Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) has been a major cause of nosocomial infection in Irish hospitals for 4 decades, and replacement of predominant MRSA clones has occurred several times. An MRSA isolate recovered in 2006 as part of a larger study of sporadic MRSA exhibited a rare spa (t878) and multilocus sequence (ST779) type and was nontypeable by PCR- and DNA microarray-based staphylococcal cassette chromosome mec (SCCmec) element typing. Whole-genome sequencing revealed the presence of a novel 51-kb composite island (CI) element with three distinct domains, each flanked by direct repeat and inverted repeat sequences, including (i) a pseudo SCCmec element (16.3 kb) carrying mecA with a novel mec class region, a fusidic acid resistance gene (fusC), and two copper resistance genes (copB and copC) but lacking ccr genes; (ii) an SCC element (17.5 kb) carrying a novel ccrAB4 allele; and (iii) an SCC element (17.4 kb) carrying a novel ccrC allele and a clustered regularly interspaced short palindromic repeat (CRISPR) region. The novel CI was subsequently identified by PCR in an additional 13 t878/ST779 MRSA isolates, six from bloodstream infections, recovered between 2006 and 2011 in 11 hospitals. Analysis of open reading frames (ORFs) carried by the CI showed amino acid sequence similarity of 44 to 100% to ORFs from S. aureus and coagulase-negative staphylococci (CoNS). These findings provide further evidence of genetic transfer between S. aureus and CoNS and show how this contributes to the emergence of novel SCCmec elements and MRSA strains. Ongoing surveillance of this MRSA strain is warranted and will require updating of currently used SCCmec typing methods.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is a significant problem in hospitals and communities worldwide, and awareness of MRSA in animals and reports of its zoonotic spread have increased in recent years (1, 2). The success of MRSA is in part due to its ability to adapt rapidly to changing environments through the acquisition of mobile genetic elements (MGE) that harbor antimicrobial resistance determinants or virulence-associated genes which form part of the accessory genome (1). Resistance to methicillin and β-lactam antibiotics in staphylococci is determined by penicillin binding protein 2a (PBP2a) encoded by the methicillin resistance gene mecA (3). In MRSA, two distinct mecA gene types have been described and are carried on a large MGE termed the staphylococcal cassette chromosome mec (SCCmec) (4, 5). Both gene types were originally termed mecA; however, the second gene has recently been renamed mecC based on its significant divergence from the classical mecA gene type (5). Numerous alleles of the mecA gene type have also been described (5, 6).
The SCCmec element is highly variable, with extensive diversity identified in this cassette in different staphylococcal species, including the 11 SCCmec types and numerous subtypes from MRSA (4, 7, 8). Considerable indirect evidence has been reported for the horizontal transfer of SCCmec DNA between S. aureus and coagulase-negative staphylococci (CoNS), and SCCmec is more diverse and abundant among CoNS (9). While the mechanism(s) of transfer is unknown, similar SCCmec elements have been found in CoNS and S. aureus, in some cases from the same patient (10). CoNS may constitute a potentially significant reservoir for antibiotic resistance genes in S. aureus and may have a significant impact on the emergence of novel MRSA strains (11). SCCmec inserts into the 3′ end of the chromosomally located orfX gene and is characterized by the presence of flanking imperfect direct repeat (DR) sequences that are generated at both ends of the element following insertion into orfX. SCCmec elements harbor two fundamental components, the mec gene complex and the cassette chromosome recombinase (ccr) gene complex, and each SCCmec element is characterized by a unique combination of these genes. SCC elements harboring ccr genes but without mecA and SCC-like elements without ccr and mec genes have also been reported within orfX and flanked by DRs in staphylococci and often harbor additional virulence or antimicrobial resistance genes (9, 12).
The mec gene complex consists of mecA and, when present, the mec regulatory genes mecR1 and mecI (7). Five classes of the mec gene complex (A to E) have been reported to date in staphylococci (7) (www.sccmec.org). The SCCmec-carried ccr genes are necessary for precise integration and excision of the SCCmec element, and three genes (ccrA, ccrB, and ccrC) have been described. Novel ccr genes and any subsequent subtypes are assigned new designations based upon guidelines published in 2009 (7), which take the sequence similarity of any previously published or forthcoming novel ccr genes into consideration. Each ccr complex consists of either the ccrA and ccrB genes together or ccrC and an associated open reading frame (ORF), previously termed ccrAA (13), which is located directly upstream of ccrC and exhibits between 35 and 41% DNA sequence similarity to ccr genes ccrA, ccrB, and ccrC. Eight types of the ccr gene complex have been reported to date in MRSA, each with a different combination of ccrA and ccrB allotypes or ccrC (7) (www.sccmec.org). Numerous allelic variants of each of the ccr allotypes have been reported based upon this criterion; however, the nomenclature is complicated, as not all variants have been assigned allelic numbers. For example, in recent years, five alleles of the ccrA4 and ccrB4 allotypes have been reported without designated allelic prefixes and 10 alleles of the ccrC1 allotype (ccrC1 to ccrC10) have been assigned in both MRSA and CoNS (9, 13–20).
MRSA has now been endemic in Ireland for over 3 decades, and clonal replacement has occurred on several occasions during this period (21–24). Over the last decade, MRSA isolates exhibiting sequence type 22 (ST22) and harboring SCCmec type IV (ST22-MRSA-IV) have predominated, accounting for approximately 80% of MRSA isolates recovered from patients in Irish hospitals (24). In the present study, we report the detailed molecular characterization of human clinical MRSA isolates recovered in Irish hospitals between 2006 and 2011 which exhibited a rare ST (ST779) and spa type (t878). Whole-genome sequencing of a representative isolate revealed a novel composite pseudo SCCmec-SCC-SCCCRISPR element carrying a clustered randomly interspersed palindromic repeat (CRISPR) region that encodes a prokaryotic defense mechanism against foreign DNA. The novel element was subsequently identified in all 14 of the ST779 isolates investigated.
MATERIALS AND METHODS
Bacterial isolates.
