed by screening the comprehensive PA14 transposon mutant library (21). Briefly, a total number of 5,850 mutants carrying transposon insertions in 4,596 different genes were analyzed for their increased resistance or susceptibility to polymyxin B with the aim of finding either mutational or intrinsic resistance determinants, respectively. This screening was carried out by using the agar dilution method as previously described (15, 22). Thus, the library was replicated into 96-well plates containing LB and grown overnight at 37°C. The next day, these cultures were diluted and approximately 1 μl from each well was inoculated onto agar plates containing Mueller-Hinton medium supplemented with different concentrations of polymyxin B. Under these conditions, the MIC of the wild-type strain was 0.5 μg/ml. The results were read following 18 h of incubation at 37°C, and those mutants that showed changes in susceptibility of at least 2-fold compared to the wild type were selected for individual analysis. The altered susceptibility phenotype of the mutants was then confirmed by the Etest and/or broth microdilution method. The correct insertion of the transposon in the selected mutants was confirmed by PCR.
Determination of MICs to different antimicrobial peptides.
Peptide MICs were investigated by broth microdilution in accordance with CLSI guidelines by utilizing BM2-glucose medium with a high (2 mM) concentration of Mg2+ (23). The MIC determination assays were performed in polypropylene microtiter plates to avoid binding of the peptides to polystyrene, which could lead to artificially high MIC values (23). The MIC plates were incubated at 37°C for 24 h, after which the concentration with no visible bacterial growth was considered the MIC.
RT-qPCR.
RNA was purified from cultures of P. aeruginosa PA14 wild-type or mutant strains grown to mid-logarithmic phase (optical density at 600 nm [OD600] of 0.4 to 0.6) in BM2-glucose medium under different conditions, namely, with a high (2 mM) or low (20 μM) Mg2+ concentration and with or without a subinhibitory concentration (0.25 μg/ml) of polymyxin B. Additionally, aliquots taken from the wild-type cultures grown with a high Mg2+ concentration without antibiotic were subsequently challenged with a lethal dose (2 μg/ml) of polymyxin B. RNA was prepared and reverse transcribed into cDNA by using reverse transcriptase (RT) as previously described (24). The quantitative PCRs (qPCRs) were performed in an ABI Prism 7000 instrument (Applied Biosystems, Foster, CA), and the mixtures contained 2.5 μl from a 1:100 dilution of cDNA as a template and SYBR green PCR master mix (Applied Biosystems). The rpsL gene was used as a housekeeping gene for the calculation of the fold changes according to the threshold cycle (CT) method. For each condition, RNA was prepared from three independent cultures, and each one was assayed in duplicate.
Analysis of LPS and lipid A.
LPS from whole-cell lysates was prepared following the method of Hitchcock and Brown (25) and subsequently analyzed by SDS-PAGE followed by silver staining according to the modified protocol of Fomsgaard et al. (26). Lipid A isolation was performed from 50-ml cultures of P. aeruginosa strains grown in low (20 μM) Mg2+–BM2-glucose to an OD600 of 0.7. Purification was by the protocol described by Zhou et al. (27), and lipid A was then kept at 4°C until its analysis by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) with an ABI Voyager DE-STR spectrometer in the reflectron mode at the University of British Columbia facilities.
Dansyl-polymyxin binding experiments.
Dansyl-polymyxin assays were performed using whole cells as previously described (28). Briefly, cultures of the different strains were grown in BM2 minimal medium to mid-logarithmic phase and then washed twice with 5 mM HEPES (pH 7.2) supplemented with 5 mM sodium azide to inhibit energization. Cells were resuspended in the same buffer at an OD600 of 0.5. Two milliliters from each bacterial suspension was placed in a quartz cuvette, and aliquots of dansyl-polymyxin were gradually added. The emission of fluorescence was measured at 485 nm following excitation at 340 nm in a Perkin-Elmer LS-50B spectrophotometer.
Outer membrane permeability assays.
To compare the outer membrane permeabilities of different mutant strains to that of the wild type, the 1-N-phenylnaphthylamine (NPN) assay was performed as described by Loh et al. (29). Briefly, overnight cultures of the different strains were subcultured into BM2 medium and grown to mid-logarithmic phase. These cells were harvested, washed with 5 mM sodium HEPES buffer (pH 7.2), and then resuspended at a final OD600 of 0.5 in the same buffer supplemented with a 5 μM concentration of the uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP). NPN was then added at a final concentration of 15 μM to 2-ml cell suspensions. Fluorescence was monitored by using a Perkin-Elmer LS-50B spectrophotometer at excitation and emission wavelengths of 350 nm and 420 nm, respectively.
Complementation of the galU (PA2023), lptC (PA4459), and ssg (PA5001) mutants.
Each gene was amplified by PCR using genomic DNA from the PA14 wild-type strain as a template. The primers used were designed with program Primer3 (30) from the genomic sequence of P. aeruginosa PA14 (31) and were as follows: COMPgalU-F, TCATCTACCAGGGCATAGGAC; COMPgalU-R, CGACAGCTCGGGTAGAGC; COMPlptC-F, GGCTATGAGCAAGTCGCCTA; COMPlptC-R, CCTGCTTGTCGTCCAGTTC; COMPssg-F, CGTGATGCTTGGGTCGAG; and COMPssg-F, CTTCACGTCCCCATTCAAAG. The fragments resulting from PCR were cloned into the Zero-Blunt TOPO vector (Invitrogen) and then cut with NsiI. The NsiI fragments were then ligated into the PstI site of the broad-host-range vector pBBR1MCS, which has a chloramphenicol selection marker (32). Thus, the plasmids pBBR1MCS-galU, pBBR1MCS-lptC, and pBBR1MCS-ssg were generated and subsequently transformed into the respective mutants in order to obtain the complemented strains (designated the CgalU, ClptC, and Cssg strains).
RESULTS AND DISCUSSION
Identification of mutants with altered polymyxin B susceptibility phenotypes.
