Abstract
Acquired metallo-β-lactamases (MBLs) are resistance determinants of increasing clinical importance in Gram-negative bacterial pathogens, which confer a broad-spectrum β-lactam resistance, including carbapenems. Several such enzymes have been described since the 1990s. In the present study, a novel acquired MBL, named FIM-1, was identified and characterized. The blaFIM-1 gene was cloned from a multidrug-resistant Pseudomonas aeruginosa clinical isolate (FI-14/157) cultured from a patient with a vascular graft infection in Florence, Italy. The isolate belonged in the sequence type 235 epidemic clonal lineage. The FIM-1 enzyme is a member of subclass B1 and, among acquired MBLs, exhibited the highest similarity (ca. 40% amino acid identity) with NDM-type enzymes. In P. aeruginosa FI-14/157, the blaFIM-1 gene was apparently inserted into the chromosome and associated with ISCR19-like elements that were likely involved in the capture and mobilization of this MBL gene. Transfer experiments of the blaFIM-1 gene to an Escherichia coli strain or another P. aeruginosa strain by conjugation or electrotransformation were not successful. The FIM-1 protein was produced in E. coli and purified by two chromatography steps. Analysis of the kinetic parameters, carried out with the purified enzyme, revealed that FIM-1 has a broad substrate specificity, with a preference for penicillins (except the 6α-methoxy derivative temocillin) and carbapenems. Aztreonam was not hydrolyzed. Detection of this novel type of acquired MBL in a P. aeruginosa clinical isolate underscores the increasing diversity of such enzymes that can be encountered in the clinical setting.
INTRODUCTION
Acquired metallo-β-lactamases (MBLs) are resistance determinants of increasing clinical importance in Gram-negative bacterial pathogens, including Pseudomonas aeruginosa, Enterobacteriaceae, and other Gram-negative nonfermenters. These enzymes can degrade most β-lactams, including carbapenems, and escape inhibition by the conventional β-lactamase inhibitors or avibactam. Thus, they are able to confer an extended β-lactam resistance profile to the bacterial host that is not reversible by β-lactamase inhibitors (1).
Acquired MBLs were detected since the early 1990s, the first representatives being IMP- and VIM-type enzymes (2–4). Thereafter, a number of additional lineages of acquired MBLs have been described, including the SPM-, GIM-, SIM-, KHM-, NDM-, AIM-, DIM-, SMB-, and TMB-type enzymes (5, 6; see also reference 1 and references therein). By convention, different MBL types diverge from each other at by least 30% at the amino acid sequence level (7). Enzymes of different types may differ in functional properties, and the corresponding genes may be associated with different types of mobile genetic elements responsible for their dissemination, such as mobile gene cassettes inserted into integrons, ISCR elements, or composite transposons (8, 9).
We report here on the identification and characterization of a new type of acquired MBL, named FIM-1, in a multidrug-resistant clinical isolate of P. aeruginosa from Italy.
MATERIALS AND METHODS
Bacterial strains and plasmids.
P. aeruginosa FI-14/157 was isolated from an inpatient at Florence University Hospital in 2007. Identification was performed by using the Vitek 2 automated system (bioMérieux, Marcy l'Etoile, France) and confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (Vitek MS; bioMérieux).
Antimicrobial susceptibility testing.
MICs were determined by broth microdilution according to the Clinical and Laboratory Standards Institute (CLSI) guideline (10). Interpretation of results was according to the EUCAST breakpoints (http://www.eucast.org/clinical_breakpoints/).
MLST.
Multilocus sequence typing (MLST) was performed as described previously (11). Sequence type (ST) was assigned by comparison to the P. aeruginosa MLST alleles database (http://pubmlst.org/paeruginosa/).
Enzyme assays.
MBL production was screened for by meropenem-EDTA combo disk (12) and confirmed by spectrophotometric assay carried out with a crude bacterial extract as described previously (3). The assay was carried out in 50 mM HEPES buffer (pH 7.5) supplemented with 50 μM ZnSO4 at 30°C, with 150 μM imipenem as the substrate. Inhibition by EDTA was assayed by measuring the residual carbapenemase activity in the presence of 5 mM EDTA, after preincubation of the sample for 30 min at 30°C with the same EDTA concentration.
DNA manipulation and analysis techniques.