MRSA isolate M06/0171 was recovered in 2006 in an Irish pediatric hospital and was initially identified as part of an investigation into 58 sporadically occurring MRSA isolates recovered in Irish hospitals between 2000 and 2006 (Table 1). M06/0171 exhibited spa type t878, but its SCCmec type could not be determined by conventional SCCmec typing PCRs or by DNA microarray profiling. Whole-genome sequencing of M06/0171 was undertaken to determine the genetic organization of its SCCmec element. The database of isolates submitted to the Irish National MRSA Reference Laboratory (NMRSARL) was subsequently examined for other spa type t878 isolates. Between 2006 and 2011, a total of 4,320 MRSA isolates were investigated by the NMRSARL, and approximately 80% were characterized as non-multiantibiotic-resistant phenotype AR06, indicative of ST22-MRSA-IV (22), the pandemic strain currently circulating in Irish hospitals. Half of the non-ST22-MRSA-IV isolates were spa typed during this time period, and 13 additional spa type t878 MRSA isolates were identified among the 431 MRSA isolates that were spa typed (Table 1). These isolates were investigated by DNA microarray profiling and detailed SCCmec analysis.
Table 1.
Epidemiological, clinical, phenotypic, and genotypic characteristics of the 14 ST779 and spa type t878 MRSA isolates harboring the novel pseudo SCCmec-SCC-SCCCRISPR element recovered in Irish hospitals between 2006 and 2011
| Hospital no. | Isolate no. | Yr of isolation | Agea | Clinical details (sex) | Antimicrobial resistance patternb,c | dru type | DNA microarray analysise |
||
|---|---|---|---|---|---|---|---|---|---|
| SCCmec genese | Antimicrobial resistance genese | Virulence-associated genese,f | |||||||
| H1 | M06/0171 | 2006 | 3 y | Burn unit (female) | AMP, COP, FUS, MUP, NEO, TOB | dt8af | mecA, ugpQ, ccrAA, ccrC,g ccrA4, ccrB4 | blaZ, fusC, sdrM, aadD, mupA | seb, sak, chp, scn, etD, edinB, clfB, sdrD |
| H2 | E4233 | 2009 | 45 y | BSIi (female) | AMP, COP, FUS | dt8af | mecA, ugpQ, ccrB4g | blaZ, fusC, sdrM | seb,g sed, sej, ser, sak, chp, scn, etD, edinB, clfB,g sdrDg |
| H3 | M11/0114 | 2011 | 5 d | Screening sample, baby of patient from whom M11/0118 was recovered (N/A)h | AMP, COP, FUS | dt8af | mecA, ugpQ, ccrAA,g ccrB4g | blaZ, fusC, sdrM | seb,g sak, chp, scn, etD, edinB, clfB,g sdrDg |
| H3 | M11/0118 | 2011 | 30 y | Screening sample, mother of baby from whom M11/0114 was recovered (female) | AMP, COP, FUS | dt8af | mecA, ugpQ, ccrAA, ccrC, ccrB4 | blaZ, fusC, sdrM | seb, sak, chp, scn, etD, edinB, clfB, sdrDg |
| H4 | E4449 | 2010 | 39 y | BSI (male) | AMP, COP, CAD,d FUS | dt11y | mecA, ugpQ, ccrAA,g ccrC,g ccrB4 | blaZ, fusC, sdrM | seb,g sed, sej, ser, sak, chp, scn, etD, edinB, clfB, sdrD |
| H4 | E2998 | 2006 | 54 y | BSI (male) | AMP, COP, FUS | dt11y | mecA, ugpQ, ccrAA, ccrB4 | blaZ, fusC, sdrM | seb, sak, chp, scn, etD, edinB, clfB, sdrD |
| H5 | E4550 | 2010 | 55 y | BSI (female) | AMP, COP, CAD,d FUS | dt11y | mecA, ugpQ, ccrAA,g ccrC,g ccrB4 | blaZ, fusC, sdrM | seb, sed, sej, ser, sak, chp, scn, etD, edinB, clfB, sdrD |
| H6 | M11/0208 | 2011 | 18 y | Dermatology clinic (male) | AMP, COP, CAD,d FUS | dt11y | mecA, ugpQ, ccrB4g | blaZ, fusC, sdrM | seb,g sed, sej, ser, sak, chp, scn, etD, edinB, sdrD,g clfBg |
| H7 | M08/0422 | 2008 | 24 y | Screening sample (female) | AMP, COP, CAD, FUS | dt11y | mecA, ugpQ | blaZ, fusC, sdrM | seb,g sak, chp, scn, etD, edinB, clfB,g sdrD |
| H8 | M07/0307 | 2007 | Stillborn | Stillborn baby postmortem (N/A)h | AMP, COP, FUS | dt11y | mecA, ugpQ, ccrAA, ccrC,g ccrB4 | blaZ, fusC, sdrM | seb,g sak, chp, scn, etD, edinB, clfB,g sdrD |
| H9 | M09/0295 | 2009 | 41 y | Screening sample (male) | AMP, COP, FUS | dt11y | mecA, ugpQ, ccrAA,g ccrB4g | blaZ, fusC, sdrM | seb,g sed, sej, ser, sak, chp, scn, etD, edinB, sdrD,g clfBg |
| H10 | E4709 | 2010 | 54 y | BSI (female) | AMP, COP, FUS | dt11y | mecA, ugpQ, ccrAA,g ccrC, ccrB4 | blaZ, fusC, sdrM | seb,g sed, sej, ser, sak, chp, scn, etD, edinB, clfB, sdrD |
| H11 | M09/0302 | 2009 | 46 y | Screening sample (male) | AMP, COP, FUS | dt10aj | mecA, ugpQ, ccrAA,g ccrB4g | blaZ, fusC, sdrM | seb,g sak, chp, scn, etD, edinB, sdrD,g clfBg |
| H12 | E4217 | 2009 | 59 y | BSI (male) | AMP, COP, FUS | dt11bm | mecA, ugpQ, ccrAA,g ccrC, ccrB4 | blaZ, fusC, sdrM | seb,g sak, chp, scn, etD, edinB, clfB, sdrD |
Age of patient; y, years; d, days.
Antimicrobial resistance was determined by antibiogram-resistogram typing against a panel of 23 antimicrobial agents including amikacin, ampicillin (AMP), cadmium acetate (CAD), chloramphenicol, ciprofloxacin, erythromycin, ethidium bromide, fusidic acid (FUS), gentamicin, kanamycin, lincomycin, mercuric chloride, mupirocin (MUP), neomycin (NEO), phenyl mercuric acetate, rifampin, spectinomycin, streptomycin, sulfonamide, tetracycline, tobramycin (TOB), trimethoprim, and vancomycin (25).