Screening of the transposon mutant library for mutations that confer increased resistance or susceptibility to polymyxin B led to the identification of 18 mutant strains (Table 1). Of these genes, 17 were deemed intrinsic resistance determinants and conferred a supersusceptible phenotype upon mutation. Only one of these mutants, which harbors the well-known phoQ mutation (12), was involved in increased, mutational resistance. Overall, this was a remarkably small number of genes in comparison with the results obtained in other screenings performed with antibiotics from other classes, such as β-lactams, aminoglycosides, and fluoroquinolones, which identified a considerably larger number and diversity of genes (14, 15, 17, 19). Although these results seem to indicate that the mechanisms involved in resistance to polymyxins are quite limited, it must be taken into account that transposon mutant library screenings are limited to the analysis of nonessential genes. Thus, it is possible that some of the genes that participate in intrinsic or mutation-derived resistance to polymyxins play essential roles and would therefore be missed in this screening.
Table 1.
Mutants identified in the screening and changes in susceptibility to different cationic antimicrobial peptides
| PAO1 ortholog | Gene name and/or description | Fold change in susceptibility of mutant compared to that of wild-type PA14 as determined bya: |
|||||
|---|---|---|---|---|---|---|---|
| Etest (PxB) | Broth microdilution |
||||||
| PxB | COL | CP28 | IND | LL37 | |||
| PA0401 | Noncatalytic dihydroorotase-like protein | −1.5 | — | — | — | — | — |
| PA0402 | pyrB; aspartate carbamoyltransferase | −3.0 | — | — | — | — | — |
| PA1180 | phoQ; two-component sensor protein | +4.0 | >+8 | >+8 | NC | NC | +2 |
| PA1375 | pdxB; erythronate-4-phosphate dehydrogenase | −1.5 | — | — | — | — | — |
| PA1588 | sucC; succinyl-coenzyme A synthetase beta chain | −1.5 | −2 | −2 | −2 | NC | −2 |
| PA1799 | parR; two-component response regulator | — | −2 | −2 | NC | NC | NC |
| PA2023 | galU; UTP-glucose-1-phosphate uridylyltransferase | −2.0 | −2 | −2 | −4 | −2 | −4 |
| PA3050 | pyrD; dihydroorotate dehydrogenase | −1.5 | |||||
| PA4020 | mpl | −1.5 | |||||
| PA4069 | Hypothetical protein, (rmlD homolog) | −1.5 | −2 | −2 | −2 | NC | NC |
| PA4109 | ampR; transcriptional regulator | — | −2 | −2 | −2 | NC | −2 |
| PA4459 | lptC; conserved hypothetical protein | −3.0 | −4 | −4 | −2 | −8 | −8 |
| PA4748 | tpiA; triosephosphate isomerase | −1.5 | — | — | — | — | — |
| PA4776 | pmrA; two-component response regulator | — | −2 | −2 | −2 | NC | −2 |
| PA5000 | wapR; putative glycosyl transferase | −3.0 | −2 | −2 | −2 | NC | −4 |
| PA5001 | ssg; conserved hypothetical protein | −3.0 | −2 | −2 | −2 | −4 | −4 |
| PA5038 | aroB; 3-dihydrokinate synthase | −3.0 | — | — | — | — | — |
| PA5199 | amgS; two-component sensor protein | −1.5 | −2 | −2 | NC | NC | NC |
Fold changes were calculated based on the modal MICs calculated for each strain. PxB, polymyxin B; COL, colistin; IND, indolicidin; —, not tested; NC, no change observed.
As mentioned, only the phoQ mutant displayed a greater (4- to 8-fold-increased) level of resistance than the wild-type PA14. This is strikingly different from results of other screenings such as those carried out with β-lactams (14), ciprofloxacin (15), and tobramycin (19), which found 41, 79, and 135 mutants, respectively, with decreased susceptibility. Furthermore, a high proportion of the resistant mutants described in those studies displayed low-level resistance, which is considered the driving force of stepwise increase in resistance (20, 33), while no such mutations were identified in the screening for polymyxin resistance. At first glance, this might imply that mutational resistance to polymyxins would be extremely rare in the clinic. However, there is evidence that gain-of-function mutations affecting the activation of the response regulators PmrA, PhoP, and ParR can, like the phoQ mutation (34), result in constitutive expression of the LPS modification (arn) operon and, consequently, in increased peptide resistance (10, 35). These results, therefore, emphasized the importance of understanding the regulatory pathways involved in adaptive resistance to antimicrobial peptides in this microbe, as alterations in the regulation of these adaptations appear to provide the major background for polymyxin clinical resistance.
Among the genes that are disrupted in the 18 polymyxin-resistant mutants, five encode proteins with a regulatory function (Table 1). Three are the aforementioned parR, pmrA, and phoQ, all of which play key roles in adaptive peptide resistance by regulating the expression of the LPS modification operon. Presumably, the mutations in parR or pmrA led to a slight increase in susceptibility by limiting the acquisition of adaptive resistance upon exposure to polymyxin B, as shown previously (11). Additionally, mutants with mutations in the genes encoding AmpR and the two-component sensor protein AgmS were also more susceptible to polymyxin B than the wild-type strain.
The screening also identified genes predicted to encode proteins that participate in metabolism, including PA0401, pyrB, pdxB, sucC, tpiA, and aroB. Although many of these mutants (except the sucC mutant) exhibited lower growth rates, other growth-defective mutants were not identified in the screen, and it should be noted that metabolism-related genes have been consistently identified in all comprehensive screenings for determinants of resistance against other antibiotics in P. aeruginosa (14, 15, 17, 19), as well as being involved in biofilm formation and swarming motility, which lead to multiresistant states (20). Consequently, we hypothesize that metabolic pathways that are affected in these mutants are likely to play a role in the ability of P. aeruginosa to withstand antibiotic and particularly polymyxin challenges.
Effects of specific LPS mutations.