Basic recombinant DNA methodology was performed as described by Sambrook et al. (13). PCR for the detection of MBL genes was performed as described previously (14). Genomic DNA was extracted from P. aeruginosa as described previously (15). The genomic library from P. aeruginosa FI-14/157 was constructed by cloning a partial Sau3AI chromosomal digest of this strain in the E. coli pBC-SK plasmid vector, as described previously (16). Sequencing was carried out on both strands using a DNA sequencer (Applied Biosystems, Carlsbad, CA) and a primer walking strategy. Nucleotide primers used for subcloning of the blaFIM-1 gene and for confirmatory mapping of the cloned DNA fragment are listed in Table 1. Database searches were performed using the BLAST software available at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST). DNA and protein sequence alignments were performed with the CLUSTAL W2 software, available at the European Molecular Biology Laboratory website (http://www.ebi.ac.uk/Tools/msa/clustalW2/) (17). A phylogenetic tree was constructed using the software Phylogeny.fr (http://www.phylogeny.fr/version2_cgi/index.cgi). The signal peptide cleavage site was predicted using SignalP (version 3.0). Southern blot analyses after genomic DNA digestion with I-CeuI endonuclease and S1 nuclease and were carried out as described previously (18, 19).
Table 1.
Nucleotide primers used in this study
| Primer | Function | Sequence (5′–3′)a |
|---|---|---|
| 14/157_F1 | PCR mapping of blaFIM-1-carrying DNA fragment with genomic DNA of P. aeruginosa FI-14/157 | ACTTCCACATGCTGTGGCTC |
| 14/157_R1 | PCR mapping of blaFIM-1-carrying DNA fragment with genomic DNA of P. aeruginosa FI-14/157 | CTCCGGGTACAACAACTGC |
| FIM-1_F | PCR mapping of blaFIM-1-carrying DNA fragment with genomic DNA of P. aeruginosa FI-14/157 | GAAGCACATGGAAAACTGGG |
| FIM-1_R | PCR mapping of blaFIM-1-carrying DNA fragment with genomic DNA of P. aeruginosa FI-14/157 | GATGGGCGAATGAGACAGC |
| FIM-1exp_F | Amplification of the blaFIM-1 ORF for cloning in pET9a to obtain pET-FIM-1 | GGAATTCCATATGCGCCCCTTACCCCATTC |
| FIM-1exp_R | Amplification of the blaFIM-1 ORF for cloning in pET9a to obtain pET-FIM-1 | CGGGATCCTCAGGGTGTGGACGGTATG |
The restriction sites used for cloning amplification products are underlined.
Gene transfer experiments.
Conjugation experiments were performed in solid medium as previously described (20), using either the E. coli strain MKD-135 (argH, rpoB18, rpoB19, recA, and rpsL) or the P. aeruginosa strain 10145/3 (an rpoB, his derivative of the reference strain ATCC 10145T) as recipients. Ceftazidime (at 5 μg/ml for E. coli or at 50 μg/ml for P. aeruginosa) was used for selection of transconjugants and rifampin (250 μg/ml) as a counterselection for the donor. Electroporation experiments were performed with electrocompetent E. coli DH5α and P. aeruginosa PAO1 cells and genomic DNA (1 μg) extracted from P. aeruginosa FI-14/157, as described previously (20). Transformants were selected on LB agar containing ceftazidime as described for conjugation experiments.
Protein expression and purification.
The FIM-1 protein was expressed in the E. coli Rosetta strain (Novagen EMD Millipore Corp., Billerica, MA) transformed with recombinant plasmid pET-FIM-1. This plasmid was constructed by subcloning the blaFIM-1 open reading frame (ORF), amplified with primers FIM-1exp_F and FIM-1exp_R (Table 1), into the pET-9a expression vector using the NdeI and BamHI restriction sites. For protein expression, E. coli Rosetta(pET-FIM-1) was grown in Zyp-5052 medium for 7 h at 37°C. Bacterial cells were then harvested by centrifugation (10,000 × g for 30 min at 4°C), resuspended in 20 mM Tris-HCl (pH 8.0), and lysed by sonication (Labsonic L sonicator; B. Braun, Melsungen, Germany). The cleared lysate, obtained after centrifugation at 77,000 × g for 60 min, was then loaded onto a Q-Sepharose high-performance column (bed volume, 75 ml; GE Healthcare, Uppsala, Sweden), previously equilibrated with 20 mM Tris-HCl (pH 8.0). Bound proteins were eluted using a linear NaCl gradient (0 to 0.25 M, in 750 ml). The β-lactamase-containing fractions were pooled and then desalted using a HiPrep 26/10 desalting column (GE Healthcare) with 20 mM Bis-Tris buffer (pH 6.5) for protein elution. To achieve a higher purity, a second anion-exchange chromatography step was performed. The desalted sample was loaded onto a Source 15Q column (bed volume, 1 ml; GE Healthcare) equilibrated with 20 mM Bis-Tris buffer (pH 6.5), and bound proteins were eluted with a linear NaCl gradient (0 to 0.2 M, in 50 ml). β-Lactamase-containing fractions were pooled and stored at −20°C in 100 mM HEPES buffer (pH 7.5) supplemented with 50 μM ZnSO4. The purity of the preparation was estimated by SDS-PAGE (21).