Isolate M06/0171 was tested for susceptibility to copper sulfate (COP) by the CLSI agar plate dilution methodology (26). Copper resistance in the remaining 13 ST779 MRSA isolates was confirmed by the CLSI disk diffusion methodology (26).
These isolates exhibited intermediate resistance to cadmium acetate.
SCCmec, antimicrobial resistance and virulence-associated genes were detected using the StaphyType DNA microarray kit (Alere, Germany) (34). ccrAA is a known ccrC-linked gene with 35 to 41% DNA sequence homology to other ccr genes.
The following MSCRAMM, adhesion, and biofilm formation genes were detected in all 14 ST779/t878 MRSA isolates by DNA microarray analysis: icaA, icaC, icaD, bbp, clfA, ebh, ebpS, eno, fib, fnbA, fnbB, sdrC, vwb, and sasG.
Ambiguous or negative DNA microarray signals were obtained for the genes and isolates indicated. The presence of seb, clfB, and sdrD was confirmed in all 14 MRSA isolates by PCR.
N/A, information not available.
BSI, bloodstream infection.
All isolates were identified as S. aureus using the tube coagulase test, and methicillin resistance was detected using 10-μg and 30-μg cefoxitin disks (Oxoid Ltd., Basingstoke, United Kingdom).
AR typing.
All isolates were subjected to antibiogram-resistogram (AR) typing as described previously (25).
Copper resistance.
All isolates were tested for susceptibility to copper sulfate (Sigma-Aldrich Chemical Company, Tallaght, Dublin, Ireland). One isolate, M06/0171, was tested using 0.125, 0.250, 0.5, 1, 2, 4, 8, and 16 mM concentrations and the Clinical and Laboratory Standards Institute (CLSI) agar plate dilution methodology (26). All 14 MRSA isolates were tested for copper sulfate resistance using the CLSI disk diffusion methodology using 4 mM copper sulfate antibiotic disks. The copper-susceptible S. aureus reference strain RN4220 (27) and the copper-resistant MRSA strain MRSA252 (12) were used as controls.
Molecular typing.
All isolates underwent direct repeat unit (dru) typing, while M06/0171 was also subjected to multilocus sequence typing (MLST) and SCCmec typing, all as described previously (24, 28–30). SCCmec typing involved the use of previously described multiplex PCRs to detect (i) the class A, B, and C mec complexes (31); (ii) the type 1 to 5 ccr complexes (31); and (iii) the joining or “J” regions (32). An additional simplex PCR using alternative ccrAB4 primers described previously by Ruppe et al. (33) was undertaken for the detection of additional ccrAB4 alleles that are not detected using the ccrAB4 primers described by Kondo et al. (31). Previously described MRSA reference strains were used as positive controls for these PCR assays (29). PCRs were performed using GoTaq Flexi DNA polymerase (Promega Corporation, Madison, WI) according to the manufacturer's instructions. PCR amplifications were performed in a G-storm GS1 thermocycler (Applied Biosystems, Foster City, CA). PCR products were visualized by conventional agarose gel electrophoresis and purified with the GenElute PCR cleanup kit (Sigma-Aldrich). Sequencing was performed commercially by Geneservice Limited (Source Bioscience, Guinness Enterprise Centre, Dublin, Ireland) using an ABI 3730xl Sanger sequencing platform.
DNA microarray analysis using the StaphyType kit.
The StaphyType kit detects 333 S. aureus gene sequences and alleles, including species-specific, antimicrobial resistance genes; virulence-associated genes; and typing markers and SCCmec-associated gene sequences and can assign S. aureus isolates to an MLST sequence type (ST) and/or clonal complex (CC) (34, 35). Array procedures were performed according to the manufacturer's instructions.
Whole-genome sequencing of MRSA isolate M06/0171.
The whole-genome sequence of one MRSA isolate, M06/0171, was determined in order to investigate the genetic organization of a possible novel SCCmec element. High-throughput de novo sequencing was undertaken commercially by Geneservice (Source BioScience plc, Nottingham, United Kingdom) using the Illumina genome analyzer system (Illumina HiSeq 2000 platform; Illumina, Essex, United Kingdom). The average coverage across the genome was 111×. The reads were assembled into contigs using a Velvet de novo genome assembler (version 1.0.15; Illumina). Contigs were analyzed using the Artemis DNA sequence viewer and annotation tool (36) and BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (37). Any contig gaps identified between SCCmec-associated sequences were closed by primer walking using PCR with primers based on the surrounding contigs and GoTaq Flexi DNA polymerase (Promega) followed by amplimer sequencing and analysis using BioNumerics software version 5.1 (Applied Maths, Ghent, Belgium) and Artemis. Open reading frames (ORFs) were predicted using Artemis and prodigal (http://prodigal.ornl.gov/), and all ORFs were analyzed using the BLAST software package. Open reading frames were aligned with the best-fit matches in GenBank, and the locations of start codons, stop codons, and potential ribosomal binding sites were checked for consistency.
Confirmation of the genetic organization and location of the novel composite element in M06/0171.
The genetic organization of the novel composite pseudo SCCmec-SCC-SCCCRISPR element in M06/0171 determined from the whole-genome sequence was confirmed using eight overlapping primer pairs to amplify the entire element (see Table S1 in the supplemental material). These PCR assays were performed by amplifying chromosomal DNA using the Expand long-template PCR system (Roche Diagnostics GmdH, Lewes, East Sussex, United Kingdom). PCR products were visualized by agarose gel electrophoresis, and the sizes of the amplimers obtained were compared to the expected size of the amplimers based on the whole-genome sequence.
PCRs to confirm the presence of the pseudo SCCmec-SCC-SCCCRISPR element in additional t878 MRSA isolates and to confirm ambiguous DNA microarray results.
The presence of the novel pseudo SCCmec-SCC-SCCCRISPR element was investigated in the remaining 13 t878 MRSA isolates using previously described primers to amplify ccrAB4 and ccrC (31, 33) and novel primers to detect the CRISPR region and the novel mec complex of M06/0171 (see Table S1 in the supplemental material). Amplimers obtained from all 13 isolates using ccrAB4-specific and ccrC-specific primers and amplimers obtained using CRISPR primers for 5/13 isolates (isolates M09/0295, M08/0422, M11/0208, M09/0302, and E4449) were sequenced and compared to the corresponding sequences of M06/0171 using BioNumerics and Artemis. The online tool CRISPRfinder (38) (http://crispr.u-psud.fr/Server/) was used for CRISPR sequence analysis. The presence of the genes encoding clumping factor B (clfB), serine aspartate repeat protein D (sdrD), and staphylococcal enterotoxin B (seb) was confirmed by PCR (see Table S1) due to ambiguous DNA microarray results.