Significantly, five of the 17 supersusceptible mutants contained insertions in genes predicted to be involved in the synthesis and transport of LPS. For example, the protein GalU is a UDP-glucose pyrophosphorylase that participates in the synthesis of UDP-d-glucose, which is the precursor of different cell surface structures. Significantly, d-glucose is the most abundant sugar in the outer core oligosaccharide of LPS (36, 37). The involvement of galU in polymyxin resistance has already been observed in other species such as Campylobacter jejuni (38), Proteus mirabilis (39), and Yersinia pestis (40). The product of PA5000, WapR, is a rhamnosyltransferase implicated in the synthesis of the capped core oligosaccharide, which is covalently linked to long-chain O polysaccharides of LPS (41, 42). Another gene related to the synthesis of the LPS core is PA5001, encoding a putative glycosyltransferase that, like wapR, is located in the LPS core biosynthesis cluster (42). PA5001 is homologous to the gene ssg (cell surface sugar biosynthetic glycosyltransferase) of Pseudomonas alkylphenolia, with 79% identity (43). By homology, the P. aeruginosa PA5001 gene is designated ssg here. PA4459 shares high similarity with the gene encoding LptC of E. coli, which is part of the transmembrane machinery necessary for the transport of LPS from the inner membrane to the outer membrane (44). Finally, the product of PA4069 shares 39% similarity with RmlD (PA5162), which is one of the enzymes required for the synthesis of dTDP-l-rhamnose, which is a precursor for the L-rhamnose residue that is found in the outer core of P. aeruginosa LPS (45). The discovery of these mutants in which the disrupted genes play essential roles in LPS biosynthesis correlated well with the known role of LPS in self-promoted uptake of polymyxin and antimicrobial peptides. It must be noted that the number of LPS-related genes actually involved in polymyxin resistance might be even greater, since many of them are essential for cell survival and growth and, as mentioned above, would not be identified in a mutant screening. However, these data do not per se imply that a particular length of LPS core is required for polymyxin susceptibility, since mutants with mutations in the O-antigenic polysaccharide or rough core transferases, which are viable, did not demonstrate polymyxin resistance in our screen.
Resistance to other antibiotics and antimicrobial peptides.
Interestingly, there was a significant overlap between the genes identified in our polymyxin B screening and genes involved in β-lactam resistance (14), although the impacts were often opposite. Thus, while the PA0401, pyrB, and pyrD mutants were more susceptible to β-lactams than the wild type, strains with mutations in galU, ampR, lptC, aroB, wapR, and ssg, in contrast, all displayed decreased susceptibility to β-lactams while being more sensitive to polymyxin B. Further overlaps with comprehensive mutant library screenings include sucC, which showed low-level resistance to the aminoglycoside tobramycin (19), as well as pdxB, sucC, and lptC, which were all more susceptible to ciprofloxacin (15). The fact that many of the same genes influence the susceptibility of Pseudomonas to different antibiotic classes is interesting as it suggests that certain genes coordinate multidrug resistance possibly by affecting regulatory pathways. Understanding these resistance networks could be the key for the development of novel antimicrobial therapy strategies as well as our understanding of microbial adaptations such as biofilm formation and swarming motility, which lead to broad-spectrum resistance, in susceptibility to antibiotics.
We also determined the MICs of these strains to other cationic antimicrobial peptides, namely, colistin, CP28, indolicidin, and LL37, excluding from this assay 6 strains with defective growth (the pyrC, pyrB, pdxB, pyrD, tpiA, and aroB mutants). All of the tested transposon mutants had altered susceptibility to colistin, and all except the parR and agmS mutants were additionally altered in susceptibility to one or more antimicrobial peptides (Table 1). The galU, lptC, and ssg mutants were supersusceptible to all peptides tested, whereas the sucC, ampR, pmrA, and wapR mutants were less resistant to CP28 and LL37 but not to indolicidin. The PA4069 mutant showed the same level of supersusceptibility to indolicidin and LL37 as the wild type, but it was more susceptible to CP28. The increased resistance displayed by the phoQ mutant was almost exclusively to polymyxins, while only a modest 2-fold increase in resistance to the human cathelicidin LL37 was observed. These results suggest that the mutations analyzed in this study differently affected the antimicrobial activity of cationic peptides depending, presumably, on the specific properties of each peptide.
Gene expression in the presence of polymyxin B.
The dysregulation of resistance determinants is thought to be an important factor in adaptive resistance. Therefore, we set out to determine whether any of the genes identified in this screening are dysregulated when the cells are grown with subinhibitory polymyxin B (0.25 μg/ml) or upon exposure to a lethal concentration of polymyxin B (2 μg/ml) for 30 min after reaching mid-logarithmic phase (Table 2). As a control, we evaluated the expression of the gene arnB, which is part of the LPS modification operon and is known to be upregulated by polymyxin B (11). As expected, arnB was induced by both subinhibitory and lethal polymyxin B concentrations (4- and 16-fold, respectively). The pmrA gene was also induced by subinhibitory polymyxin B (approximately 3-fold) but only slightly by a lethal treatment (approximately 2-fold). Additionally, we observed a 2-fold and an 8-fold induction of the gene galU by sublethal and lethal polymyxin B, respectively, as well as increased transcription of lptC (approximately 3-fold) following a challenge with lethal polymyxin B. Expression of galU is also induced by polymyxin B in P. mirabilis through activation by the response regulator RppA (39).
Table 2.
Analysis by RT-qPCR of the dysregulation of the genes identified in the screening by a subinhibitory (0.25-μg/ml) or a lethal (2-μg/ml) concentration of polymyxin B
| PAO1 ortholog | Gene | Fold expression change (mean ± SD)a with polymyxin B concn of: |
|
|---|---|---|---|
| 0.25 μg/ml (sublethal)b | 2 μg/ml (lethal)c | ||
| PA0401 | 0.8 ± 0.2 | 0.7 ± 0.1 | |
| PA1375 | pdxB | 0.8 ± 0.4 | 0.7 ± 1.6 |
| PA1558 | sucC | 1.2 ± 0.1 | 1.4 ± 0.5 |
| PA2023 | galU | 2.0 ± 0.8 | 8.0 ± 1.3 |
| PA3050 | pyrD | 0.9 ± 0.1 | 0.8 ± 0.2 |
| PA3552d | arnB | 4.4 ± 2.8 | 16.3 ± 6.8 |
| PA4020 | mpl | 1.21 ± 0.20 | 1.48 ± 0.1 |
| PA4069 | 1.0 ± 0.2 | 1.5 ± 0.1 | |
| PA4109 | ampR | 1.4 ± 0.2 | 1.1 ± 0.1 |
| PA4459 | lptC | 1.1 ± 0.2 | 2.9 ± 0.4 |
| PA4748 | tpiA | 0.9 ± 0.1 | 1.7 ± 0.3 |
| PA5000 | wapR | 1.4 ± 0.1 | 1.0 ± 0.7 |
| PA4776 | pmrA | 2.8 ± 0.5 | 1.9 ± 0.1 |
| PA5001 | ssg | 1.4 ± 0.4 | 1.1 ± 0.2 |
| PA5038 | aroB | 0.8 ± 0.1 | 1.1 ± 0.2 |
| PA5199 | amgS | 1.0 ± 0.4 | 0.6 ± 0.1 |
Boldface indicates that the gene was more than 2-fold dysregulated under the assay conditions.