Determination of kinetic parameters.
The kinetic parameters for the hydrolysis of β-lactam substrates by the purified FIM-1 enzyme were determined spectrophotometrically using a Cary 100 UV-Vis spectrophotometer (Varian, Walnut Creek, CA) at 30°C in 50 mM HEPES buffer (pH 7.5) containing 50 μM ZnSO4 in a final reaction volume of 500 μl. The purified FIM-1 enzyme was diluted in the same buffer supplemented with 20 μg of bovine serum albumin/ml to prevent enzyme denaturation. The steady-state kinetic parameters (kcat and Km) were calculated after direct fit of the initial reaction rates to the Henri-Michaelis-Menten equation or using the Hanes-Woolf linearization (22). Km values for ampicillin, meropenem, and ertapenem were measured as inhibition constants using a competitive inhibition model (22), and 200 μM cefoxitin was used as the reporter substrate. When pseudo-first-order kinetics were observed in the range of tested concentrations, only kcat/Km could be calculated, along with the lower limit value exceeded by each individual kinetic parameter.
Nucleotide sequence accession number.
The nucleotide sequence described here has been submitted to the GenBank/EMBL database under accession number JX570731.
RESULTS AND DISCUSSION
Features of P. aeruginosa FI-14/157.
P. aeruginosa FI-14/157 was isolated in June 2007 from blood of a patient with infection of a previously implanted aortic vascular graft, who underwent reintervention for replacement of the infected graft. The isolate was resistant to all antipseudomonas agents except colistin (Table 2). According to the laboratory and clinical records, additional isolates with the same resistance profile were cultured from tissue samples taken from the explanted graft. However, these additional isolates had not been stored and could not be further investigated. Once results of microbiology became available the patient was given colistin (1.5 million units every 12 h) plus rifampin (600 mg every 24 h), with a favorable outcome. The patient was an Italian citizen resident in Tuscany and reporting no recent history of travel abroad.
Table 2.
Antimicrobial susceptibility of P. aeruginosa FI-14/157 and of E. coli DH5α(pSPo1), producing the FIM-1 enzyme
| Antibiotic | MIC (μg/ml)a |
||
|---|---|---|---|
| FI-14/157 | DH5α(pSPo1) | DH5α(pBC-SK) | |
| Ampicillin | >128 | 128 | 2 |
| Amoxicillin-clavulanate | >128 | >128 | 4 |
| Piperacillin | >128 | 16 | 1 |
| Piperacillin-tazobactam | 128 | 8 | 1 |
| Temocillin | >128 | 8 | 8 |
| Cefotaxime | >128 | 32 | ≤0.06 |
| Ceftazidime | 128 | 64 | 0.25 |
| Cefepime | 32 | 0.25 | ≤0.06 |
| Cefoxitin | >128 | 16 | 4 |
| Aztreonam | 16 | 0.06 | ≤0.06 |
| Ertapenem | >32 | 0.5 | ≤0.06 |
| Imipenem | 128 | 4 | 0.25 |
| Meropenem | 32 | 1 | ≤0.06 |
| Ciprofloxacin | 32 | ≤0.03 | ≤0.03 |
| Gentamicin | 128 | 0.25 | 0.25 |
| Tobramycin | >128 | 1 | 1 |
| Amikacin | 64 | 1 | 1 |
| Colistin | 1 | 0.25 | 0.12 |
MICs for DH5α carrying the cloning vector are also shown for comparison.
MLST analysis revealed that FI-14/157 belonged in ST-235, a clonal lineage associated with the dissemination of several clinically relevant β-lactamase determinants (e.g., blaVIM, blaIMP, blaGES, blaPER, and blaBEL) in several countries, and a typical representative of the so-called high-risk multiresistant clones (24–26; see also reference 23 and references therein).