Nucleotide sequence accession number.
The nucleotide sequence of the novel pseudo SCCmec-SCC-SCCCRISPR element harbored by M06/0171 has been deposited in GenBank under accession number HE980450.
RESULTS
Phenotypic and genotypic characteristics of isolates.
Fourteen spa type t878 MRSA isolates recovered from separate patients in 12 different Irish hospitals between 2006 and 2011 were investigated (Table 1). These represented 0.32% (14/4,320) of all MRSA isolates submitted to the Irish NMRSARL between 2006 and 2011 and 3.2% (14/431) of non-AR06 isolates (indicative of ST22-MRSA-IV, the predominant MRSA clone in Irish hospitals since 2002) spa typed by the Irish NMRSARL during the same period. All isolates exhibited resistance to ampicillin and fusidic acid. M06/0171 was also resistant to mupirocin, neomycin, and tobramycin and was copper resistant with a copper MIC of 4 mM as determined by agar dilution. The remaining 13 isolates were also resistant to copper as determined by disk diffusion (Table 1). Four isolates exhibited resistance to cadmium (Table 1). The isolates exhibited four dru types, were assigned to ST779, and belonged to agr type III and capsule type 5 (Table 1). All isolates harbored the beta-lactamase resistance gene blaZ, the fusidic acid resistance gene fusC, and the multidrug-efflux pump gene sdrM. The mupirocin and aminoglycoside resistance genes, mupA and aadD, respectively, were detected in M06/0171 only (Table 1). All isolates harbored the exfoliative toxin gene etD; the epidermal cell differentiation inhibitor gene edinB; the enterotoxin gene seb; the immune evasion cluster (IEC) genes sak, chp, and scn (IEC type B) (39); and genes for microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), adhesion, and biofilm formation. The enterotoxin genes sed, sej, and ser were detected in six isolates (Table 1).
SCCmec typing.
SCCmec typing PCRs were performed on isolate M06/0171 only, while SCCmec analysis of the other 13 t878 isolates was performed by DNA microarray profiling (Table 1). Isolate M06/0171 was found to harbor mecA by SCCmec typing PCR and DNA microarray profiling, but no mec regulatory genes were detected by either method. The mec complex-associated gene ugpQ was detected in M06/0171 using the DNA microarray (Table 1). The ccrC gene was detected in M06/0171 following SCCmec typing PCR but was ambiguous by DNA microarray, and ccrAA was detected by DNA microarray analysis only (Table 1). The ccrAB4 gene was detected in M06/0171 following SCCmec typing PCR using the primers designed by Ruppe et al. and the DNA microarray (Table 1) but was not detected using the primers described by Kondo et al. (31, 33).
For the remaining 13 t878 isolates, the DNA microarray detected the following SCCmec genes: mecA (13/13 isolates), ugpQ (13/13 isolates), ccrC (7/13 isolates, including four yielding ambiguous signals), ccrB4 (12/13 isolates, including five yielding ambiguous signals), and ccrAA (10/13 isolates, including seven yielding ambiguous signals) (Table 1).
Identification of a novel pseudo SCCmec-SCC-SCCCRISPR element in MRSA isolate M06/0171.
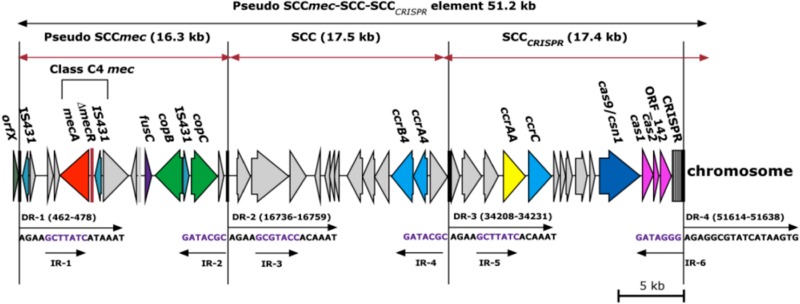
Whole-genome sequencing of the ST779/t878 MRSA isolate M06/0171 yielded 89 contigs ranging in size from 216 bp to 226 kb, and 25 of these were >40 kb. Six contigs were identified with SCCmec-associated DNA sequences. A novel composite SCC element, which we termed a pseudo SCCmec-SCC-SCCCRISPR element, was identified. The novel element was ca. 51 kb in size, consisted of 43 ORFs (see Table S2 in the supplemental material), was located at the 3′ end of the orfX gene, and was flanked by imperfect direct repeat (DR) and inverted repeat (IR) sequences (Fig. 1, DR-1 and DR-4 and IR-1 and IR-6). Two additional DRs and four additional IRs were identified within the element (Fig. 1, DR-2 and DR-3 and IR-2, IR-3, IR-4, and IR-5) demarcating a three-domain composite element (Fig. 1).
Fig 1.
Schematic diagram showing the genetic organization of the novel composite pseudo SCCmec-SCC-SCCCRISPR element harbored by the ST779/t878 MRSA isolate M06/0171 (GenBank accession number HE980450). The 51-kb composite pseudo SCCmec-SCC-SCCCRISPR element, as well as each of the individual SCC elements of this composite island, is flanked by direct repeat (DR) and inverted repeat (IR) sequences. The methicillin, fusidic acid, and copper resistance genes mecA, fusC, and copB/copC are shown in red, purple, and green, respectively. The ccrAB4 and ccrC genes are shown in blue, the ccrAA gene is shown in yellow, and the clustered regularly interspaced short palindromic repeats (CRISPRs) and the genes encoding CRISPR-associated proteins (cas9/csn1, cas1, cas2, and ORF_142) are shown in pink. The direction of transcription for each ORF is indicated.