Cells were grown to mid-log phase in BM2-glucose with a high Mg2+ concentration (2 mM) supplemented with sublethal (0.25 μg/ml) polymyxin B; gene expression is described relative to that of no-antibiotic control cells.
Cells were grown to mid-log phase in BM2-glucose with a high Mg2+ concentration (2 mM) and exposed for 30 min to the antibiotic; gene expression is described relative to that of no-antibiotic control cells.
The gene arnB (LPS modification operon) was used as a control as it is known to be upregulated by antimicrobial peptides.
Analysis of LPS from selected susceptible mutants.
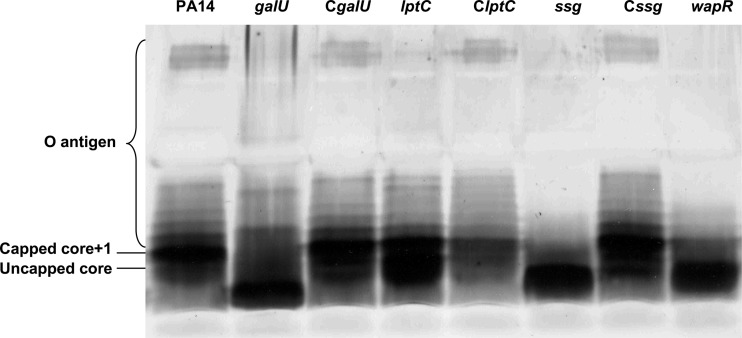
The polymyxin B-supersusceptible mutants carrying insertions in genes related to LPS biosynthesis, as well as other mutants, were further analyzed by SDS-PAGE to observe if they indeed had altered LPS patterns. The identities of the uncapped core and the capped core-plus-one bands were confirmed by Western blotting (data not shown). LPS was prepared from the amgS, mpl, ampR, PA4069, galU, lptC, wapR, and ssg mutants and compared to LPS from the PA14 wild-type strain. In such analyses, the most predominant band was the capped core substituted with one unit of O polysaccharide (capped core plus one), in contrast to strain PAO1, in which the most intense band is the fastest-migrating band, which represents the uncapped core. Visible differences were observed for the galU, lptC, wapR, and ssg mutants (Fig. 1), whereas the envZ, mpl, ampR, and PA4069 mutants showed no clear differences (data not shown). Three mutants, namely, the galU, wapR, and ssg mutants, demonstrated a prominent leading band with much higher mobility than the uncapped core band, consistent with a truncation in LPS yielding a rough core, and these mutants also lacked slower-migrating bands capped with O polysaccharide (Fig. 1). In contrast, the LPS of the lptC mutant was characterized by a greater abundance of uncapped core compared to that in the wild type, but it did not lack O-polysaccharide-capped species (Fig. 1). This was in part consistent with prior studies on galU and wapR mutants demonstrating a truncated core and lack of O polysaccharide (42, 46, 47). These observations were consistent with the predicted functions of the proteins encoded by these two genes. Thus, in the galU mutant, the lipid A-core would lack d-glucose, resulting in a truncated core. Therefore, as stated above, such a core oligosaccharide would not be capped (or substituted) by either O polysaccharide (46). In the case of wapR, its product is essential for the addition of the α-1,3-linked l-rhamnose to which both A- and B-band O-antigen polysaccharides are attached (41, 42). In contrast, no study had previously shown a similar phenotype produced by a mutation in PA5001 in this microorganism. However, the ssg mutant of P. alkylphenolia has an LPS with a truncated core and without O antigen (43).
Fig 1.
LPS profiles obtained by SDS-PAGE analysis of wild-type strain PA14 and the mutants with mutations in the genes galU, lptC, wapR, and ssg, as well as the complemented CgalU, ClptC, and Cssg strains.
To demonstrate that the observed phenotypes were due to the predicted mutations, complementation of three of the LPS-related genes was performed. These genes were cloned in the low-copy vector pBBR1MCS and then transformed into the respective mutant strains. All three complemented strains showed a wild-type LPS banding pattern as visualized in a silver-stained SDS-polyacrylamide gel (Fig. 1). Peptide susceptibility also decreased in the complemented strains compared to the mutants, although in a few cases, resistance did not reach wild-type levels (Table 3). As a result, we can conclude that the phenotypes displayed by these three mutants were caused by the transposon insertions in the genes galU, lptC, and ssg and were not due to secondary mutations.
Table 3.
MICs to different peptides of the wild-type PA14, the galU, lptC, and ssg mutants, and their respective complemented strains
| Strain | MIC (μg/ml) |
||||
|---|---|---|---|---|---|
| Polymyxin B | Colistin | CP28 | Indolicidin | LL-37 | |
| PA14 | 0.5 | 1 | 16 | 64 | 64 |
| galU | 0.25 | 0.5 | 4 | 16 | 16 |
| CgalU | 0.5 | 1 | 8 | 64 | 32 |
| lptC | 0.125 | 0.25 | 8 | 8 | 8 |
| ClptC | 0.5 | 1 | 16 | 64 | 16 |
| ssg | 0.25 | 0.5 | 8 | 32 | 16 |
| Cssg | 0.5 | 1 | 16 | 64 | 64 |
Previous studies had reported contradictory results about the link between rough LPS and resistance to cationic compounds, such as aminoglycosides and polymyxins. For example, Yokota and Fujii (48) showed that only deep-rough mutants were more susceptible, while other rough mutants demonstrated normal to decreased susceptibility. Below we describe experiments aimed at understanding the mechanism by which these mutants acquire increased susceptibility. However, as it is the lipid A region of LPS that binds polymyxin, we assume that the lack of O antigen and core truncations were not per se responsible for the polymyxin supersusceptibility.