The extended β-lactam resistance profile, including high-level carbapenem MICs (Table 2), suggested the possibility of carbapenemase production. An EDTA plus meropenem combo-disk test suggested production of MBL activity by FI-14/157 (increase of inhibition zone by 6 mm in the presence of EDTA). Carbapenemase activity was detected in a crude extract of the strain by a spectrophotometric assay (specific imipenemase activity, 628 ± 70 nmol/min·mg of protein). This activity was inhibited by >95% in the presence of EDTA, suggesting that it was due to an MBL.
Analysis of the isolate for several known acquired MBL determinants (including those of blaIMP, blaVIM, blaSPM, blaGIM, blaSIM, blaNDM, blaDIM, and blaAIM types) by PCR (14) did not yield any positive result.
Cloning and characterization of the MBL determinant from P. aeruginosa FI-14/157.
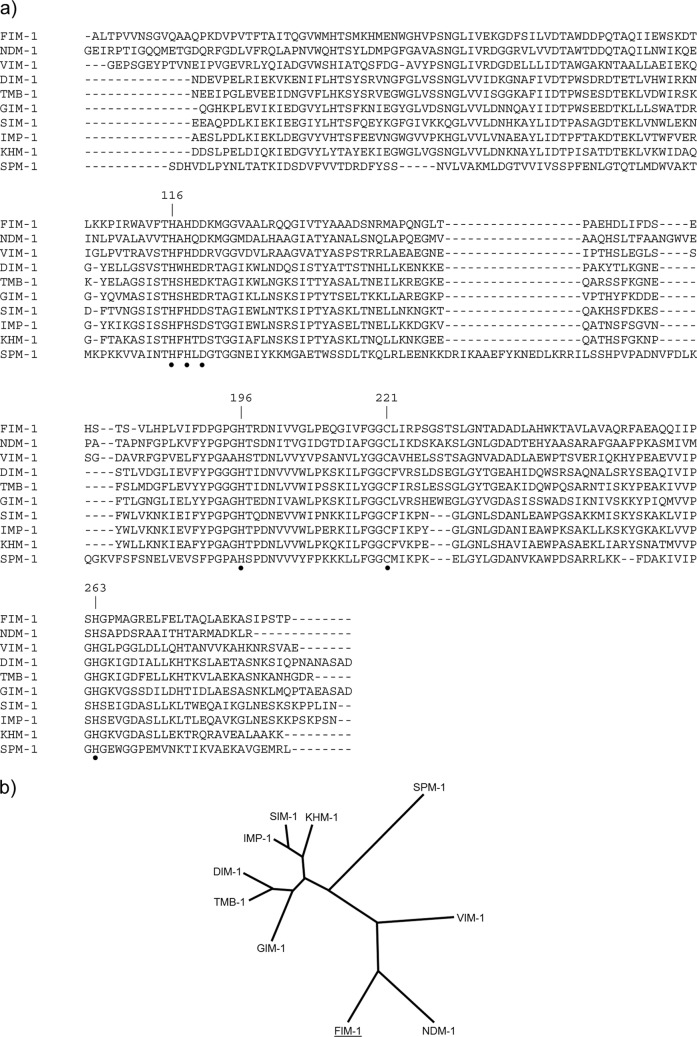
A genomic library of FI-14/157, constructed in the plasmid vector pBC-SK and transformed in E. coli DH5α, was replica plated on medium containing chloramphenicol (85 μg/ml) and imipenem (5 μg/ml). One clone growing on this medium was obtained, whose crude extract exhibited MBL activity (data not shown). The clone contained a recombinant plasmid, named pSPo1, with an ∼7-kb insert (Fig. 1). Sequencing of the insert revealed the presence of a 789-bp ORF encoding a protein with significant sequence similarity to other MBLs of subclass B1. The protein, named FIM-1 (after Florence IMipenemase), contained the conserved zinc-binding residues typical of subclass B1 MBLs, namely, His116, His118 and His196 for the first metal-binding site, and Asp120, Cys221 and His263 for the second metal-binding site (Fig. 2a). According to the SignalP prediction software, a signal peptide (amino acids 1 to 21) was identified in the N-terminal region of the FIM-1 protein, consistent with a possible cleavage site after residue 21 and yielding a mature protein of 241 amino acids (theoretical mass, 25,888.23 g/mol; predicted pI, 5.4).