The first SCC region of the novel element consisted of a 16.3-kb pseudo SCCmec element located immediately downstream of orfX and flanked by DR-1 and DR-2. It consisted of 15 ORFs and was termed a pseudo SCCmec element because while a mec complex was identified in this 16.3-kb region, there were no ccr genes (Fig. 1). The mec complex genes exhibited 100% DNA sequence identity to the class C1-like mec complex previously identified in SCCmec X in MRSA isolate JCSC6945 (GenBank accession number AB505630). However, the mec complex genes were transcribed divergently from those in SCCmec X (8) but in the same direction as all other mec regions described to date (Fig. 1). Additionally, variation was exhibited within the intergenic region between ΔmecR1 and IS431 (17-bp deletion in M06/0171), suggesting that two separate insertions of IS431 had occurred in these two SCCmec elements. This mec complex consists of mecA, a 17-bp ΔmecR1, and flanking IS431 sequences (Fig. 1). The presence of flanking IS431 sequences as well as the DNA sequence identity to the class C1-like mec complex of SCCmec X indicated that this mec complex should be assigned to class C mec. To date, three subtypes of the class C mec complex have been reported, class C1 (40), class C2 (41), and class C1-like (8). The ΔmecR1 in the class C1 mec complex has a different truncation site resulting in a different ΔmecR1 length (73 bp), indicating a separate genetic event from that of the class C1-like mec complex; therefore, we propose that the class C1-like mec complex be renamed class C3 mec. Since the novel mec complex in M06/0171 has the same genetic organization as that of the class C3 mec complex but (i) is transcribed divergently and (ii) exhibits variation within the intergenic region, we propose that the novel subtype of the class C mec complex identified in the present study in M06/0171 be designated class C4 mec complex.
In addition to the mec complex, genes encoding fusidic acid (fusC) and copper resistance were also identified within the pseudo SCCmec element. The fusC gene exhibited 100% amino acid sequence identity to fusC previously identified in SCCfus in methicillin-susceptible S. aureus (MSSA) isolate MSSA476 (YP_042173) (12). Two ORFs associated with copper resistance, which we have designated copB and copC, were located downstream of fusC. The copB gene exhibited 99% amino acid sequence similarity to an annotated ORF encoding a copper-exporting ATPase in Staphylococcus epidermidis strain VCU120 (EHR82803), and the copC gene exhibited 100% amino acid sequence identity to an unannotated copper transport gene previously identified in an SCCmec X element in the MRSA strain JCSC6945 (BAK53188) (8) (Fig. 1).
The second SCC region, located immediately downstream from the pseudo SCCmec element and flanked by direct repeats DR-2 and DR-3, consisted of a 17.5-kb SCC element with 13 ORFs, including ccrAB4 (Fig. 1). The ccrA4 gene exhibited 93% amino acid sequence identity to ccrA4 harbored by the S. aureus strain CHE482 (ABL75417), and the ccrB4 gene exhibited 98% amino acid sequence identity to ccrB4 harbored by the Staphylococcus haemolyticus strain MCS13 (BAJ53095). We have designated the ccrA4 and ccrB4 genes as allele 6 in each case, considering that five alleles of the ccrA4 and ccrB4 genes have already been described in S. aureus and CoNS (15, 19, 20, 42). We recommend assigning each of these previously described ccrA4 and ccrB4 alleles an allelic number 1 to 5 in order of publication.
The third SCC region, located immediately downstream of the SCC element and flanked by DR-3 and DR-4, consisted of a 17.4-kb SCC element with 14 ORFs (Fig. 1). This SCC region harbored a ccrC1 gene with 95% amino acid sequence identity to ccrC1 harbored by S. aureus strain UMCG-M4 (ADC79473), S. aureus strain S0385 (YP_005732860), and Staphylococcus pseudintermedius strain AVDL-32616 (ACT82836). We have designated this as allotype ccrC1 and allele ccrC11, considering that alleles ccrC1 to -10 of the ccrC1 allotype have been previously reported (18). The final SCC region also carried a clustered regularly interspersed short palindromic repeat (CRISPR) region and four CRISPR-associated genes (cas9/csn1, cas1, cas2, and ORF_142) (Fig. 1). However, the cas genes exhibited the highest amino acid sequence similarity (46 to 70%) to those in Staphylococcus lugdunensis (NZ_AEQA01000016). The CRISPR region consists of clustered regularly interspaced short palindromic repeats that are generally segments of DNA captured from viral or plasmid sequences and are located between the conserved direct repeat sequences of the CRISPR region (43). Analysis of the DNA sequences of the variable interspersed sequences in this CRISPR region using the online tool CRISPRfinder revealed the most probable origins of each individual variable interspersed sequence (Table 2). Twelve interspersed repeats were identified, and the most common similarity detected was that to S. haemolyticus with 4/12 repeats exhibiting between 93% and 100% DNA sequence identity.
Table 2.
Highest probable matches for the 13 variable interspersed DNA sequences in the clustered regularly interspaced short palindromic repeat (CRISPR) region in ST779/t878 MRSA isolatesa generated using the online CRISPRfinder software tool
| CRISPR repeat no. | BLASTn resultb | GenBank accession no. | % DNA sequence similarity | % query coverage |
|---|---|---|---|---|
| 1 | Phenylobacterium zucineum HLK1 plasmid | CP000748 | 100 | 60 |
| 2 | Geobacillus thermoleovorans | CP003125 | 100 | 86 |
| 3 | Megamonas hypermegale | FP929048 | 100 | 66 |
| 4 | Bacteroides xylanisolvens XB1A | FP929033 | 100 | 100 |
| 5 | Staphylococcus haemolyticus | AP006716 | 100 | 93 |
| 6 | S. haemolyticus | AP006716 | 100 | 93 |
| 7 | Shewanella piezotolerans | CP000472 | 100 | 80 |
| 8 | S. haemolyticus | AP006716 | 93 | 100 |
| 9 | Escherichia blattae | CP001560 | 100 | 56 |
| 10 | Methylophaga sp. | CP003380 | 100 | 70 |
| 11 | S. epidermidis plasmid | GQ900454 | 100 | 86 |
| 12 | S. haemolyticus | AP006716 | 100 | 93 |
The CRISPR regions of six isolates were sequenced (isolates M06/0171, M09/0295, M08/0422, M11/0208, M09/0302, and E4449).
The BLASTn algorithm was used to search for similar sequences in GenBank compared to each of the variable interspersed repeats in the CRISPR region in the novel SCCCRISPR element.
Confirmation of the presence of the pseudo SCCmec-SCC-SCCCRISPR element in other ST779/t878 MRSA isolates.