Dansyl-polymyxin and NPN assays.
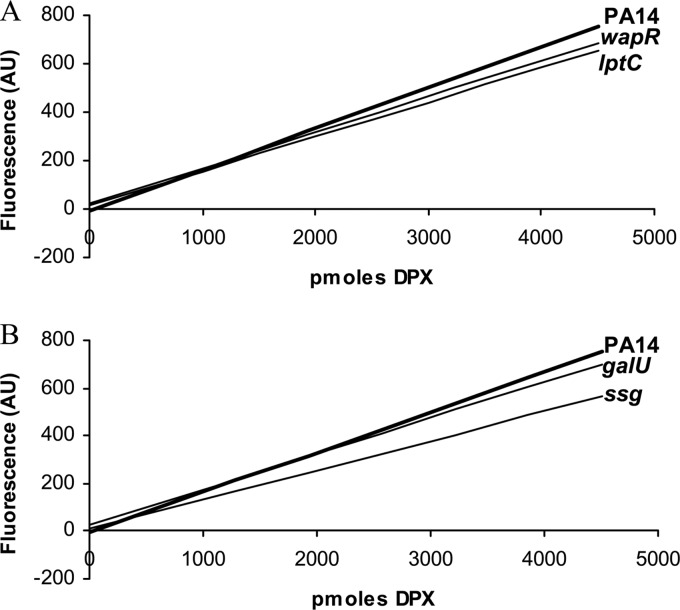
Although four of the supersusceptible mutants identified in the screening displayed changes in their LPS patterns compared to that of the wild type, this characteristic does not per se explain their increased susceptibility to polymyxins. One possible explanation for their phenotypes could be an increased binding of polymyxin B to the LPS on the cell surface of these strains, leading to enhanced uptake. To investigate this, the binding of dansyl-polymyxin to the LPS and to the cell surface was assessed. However, no significant difference was observed between any of the tested mutants (galU, lptC, wapR, and ssg) and the wild type, indicating that this was not the reason for the polymyxin susceptibility observed in these strains (Fig. 2).
Fig 2.
Binding of dansyl-polymyxin (DPX) to whole cells of different P. aeruginosa strains. (A) Wild-type PA14 and the lptC and wapR mutant strains; (B) wild-type PA14 and the galU and ssg mutant strains. The graphs represent trend lines calculated for each strain. A greater slope indicates greater affinity for dansyl-polymyxin. Previous experiments have demonstrated that dansyl-polymyxin binds principally to LPS in the outer membrane. The results shown correspond to one representative experiment out of three with the same trends. AU, arbitrary units.
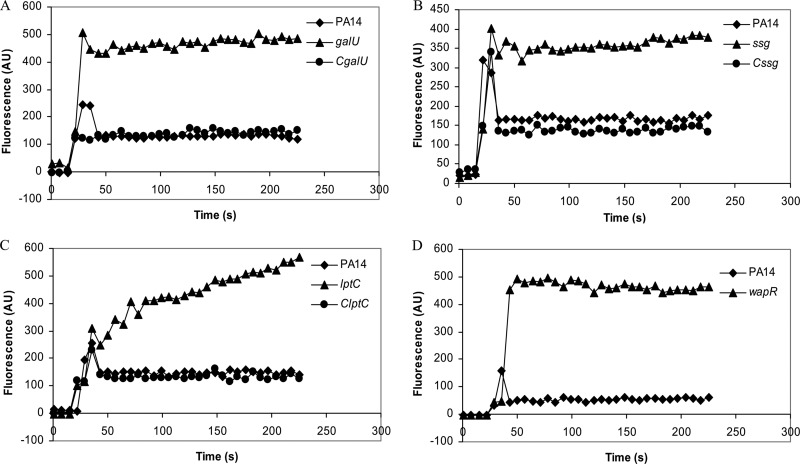
Another possibility is that defects in LPS structure increased the general permeability of the cell envelope, resulting in lower resistance to polymyxins. This can be analyzed by performing an NPN assay, in which the fluorescence emission due to partitioning of the hydrophobic fluorescent probe NPN into the outer membrane gives an indication of outer membrane permeability for the strains of interest. Thus, a greater emission of fluorescence would indicate greater permeability of the outer membrane to NPN. The results of this assay revealed that all four tested mutants, namely, the galU, lptC, wapR, and ssg mutants, exhibited greater outer membrane permeability than the wild-type PA14 (Fig. 3). In each case examined, the increased permeability to NPN was reduced in the complemented strains (Fig. 3). This provides a tenable explanation for their polymyxin B supersusceptibility, as the peptide would be able to translocate across the outer membrane more easily than in the wild type.
Fig 3.
Partitioning of NPN into the outer membranes of different P. aeruginosa strains. (A) Wild-type PA14, galU mutant, and complemented CgalU strain; (B) wild-type PA14, ssg mutant, and complemented Cssg strain; (C) wild-type PA14, lptC mutant, and complemented ClptC strain; (D) wild-type PA14 and wapR mutant. Greater emission of fluorescence corresponds to greater outer membrane permeability to NPN, which fluoresces only after it partitions into the membrane and is excluded by the outer membrane barrier of wild-type cells. The graphs show results from one representative experiment out of at least three with the same trends. AU, arbitrary units.
Analysis of lipid A modifications.