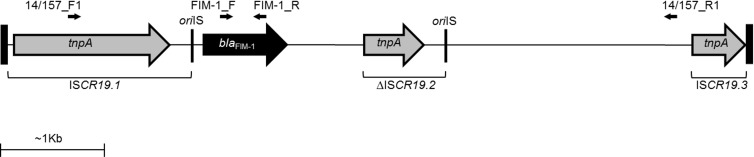
Fig 1.
Structure of the blaFIM-1-carrying DNA fragment cloned in the recombinant plasmid pSPo1. Genes are represented by arrows. The ISCR19-like origin of replication (oriIS) is indicated by thin vertical lines. Thick vertical lines indicate the cloned fragment boundaries. Targets of primers used for PCR mapping (Table 1) are also indicated.
Fig 2.
(a) Amino acid sequence alignment of FIM-1 with other acquired subclass B1 MBLs. Mature protein sequences are shown, either experimentally determined (for IMP-1, VIM-1, and SPM-1) (27–29) or deduced using the SignalP software. The sequence sources were as follows: FIM-1 (the present study), NDM-1 (GenBank/EMBL accession no. CAZ39946), DIM-1 (ADD91577), TMB-1 (CBY88906), GIM-1 (CAF05908), SIM-1 (AAX76774), KHM-1 (BAH16555), VIM-1 (CAC35170), IMP-1 (ADI87504), and SPM-1 (CAD37801). The positions are numbered according to the standard scheme proposed for class B β-lactamases (30); the conserved zinc-binding residues typical of subclass B1 MBLs are indicated by black dots. (b) Phylogenetic tree of acquired subclass B1 MBLs, based on the sequences used for multiple alignment.
Among known acquired MBLs, the closest relatives of FIM-1 are the NDM-type enzymes (39 to 40% amino acid identity), while other enzymes are more distantly related (Fig. 2b). Interestingly, when the FIM-1 sequence was compared to other protein sequences present in the GenBank database, the closest FIM-1 homologs were β-lactamases encoded by the genomes of the halophilic myxobacterium Haliangium ochraceum (GenBank/EMBL no. CP001804) and by the alphaproteobacterium Hirschia baltica (GenBank/EMBL no. CP001678) and Erythrobacter litoralis (GenBank/EMBL no. CP000157), with 46, 44, and 42% amino acid identities, respectively.
Genetic context of blaFIM-1.
Analysis of the blaFIM-1-flanking regions in the cloned DNA fragment revealed that the MBL gene was associated with an array of ISCR19-like elements or remnants thereof (Fig. 1). In particular, an ISCR19-like element 89% identical to ISCR19 (31), named ISCR19.1, was present upstream of blaFIM-1. This element was interrupted at the 5′-end (between the terIS site and the tnpA ORF) by the cloning junction. A second ISCR19-like element was present downstream of blaFIM-1, in the same orientation. This second element, named ΔISCR19.2, was deleted at the 5′-end (including terIS and the beginning of the tnpA gene) and also within the remnant of the tnpA gene, that was actually a hybrid of two different tnpA moieties of which one (at the 5′ end) was 93% identical to the corresponding region of ISCR19.1, while the other (at the 3′ end and also including the downstream region until oriIS) was 98% identical to the corresponding region of ISCR19.1. A third ISCR19-like element was present further downstream, in the same orientation. This third element, named ISCR19.3, was interrupted at the 3′ end (within the tnpA gene) by the cloning site and exhibited 93 and 95% identities with the corresponding regions of ISCR19.1 and ΔISCR19.2, respectively. The blaFIM-1 flanking sequences located between ISCR19.1 and ΔISCR19.2 did not show any homology with known sequences, while the sequence located between ΔISCR19.2 and ISCR19.3 contained regions of homology with plasmids from the methylotropic alphaproteobacterium Methylobacterium extorquens (GenBank/EMBL no. CP001511) and the alphaproteobacterium Ochrobactrum anthropi (GenBank/EMBL no. CP000760).
PCR mapping of this region, carried out with the genomic DNA of FI-14/157 and the primers pairs 14/157_F1/FIM-1_R and FIM-1_F/14/157_R1 (Table 1 and Fig. 1), confirmed the authenticity of the structure of the cloned insert.