The presence of the novel pseudo SCCmec-SCC-SCCCRISPR element identified in M06/0171 was confirmed in the 13 additional ST779/t878 MRSA isolates by PCR using previously described primers to amplify ccrAB4 and ccrC and novel primers to amplify CRISPR and the mec complex (see Table S1 in the supplemental material). All isolates yielded amplimers of the expected size compared to M06/0171. Sequencing of the amplimers obtained for ccrAB4 and ccrC revealed that the 13 additional ST779/t878 MRSA isolates harbored ccrAB4 and ccrC genes identical to each other and to those of M06/0171. Sequencing of amplimers obtained following amplification of the CRISPR region in 5/13 isolates revealed that they harbored CRISPR regions identical to each other and to that of M06/0171.
DISCUSSION
The present study reports the emergence of ST779/t878 MRSA harboring a novel 51-kb pseudo SCCmec composite island (CI) in Ireland. In-depth molecular analysis revealed that the novel CI consisted of three distinct and unique domains, each demarcated by direct repeat sequences. The first domain was a pseudo SCCmec with a novel mec complex, a fusidic acid resistance gene (fusC), and two copper resistance genes but lacking ccr genes. The second domain was an SCC with a novel ccrAB4 allele, whereas the third element was an SCC with a novel ccrC allele and a CRISPR region. Comparative sequence analysis of the novel pseudo SCCmec-SCC-SCCCRISPR element suggested that this CI may have originated in bacterial species and genera other than S. aureus and Staphylococcus, respectively. First, for some of the ORFs identified within the CI the highest amino acid identity was to ORFs from non-S. aureus staphylococcal species. In addition, the interspersed sequences of the CRISPR region located within the CI exhibited the highest DNA sequence identity to CoNS and to other genera. Taken together, these data provide further evidence for SCCmec diversity and indicate that genetic transfer between S. aureus, CoNS, and possibly other bacterial genera contributes to the emergence of novel SCCmec/SCC elements and CIs and ultimately to the emergence of novel MRSA strains.
While SCC elements lacking mecA have been reported previously (29), to the best of our knowledge there have been only two previous reports of SCC elements harboring mecA but lacking ccr genes in MRSA (44, 45). The presence of ccr genes on the adjacent SCC elements in ST779 MRSA suggests that even though the pseudo SCCmec lacks ccr genes, it and possibly the entire CI may be mobilized using the ccr genes on either of the adjacent SCC elements. This type of mechanism has been suggested previously as a means of mobilization of the arginine catabolic mobile element (ACME) (46).
The present study highlights the difficulties associated with SCCmec typing both using DNA microarray profiling and using conventional SCCmec typing PCRs. The distant amino acid sequence similarity (93 to 95%) of ccrA4, ccrB4, and ccrC to their closest counterparts resulted in negative and/or ambiguous results by DNA microarray analysis for ccrAB4 and ccrC genes among the 14 ST779 MRSA isolates as well as no amplimers for ccrAB4 using the multiplex SCCmec typing PCR of Kondo et al. (31). It was only by using ccrAB4 primers and PCRs that detect additional alleles that the ccrAB4 allele of the novel CI was detected. Updating currently used multiplex SCCmec typing PCRs to detect all ccr alleles identified to date would enhance detection of this and other recently described SCCmec elements (4, 8). The abundance of SCCmec elements in S. aureus and other staphylococci and the diversity evident within SCCmec elements constitute a challenge for SCCmec nomenclature, for which guidelines have been published by the IWG-SCC (5, 7), but these are not always adhered to.
An unusual feature within the pseudo SCCmec-SCC-SCCCRISPR was the presence of the CRISPR/cas region. The CRISPR region is a recently described class of repetitive DNA element, and it and the CRISPR-associated genes (cas) are involved in the protection of the bacterial genome against foreign invading DNA, i.e., viral and plasmid DNA (43, 47). The CRISPR region has been identified in approximately 40% of prokaryotes and 90% of archaea, and multiple distinct CRISPR loci have been located on prokaryotic genomes (47). The CRISPR region consists of multiple short nucleotide repeat sequences, varying from 21 to 37 bp in length, separated by unique variable spacer sequences which originate from phage DNA and provide a record of genetic encounters (38). The cas genes are involved in cleavage of the CRISPR RNA precursor in each repeat, and the resulting cleaved products act as leaders for other cas gene protein products, guiding them to foreign invading DNA (47). It remains unclear how segments of foreign invading DNA are incorporated into the CRISPR region (47). The relatedness of the cas genes can vary within a bacterial species as well as between different species, and particular CRISPR/cas loci are associated with particular strains within a species (47). Interestingly, all ST779 MRSA isolates investigated in the present study appeared to harbor the same CRISPR region. The CRISPR/cas region is uncommon among staphylococci, though it has been detected in one CC75/ST1850-MRSA-IVa isolate, an S. epidermidis isolate harboring SCCmec II, and in several S. lugdunensis isolates (43, 48, 49), and in each case the CRISPR region was located downstream of the SCCmec element. CRISPR has also been detected previously in a novel SCCmec V subtype harbored by four ST398 MRSA isolates (50). The same cas region gene organization detected in the ST779 MRSA isolate M06/0171 in the present study is present in S. lugdunensis strain M23590, i.e., cas9, cas1, cas2, and a cas-associated gene (ORF_142). However, they exhibit low amino acid sequence similarity, and the CRISPR region is upstream of the cas genes in the S. lugdunensis strain.
Comparison of the CRISPR region identified in the ST779 MRSA isolates with available staphylococcal CRISPR region sequences in GenBank revealed that the ST779 MRSA CRISPR region has a significantly higher number of variable spacer regions than those described previously (12 compared with 2 to 6 spacer regions). Additionally, each spacer sequence was unique to the staphylococcal strain in which it was reported. The genetic diversity of the CRISPR spacer sequences has been reported previously, and it has been suggested that CRISPR loci have potential for typing of strains and microbial populations. However, the use of a given CRISPR locus for typing and epidemiological analysis has to be critically assessed due to its rarity in staphylococci and the various rates of polymorphisms within this region (47). The role of the CRISPR/cas locus in ST779 MRSA requires further investigation, to determine which of the cas genes are responsible for acquiring additional variable spacer regions and which of the cas genes are responsible for spacer lead targeted defense against foreign DNA.