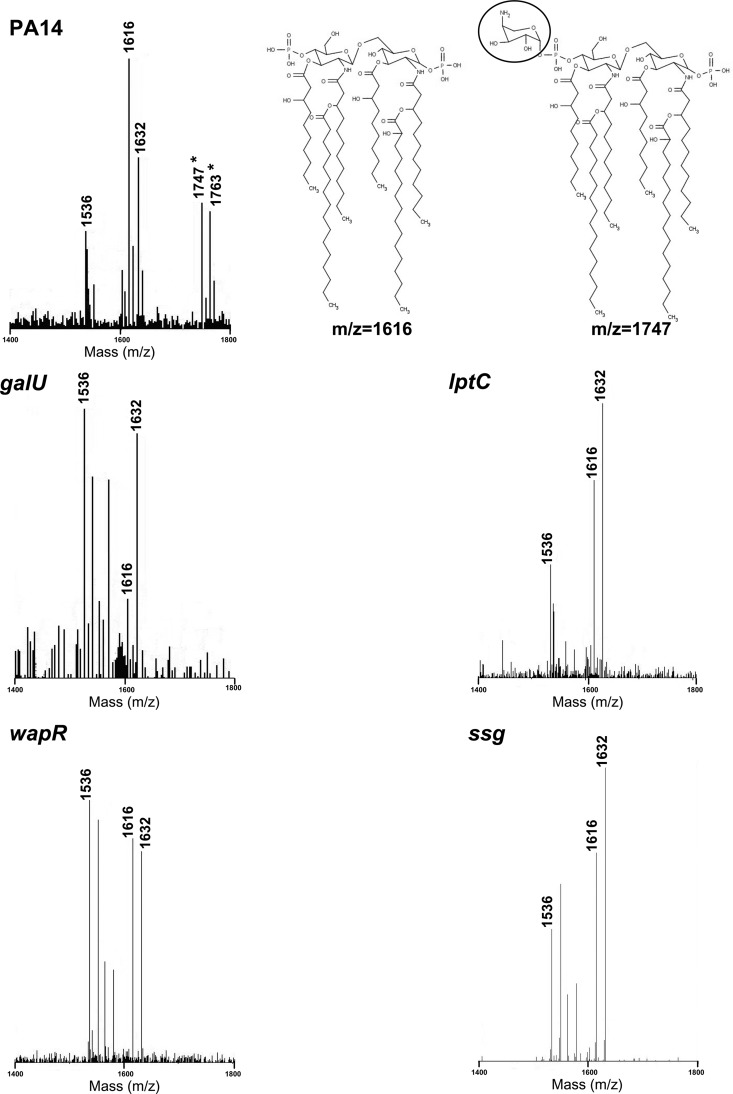
A previous study had shown that the level of resistance to polymyxins observed in vitro is related to the induction of lipid A modifications with aminoarabinose by polymyxins themselves (11). Using knockout mutants, it was demonstrated that the lack of aminoarabinose modification of LPS rendered the bacteria more susceptible to the inducing cationic antibiotic. Therefore, those mutants defective in the acquisition of such adaptation, such as the parR regulatory mutant and the arn operon effector mutants, exhibit increased susceptibility to this type of antibiotic (11, 13). To test whether lipid A modification was hampered in the four LPS mutants of interest (galU, lptC, wapR, and ssg), the lipid A species present in these strains were analyzed under low-Mg2+ growth conditions. These conditions are known to trigger LPS modification in P. aeruginosa, and, in agreement with this, lipid A analysis of the wild-type PA14 showed the presence of aminoarabinosylated ion peaks with m/z values of 1747 and 1763, which correspond, respectively, to the hexa-acylated species with m/z values of 1616 and 1632 with the addition of one aminoarabinose molecule (Fig. 4). In contrast, lipid A analysis by MALDI-TOF MS indicated that all four LPS mutants lacked the aminoarabinosylated forms observed in the wild type (Fig. 4). A decrease in the presence and/or inducibility of aminoarabinose-modified lipid A species would, therefore, provide an additional explanation for the supersusceptible phenotype of these mutants.
Fig 4.
MALDI-TOF MS reflectron mode analysis of lipid A from the PA14, galU, lptC, wapR, and ssg strains and structures of selected lipid A species, showing the aminoarabinose in the modified species inside a circle. Only selected peaks of interest are labeled for the sake of clarity. Differences in m/z units of 16 and 80 correspond to the loss or addition of a hydroxyl or a phosphate group, respectively. Asterisks represent aminoarabinosylated lipid A species.
To determine if the loss of lipid A modification was due to a lower level of expression of the LPS modification (arn) operon under low-Mg2+ growth conditions, we performed RT-qPCR experiments. RNA was prepared from cultures of the wild-type PA14 as well as the mutants with mutations in the genes galU, lptC, wapR, and ssg grown in a low (20 μM)-Mg2+ BM2 medium. However, this did not reveal any defect in the transcription of the arnB gene (the first gene in the arn operon) in any of the mutants compared to the wild type (Table 4). Therefore, it seems likely that the mechanisms determining this altered phenotype occur at the posttranscriptional level, possibly in the modification, with aminoarabinose, of the structurally altered mutant LPS.
Table 4.
RT-qPCR analysis of the expression of the arn operon under low-magnesium conditions in selected mutant strains relative to wild-type PA14
| Mutant | Fold change (mean ± SD) in arnB expression |
|---|---|
| galU | 1.51 ± 0.30 |
| lptC | 1.94 ± 0.25 |
| wapR | 1.28 ± 0.22 |
| ssg | 1.45 ± 0.47 |
In the case of galU, previous studies with P. mirabilis (39) and Y. pestis (40) revealed an impact of the galU mutation on lipid A modification and polymyxin B susceptibility. The product of galU, UDP-glucose phosphorylase, catalyzes the biosynthesis of UDP-glucose (49), a precursor of aminoarabinose, so the most likely explanation for the loss of lipid A modification would be the inability of this mutant strain to synthesize aminoarabinose. Further experiments will be necessary to explain why the other three mutants, namely, the lptC, wapR, and ssg mutants, did not exhibit aminoarabinosylated lipid A species under low-magnesium conditions. Nonetheless, it can be speculated that the altered LPS structure observed in these mutants might hinder the addition of aminoarabinose to the lipid A.
Concluding remarks.
P. aeruginosa exhibits a quite small intrinsic polymyxin B resistome, as well as a relatively limited capacity for mutational resistance through gene knockout, compared to other types of antimicrobials. However, it must be noted that, due to the limitations associated with screening a transposon mutant library, not all P. aeruginosa genes could be evaluated for their participation in polymyxin B resistance. For example, the library does not have transposon mutants corresponding to essential genes (for instance, waaF and waaC, encoding heptosyltransferases, and waaP, encoding a sugar kinase involved in phosphorylation of the heptose residues in the inner core [50]), as these would not be viable. However, the screen did identify a transposon insertion in the gene encoding the sensor kinase PhoQ that conferred high-level polymyxin resistance. This correlates with previous results showing that mutations that inactivate phoQ confer resistance to polymyxin B and other peptides. The identification of clinical isolates harboring mutations that constitutively activate PhoP (10, 35) emphasizes the importance and clinical relevance of understanding the pathways regulating modification of lipid A by aminoarabinose in P. aeruginosa.
Maintenance of the structure and stability of LPS appears to be the prevalent mechanism of polymyxin B intrinsic resistance in this microorganism, and alterations in this structure lead to susceptibility to polymyxins and, in many cases, also to other antimicrobial peptides. Certain mutations affecting LPS render the bacteria more susceptible by increasing outer membrane permeability and/or inhibiting the addition of aminoarabinose to lipid A. Interestingly, many of the mutations leading to supersusceptibility to polymyxins were found to also confer increased resistance to β-lactams. Furthermore, overlaps with aminoglycoside and fluoroquinolone resistance screenings were also observed. The finding of such overlapping mutants among these screenings highlights the importance of defining the P. aeruginosa resistome. This information will be very useful in defining critical overlaps in adaptive resistance that might drive multidrug resistance and for the design of improved therapies that exploit redundancies between the resistance mechanisms to different antibiotic classes.
Overall, these results provide us with a clearer picture of the mechanisms of resistance to polymyxins and other antimicrobial peptides in P. aeruginosa. The apparently well-defined intrinsic resistance determinants together with the relatively limited possibilities for mutational resistance make this type of antibiotic very useful as an antipseudomonal agent, as long as it is used appropriately. Moreover, our data give an indication of which genes should be analyzed to follow the evolution of polymyxin resistance in the clinic.
ACKNOWLEDGMENTS
This work was funded by grants from Cystic Fibrosis Canada (CFC) to R.E.W.H. and J.S.L. and grants BIO2011-25255, HEALTH-F3-2011-282004, and HEALTH-F3-2010-241476 to J.L.M. L.F. received a postdoctoral fellowship from the Fundacion Alfonso Martin Escudero (Spain). I.W. was supported by the Juergen Manchot Foundation and the Mukoviszidose e.V., Bonn, Germany (German Cystic Fibrosis Association). J.O. is the recipient of a fellowship from Programa Beca Chile, CONICYT. D.K. received a postdoctoral fellowship from CFC. R.E.W.H. holds a Canada Research Chair in Microbiology, and J.S.L. holds a Canada Research Chair in Cystic Fibrosis and Microbial Glycobiology.
We thank Suzanne Perry, M. Stephen Trent, and Shaan Gellatly for their help and advice with the LPS and lipid A experiments.
Footnotes
Published ahead of print 15 October 2012
REFERENCES
- 1. Bonomo RA, Szabo D. 2006. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43:SS49–SS56 [DOI] [PubMed] [Google Scholar]
- 2. Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051–1060 [DOI] [PubMed] [Google Scholar]
- 3. Rowe SM, Miller S, Sorscher EJ. 2005. Cystic fibrosis. N. Engl. J. Med. 352:1992–2001 [DOI] [PubMed] [Google Scholar]
- 4. Hancock REW, Speert DP. 2000. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist. Updat. 3:247–255 [DOI] [PubMed] [Google Scholar]
- 5. Falagas ME, Kasiakou SK. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40:1333–1341 [DOI] [PubMed] [Google Scholar]
- 6. Zhang L, Dhillon P, Yan H, Farmer S, Hancock REW. 2000. Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3317–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Falagas ME, Rafailidis PI, Matthaiou DK. 2010. Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist. Updat. 13:132–138 [DOI] [PubMed] [Google Scholar]
- 8. Miller AK, Brannon MK, Stevens L, Johansen HK, Selgrade SE, Miller SI, Høiby N, Moskowitz SM. 2011. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob. Agents Chemother. 55:5761–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moskowitz SM, Brannon MK, Dasgupta N, Pier M, Sgambati N, Miller AK, Selgrade SE, Miller SI, Denton M, Conway SP, Johansen HK, Høiby N. 2012. PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients. Antimicrob. Agents Chemother. 56:1019–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Muller C, Plésiat P, Jeannot K. 2011. A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and β-lactams in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55:1211–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fernández L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock REW. 2010. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob. Agents Chemother. 54:3372–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Macfarlane ELA, Kwasnicka A, Ochs MM, Hancock REW. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34:305–316 [DOI] [PubMed] [Google Scholar]
- 13. McPhee JB, Lewenza S, Hancock REW. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50:205–217 [DOI] [PubMed] [Google Scholar]
- 14. Alvarez-Ortega C, Wiegand I, Olivares J, Hancock REW, Martínez JL. 2010. Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to beta-lactam antibiotics. Antimicrob. Agents Chemother. 54:4159–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Breidenstein EBM, Khaira BK, Wiegand I, Overhage J, Hancock REW. 2008. Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob. Agents Chemother. 52:4486–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Groote VN, Verstraeten N, Fauvart M, Kint CI, Verbeeck AM, Beullens S, Cornelis P, Michiels J. 2009. Novel persistence genes in Pseudomonas aeruginosa identified by high-throughput screening. FEMS Microbiol. Lett. 297:73–79 [DOI] [PubMed] [Google Scholar]
- 17. Dötsch A, Becker T, Pommerenke C, Magnowska Z, Jänsch Häussler LS. 2009. Genomewide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:2522–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gallagher LA, Shendure J, Manoil C. 2011. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2(1):e00315–10 doi:10.1128/mBio.00315–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schurek KN, Marr AK, Taylor PK, Wiegand I, Semenec L, Khaira BK, Hancock REW. 2008. Novel genetic determinants of low-level aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52:4213–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fernández L, Breidenstein EBM, Hancock REW. 2011. Creeping baselines and adaptive resistance to antibiotics. Drug Resist. Updat. 14:1–21 [DOI] [PubMed] [Google Scholar]
- 21. Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103:2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fajardo A, Martínez-MartíN N, Mercadillo M, Galán JC, Ghysels B, Matthijs S, Cornelis P, Wiehlmann L, Tümmler B, Baquero F, Martínez JL. 2008. The neglected intrinsic resistome of bacterial pathogens. PLoS One 3:e1619 doi:10.1371/journal.pone.0001619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wiegand I, Hilpert K, Hancock REW. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3:163–175 [DOI] [PubMed] [Google Scholar]