Whether ST779 MRSA will become a more widespread MRSA clone remains to be determined, but it is possible that the novel composite element harbored by this clone may confer advantageous attributes in addition to methicillin resistance, such as copper or fusidic acid resistance or resistance or immunity to foreign invading DNA encoded by CRISPR. Several other isolates exhibiting CC779/ST779 or closely related STs have been reported previously, indicating their sporadic presence in Australia (WA-MRSA-100) Canada, Germany, Thailand, the United Arab Emirates, and the United Kingdom (http://saureus.mlst.net/) (13, 51). Ongoing surveillance of ST779/t878 MRSA with the novel pseudo SCCmec-SCC-SCCCRISPR element is warranted. SCCmec typing methods will need to be updated to ensure successful detection and monitoring of this and other emerging MRSA strains. The identification of a novel pseudo SCCmec-SCC-SCCCRISPR element exhibiting sequence similarity to non-S. aureus staphylococci as well as to other genera further indicates the potential role that other organisms may play in the emergence of novel SCCmec elements in MRSA.
Supplementary Material
ACKNOWLEDGMENT
This study was supported by the Microbiology Research Unit, Dublin Dental University Hospital.
Footnotes
Published ahead of print 12 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01689-12.
REFERENCES
- 1. Chambers HF, Deleo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7:629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weese JS. 2010. Methicillin-resistant Staphylococcus aureus in animals. ILAR J. 51:233–244 [DOI] [PubMed] [Google Scholar]
- 3. Tomasz A, Drugeon HB, de Lencastre HM, Jabes D, McDougall L, Bille J. 1989. New mechanism for methicillin resistance in Staphylococcus aureus: clinical isolates that lack the PBP2a gene and contain normal penicillin-binding proteins with modified penicillin-binding capacity. Antimicrob. Agents Chemother. 33:1869–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shore AC, Deasy EC, Slickers P, Brennan G, O'Connell B, Monecke S, Ehricht R, Coleman DC. 2011. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 55:3765–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ito T, Hiramatsu K, Tomasz A, de Lencastre H, Perreten V, Holden MT, Coleman DC, Goering R, Giffard PM, Skov RL, Zhang K, Westh H, O'Brien F, Tenover FC, Oliveira DC, Boyle-Vavra S, Laurent F, Kearns AM, Kreiswirth B, Ko KS, Grundmann H, Sollid JE, John JF, Daum R, Soderquist B, Buist G. 2012. Guidelines for reporting novel mecA gene homologues. Antimicrob. Agents Chemother. 56:4997–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Monecke S, Muller E, Schwarz S, Hotzel H, Ehricht R. 2012. Rapid microarray based identification of different mecA alleles in staphylococci. Antimicrob. Agents Chemother. 56:5547–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53:4961–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li S, Skov RL, Han X, Larsen AR, Larsen J, Sorum M, Wulf M, Voss A, Hiramatsu K, Ito T. 2011. Novel types of staphylococcal cassette chromosome mec elements identified in clonal complex 398 methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 55:3046–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mongkolrattanothai K, Boyle S, Murphy TV, Daum RS. 2004. Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing pbp4 and tagF genes in a commensal staphylococcal species: a possible reservoir for antibiotic resistance islands in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:1823–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bloemendaal AL, Brouwer EC, Fluit AC. 2010. Methicillin resistance transfer from Staphylococcus epidermidis to methicillin-susceptible Staphylococcus aureus in a patient during antibiotic therapy. PLoS One 5:e11841 doi:10.1371/journal.pone.0011841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hanssen AM, Kjeldsen G, Sollid JU. 2004. Local variants of staphylococcal cassette chromosome mec in sporadic methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci: evidence of horizontal gene transfer? Antimicrob. Agents Chemother. 48:285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, Barron A, Bason N, Bentley SD, Chillingworth C, Chillingworth T, Churcher C, Clark L, Corton C, Cronin A, Doggett J, Dowd L, Feltwell T, Hance Z, Harris B, Hauser H, Holroyd S, Jagels K, James KD, Lennard N, Line A, Mayes R, Moule S, Mungall K, Ormond D, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Sharp S, Simmonds M, Stevens K, Whitehead S, Barrell BG, Spratt BG, Parkhill J. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. U. S. A. 101:9786–9791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, Chow H, Ip M, Jatzwauk L, Jonas D, Kadlec K, Kearns A, Laurent F, O'Brien FG, Pearson J, Ruppelt A, Schwarz S, Scicluna E, Slickers P, Tan HL, Weber S, Ehricht R. 2011. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936 doi:10.1371/journal.pone.0017936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oliveira DC, Milheirico C, de Lencastre H. 2006. Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob. Agents Chemother. 50:3457–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ender M, Berger-Bachi B, McCallum N. 2007. Variability in SCCmecN1 spreading among injection drug users in Zurich, Switzerland. BMC Microbiol. 7:62 doi:10.1186/1471-2180-7-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hanssen AM, Sollid JU. 2007. Multiple staphylococcal cassette chromosomes and allelic variants of cassette chromosome recombinases in Staphylococcus aureus and coagulase-negative staphylococci from Norway. Antimicrob. Agents Chemother. 51:1671–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen L, Mediavilla JR, Oliveira DC, Willey BM, de Lencastre H, Kreiswirth BN. 2009. Multiplex real-time PCR for rapid staphylococcal cassette chromosome mec typing. J. Clin. Microbiol. 47:3692–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chlebowicz MA, Nganou K, Kozytska S, Arends JP, Engelmann S, Grundmann H, Ohlsen K, van Dijl JM, Buist G. 2010. Recombination between ccrC genes in a type V (5C2&5) staphylococcal cassette chromosome mec (SCCmec) of Staphylococcus aureus ST398 leads to conversion from methicillin resistance to methicillin susceptibility in vivo. Antimicrob. Agents Chemother. 54:783–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang K, McClure JA, Elsayed S, Conly JM. 2009. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Urushibara N, Paul SK, Hossain MA, Kawaguchiya M, Kobayashi N. 2011. Analysis of Staphylococcus haemolyticus and Staphylococcus sciuri: identification of a novel ccr gene complex with a newly identified ccrA allotype. Microb. Drug Resist. 17:291–297 [DOI] [PubMed] [Google Scholar]