- 24. McPhee JB, Bains M, Winsor G, Lewenza S, Kwasnicka A, Brazas MD, Brinkman FS, Hancock REW. 2006. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 188:3995–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hitchcock PJ, Brown TM. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fomsgaard A, Freudenberg MA, Galanos C. 1990. Modification of the silver staining technique to detect lipopolysaccharide in polyacrylamide gels. J. Clin. Microbiol. 28:2627–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou Z, Lin S, Cotter RJ, Raetz CR. 1999. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-l-arabinose, phosphoethanolamine and palmitate J. Biol. Chem. 274:18503–18514 [DOI] [PubMed] [Google Scholar]
- 28. Moore RA, Bates NC, Hancock REW. 1986. Interaction of polycationic antibiotics with Pseudomonas aeruginosa lipopolysaccharide and lipid A studied by using dansyl-polymyxin. Antimicrob. Agents Chemother. 29:496–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Loh B, Grant C, Hancock REW. 1984. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 26:546–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rozen S, Skaletsky HJ. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 31. Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39:D596–D600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Elzer PH, Kovach ME, Phillips RW, Robertson GT, Peterson KM, Roop RM., II 1995. In vivo and in vitro stability of the broad host-range cloning vector pBBR1MCS in six Brucella species. Plasmid 33:51–57 [DOI] [PubMed] [Google Scholar]
- 33. Baquero F. 2001. Low-level antibacterial resistance: a gateway to clinical resistance. Drug Resist. Updat. 4:93–105 [DOI] [PubMed] [Google Scholar]
- 34. Gooderham WJ, Gellatly SL, Sanschagrin F, McPhee JB, Bains M, Cosseau C, Levesque RC, Hancock REW. 2009. The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa. Microbiology 155:699–711 [DOI] [PubMed] [Google Scholar]
- 35. Barrow K, Kwon DH. 2009. Alterations in two-component regulatory systems of phoPQ and pmrAB are associated with polymyxin B resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:5150–5154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Masoud H, Altman E, Richards JC, Lam JS. 1994. A general strategy for structual analysis of the oligosaccharide region of lipopolysaccharides. Structure of oligosaccharide component of Pseudomonas aeruginosa IATS serotype O6 mutant R5 rough-type lipopolysaccharide. Biochem. 33:10568–10578 [DOI] [PubMed] [Google Scholar]
- 37. Priebe GP, Dean CR, Zaidi T, Meluleni GJ, Coleman FT, Coutinho YS, Noto MJ, Urban TA, Pier GB, Goldberg JB. 2004. The galU Gene of Pseudomonas aeruginosa is required for corneal infection and efficient systemic spread following pneumonia but not for infection confined to the lung. Infect. Immun. 72:4224–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin J, Wang Y, Hoang KV. 2009. Systematic identification of genetic loci required for polymyxin resistance in Campylobacter jejuni using an efficient in vivo transposon mutagenesis system. Foodborne Pathog. Dis. 6:173–185 [DOI] [PubMed] [Google Scholar]
- 39. Jiang SS, Lin TY, Wang WB, Liu MC, Hsueh PR, Liaw SJ. 2010. Characterization of UDP-glucose dehydrogenase and UDP-glucose pyrophosphorylase mutants of Proteus mirabilis: defectiveness in polymyxin B resistance, swarming, and virulence. Antimicrob. Agents Chemother. 54:2000–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klein KA, Fukuto HS, Pelletier M, Romanov G, Grabenstein JP, Palmer LE, Ernst R, Bliska JB. 2012. A transposon site hybridization screen identifies galU and wecBC as important for survival of Yersinia pestis in murine macrophages. J. Bacteriol. 194:653–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kocíncová D, Ostler SL, Anderson EM, Lam JS. 2012. Rhamnosyltransferase genes migA and wapR are regulated in a differential manner to modulate the quantities of core oligosaccharide glycoforms produced by Pseudomonas aeruginosa. J. Bacteriol. 194:4295–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Poon KK, Westman EL, Vinogradov E, Jin S, Lam JS. 2008. Functional characterization of MigA and WapR: putative rhamnosyltransferases involved in outer core oligosaccharide biosynthesis of Pseudomonas aeruginosa. J. Bacteriol. 190:1857–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Veeranagouda Y, Lee K, Cho AR, Cho K, Anderson EM, Lam JS. 2011. Ssg, a putative glycosyltransferase, functions in lipo- and exopolysaccharide biosynthesis and cell surface-related properties in Pseudomonas alkylphenolia. FEMS Microbiol. Lett. 315:38–45 [DOI] [PubMed] [Google Scholar]
- 44. Sperandeo P, Lau FK, Carpentieri A, De Castro C, Molinaro A, Dehò G, Silhavy TJ, Polissi A. 2008. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J. Bacteriol. 190:4460–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rahim R, Burrows LL, Monteiro MA, Perry MB, Lam JS. 2000. Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa. Microbiology 146:2803–2814 [DOI] [PubMed] [Google Scholar]
- 46. Choudhury B, Carlson RW, Goldberg JB. 2005. The structure of the lipopolysaccharide from a galU mutant of Pseudomonas aeruginosa serogroup-O11. Carbohydr. Res. 340:2761–2772 [DOI] [PubMed] [Google Scholar]
- 47. Dean CR, Goldberg JB. 2002. Pseudomonas aeruginosa galU is required for a complete lipopolysaccharide core and repairs a secondary mutation in a PA103 (serogroup O11) wbpM mutant. FEMS Microbiol. Lett. 210:277–283 [DOI] [PubMed] [Google Scholar]
- 48. Yokota S, Fujii N. 2007. Contributions of the lipopolysaccharide outer core oligosaccharide region on the cell surface properties of Pseudomonas aeruginosa. Comp. Immunol. Microbiol. Infect. Dis. 30:97–109 [DOI] [PubMed] [Google Scholar]
- 49. Nesper J, Lauriano CM, Klose KE, Kapfhammer D, Kraiss A, Reidl J. 2001. Characterization of Vibrio cholerae O1 El tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69:435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lam JS, Taylor VL, Islam ST, Hao Hao Y, Kocíncová D. 2011. Genetics and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front. Microbiol. 2:118. [DOI] [PMC free article] [PubMed] [Google Scholar]