- 21. Humphreys H, Keane CT, Hone R, Pomeroy H, Russell RJ, Arbuthnott JP, Coleman DC. 1989. Enterotoxin production by Staphylococcus aureus isolates from cases of septicaemia and from healthy carriers. J. Med. Microbiol. 28:163–172 [DOI] [PubMed] [Google Scholar]
- 22. Rossney AS, Lawrence MJ, Morgan PM, Fitzgibbon MM, Shore AC, Coleman DC, Keane CT, O'Connell B. 2006. Epidemiological typing of MRSA isolates from blood cultures taken in Irish hospitals participating in the European Antimicrobial Resistance Surveillance System (1999–2003). Eur. J. Clin. Microbiol. Infect. Dis. 25:79–89 [DOI] [PubMed] [Google Scholar]
- 23. Shore AC, Rossney AS, Keane CT, Enright MC, Coleman DC. 2005. Seven novel variants of the staphylococcal chromosomal cassette mec in methicillin-resistant Staphylococcus aureus isolates from Ireland. Antimicrob. Agents Chemother. 49:2070–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shore AC, Rossney AS, Kinnevey PM, Brennan OM, Creamer E, Sherlock O, Dolan A, Cunney R, Sullivan DJ, Goering RV, Humphreys H, Coleman DC. 2010. Enhanced discrimination of highly clonal ST22-methicillin-resistant Staphylococcus aureus IV isolates achieved by combining spa, dru, and pulsed-field gel electrophoresis typing data. J. Clin. Microbiol. 48:1839–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rossney AS, Shore AC, Morgan PM, Fitzgibbon MM, O'Connell B, Coleman DC. 2007. The emergence and importation of diverse genotypes of methicillin-resistant Staphylococcus aureus (MRSA) harboring the Panton-Valentine leukocidin gene (pvl) reveal that pvl is a poor marker for community-acquired MRSA strains in Ireland. J. Clin. Microbiol. 45:2554–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. CLSI 2006. Performance standards for antimicrobial susceptibility testing; sixteenth informational supplement. CLSI document M100-S16. CLSI, Wayne, PA [Google Scholar]
- 27. Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712 [DOI] [PubMed] [Google Scholar]
- 28. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shore AC, Rossney AS, O'Connell B, Herra CM, Sullivan DJ, Humphreys H, Coleman DC. 2008. Detection of staphylococcal cassette chromosome mec-associated DNA segments in multiresistant methicillin-susceptible Staphylococcus aureus (MSSA) and identification of Staphylococcus epidermidis ccrAB4 in both methicillin-resistant S. aureus and MSSA. Antimicrob. Agents Chemother. 52:4407–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goering RV, Morrison D, Al-Doori Z, Edwards GF, Gemmell CG. 2008. Usefulness of mec-associated direct repeat unit (dru) typing in the epidemiological analysis of highly clonal methicillin-resistant Staphylococcus aureus in Scotland. Clin. Microbiol. Infect. 14:964–969 [DOI] [PubMed] [Google Scholar]
- 31. Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oliveira DC, de Lencastre H. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ruppe E, Barbier F, Mesli Y, Maiga A, Cojocaru R, Benkhalfat M, Benchouk S, Hassaine H, Maiga I, Diallo A, Koumare AK, Ouattara K, Soumare S, Dufourcq JB, Nareth C, Sarthou JL, Andremont A, Ruimy R. 2009. Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob. Agents Chemother. 53:442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Monecke S, Jatzwauk L, Weber S, Slickers P, Ehricht R. 2008. DNA microarray-based genotyping of methicillin-resistant Staphylococcus aureus strains from Eastern Saxony. Clin. Microbiol. Infect. 14:534–545 [DOI] [PubMed] [Google Scholar]
- 35. Monecke S, Slickers P, Ehricht R. 2008. Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol. Med. Microbiol. 53:237–251 [DOI] [PubMed] [Google Scholar]
- 36. Berriman M, Rutherford K. 2003. Viewing and annotating sequence data with Artemis. Brief. Bioinform. 4:124–132 [DOI] [PubMed] [Google Scholar]
- 37. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 38. Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35:W52–W57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Wamel WJ, Rooijakkers SH, Ruyken M, van Kessel KP, van Strijp JA. 2006. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 188:1310–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Berglund C, Ito T, Ikeda M, Ma XX, Soderquist B, Hiramatsu K. 2008. Novel type of staphylococcal cassette chromosome mec in a methicillin-resistant Staphylococcus aureus strain isolated in Sweden. Antimicrob. Agents Chemother. 52:3512–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ito T, Ma XX, Takeuchi F, Okuma K, Yuzawa H, Hiramatsu K. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oliveira DC, Tomasz A, de Lencastre H. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349–361 [DOI] [PubMed] [Google Scholar]
- 43. Holt DC, Holden MT, Tong SY, Castillo-Ramirez S, Clarke L, Quail MA, Currie BJ, Parkhill J, Bentley SD, Feil EJ, Giffard PM. 2011. A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol. Evol. 3:881–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Han X, Ito T, Takeuchi F, Ma XX, Takasu M, Uehara Y, Oliveira DC, de Lencastre H, Hiramatsu K. 2009. Identification of a novel variant of staphylococcal cassette chromosome mec, type II.5, and its truncated form by insertion of putative conjugative transposon Tn6012. Antimicrob. Agents Chemother. 53:2616–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen L, Mediavilla JR, Smyth DS, Chavda KD, Ionescu R, Roberts BR, Robinson DA, Kreiswirth BN. 2010. Identification of a novel transposon (Tn6072) and a truncated staphylococcal cassette chromosome mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 54:3347–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goering RV, McDougal LK, Fosheim GE, Bonnstetter KK, Wolter DJ, Tenover FC. 2007. Epidemiologic distribution of the arginine catabolic mobile element among selected methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates. J. Clin. Microbiol. 45:1981–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190:1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tse H, Tsoi HW, Leung SP, Lau SK, Woo PC, Yuen KY. 2010. Complete genome sequence of Staphylococcus lugdunensis strain HKU09-01. J. Bacteriol. 192:1471–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Golding GR, Bryden L, Levett PN, McDonald RR, Wong A, Wylie J, Graham MR, Tyler S, Van Domselaar G, Simor E, Gravel D, Mulvey MR. 2010. Livestock-associated methicillin-resistant Staphylococcus aureus sequence type 398 in humans, Canada. Emerg. Infect. Dis. 16:587–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Coombs G, Pearson J, Christiansen K, Nimmo G. 2011. Staphylococcus aureus programme 2010 (SAP 2010). Community survey. MRSA epidemiology and typing report on behalf of the Australian Group for Antimicrobial Resistance (AGAR). http://www.agargroup.org/publications
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.