Altogether, these findings suggested that, similar to other β-lactamases, including some acquired MBLs, namely, blaSPM-1 and blaAIM-1 (32, 33), an ISCR element was involved in the capture and mobilization of blaFIM-1 and underscored the potential role of these elements in the dissemination of acquired MBL genes. Given the location of the ISCR19-like elements flanking the blaFIM-1 gene and the mechanisms by which ISCR elements can mobilize flanking DNA regions (34), it is likely that an ancestor of ΔISCR19.2 or ISCR19.3 were originally involved in the capture of the blaFIM-1 gene from its original host and in its mobilization. ISCR19 was originally reported in association with the blaOXA-18 gene, encoding a class D β-lactamase unusually susceptible to clavulanic acid, detected in a P. aeruginosa clinical isolate from France (35). This ISCR element was considered to be at the origin of the blaOXA-18 gene capture and likely involved in its mobilization and spread (31).
Southern blot experiments, carried out with blaFIM-1 and a 16S rRNA gene probe following pulsed-field gel electrophoresis separation of genomic DNA after digestion with I-CeuI restriction endonuclease or S1 nuclease, revealed that the blaFIM-1 probe hybridized with chromosomal DNA (data not shown) and suggested that the acquired MBL gene was inserted into the chromosome. Indeed, attempts at transferring the MBL gene to E. coli or P. aeruginosa by conjugation or electrotransformation were unsuccessful.
Functional characterization of FIM-1.
In comparison to DH5α carrying the cloning vector, E. coli DH5α(pSPo1) carrying the cloned blaFIM-1 gene showed decreased susceptibility to most β-lactams, including penicillins, cephalosporins, and carbapenems. Only temocillin and aztreonam MICs were unaffected by the presence of the MBL gene (Table 2). Taken together, these results suggested that FIM-1 had a broad substrate specificity.
For biochemical characterization, the FIM-1 enzyme was produced in the E. coli strain Rosetta(pET-FIM-1) and purified from a crude lysate of the strain by means of two anion-exchange chromatography steps. The overall yield of purified enzyme was 4 mg/liter of culture, and the purity was estimated to be >95% by SDS-PAGE (data not shown).
The kinetic parameters of purified FIM-1 for the hydrolysis of a representative set of β-lactam substrates were determined. Like most other subclass B1 MBLs, FIM-1 exhibited a broad substrate profile including penicillins, cephalosporins and carbapenems (Table 3). Aztreonam hydrolysis was not detected with enzyme concentrations up to 400 nM. The highest catalytic efficiencies (kcat/Km ≥106 M−1 s−1) were observed with ampicillin, piperacillin, and carbapenems due to either very low Km values (e.g., ertapenem), relatively high turnover rates (e.g., imipenem, ampicillin, and piperacillin), or a combination thereof (e.g., meropenem). Activity against cephalosporins was overall lower, with high Km values observed for oxyimino-cephalosporins. The apparently higher impact of FIM-1 production on ceftazidime susceptibility versus cefepime (Table 2), despite an overall similar catalytic efficiency, was likely due to the slower permeant nature of the former substrate across the outer membrane. Temocillin, an α-methoxy-substituted penicillin, was also poorly recognized by the enzyme, as observed with many other MBLs, and showed the lowest catalytic efficiencies among the tested substrates (Table 3).
Table 3.
Steady-state kinetic parameters of purified FIM-1 enzymes
| Substratea | Steady-state kinetic parametersb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FIM-1 |
NDM-1 |
VIM-2 |
IMP-1 |
|||||||||
| kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) | |
| Ampicillin* | 150 | 8.3 | 1.8 × 107 | 15 | 22 | 6.6 × 105 | 125 | 90 | 1.4 × 106 | 950 | 200 | 4.8 × 106 |
| Piperacillin | 420 | 36 | 1.2 × 107 | 14 | 12 | 1.2 × 105 | 300 | 125 | 2.4 × 106 | – | – | 7.2 × 105 |
| Temocillin† | >0.3 | >200 | 1.5 × 103 | – | – | – | 7.7 | 390 | 2.0 × 104 | – | >2,000 | <100 |
| Cefoxitin | 57 | 116 | 5.0 × 105 | 1 | 49 | 2.0 × 104 | 15 | 13 | 1.2 × 106 | 16 | 8 | 2.0 × 106 |
| Ceftazidime† | >80 | >200 | 4.0 × 105 | 5 | 181 | 2.8 × 104 | 3.6 | 72 | 5.0 × 104 | 8 | 44 | 1.8 × 105 |
| Cefepime† | >25 | >200 | 1.2 × 105 | 13 | 77 | 1.7 × 105 | >40 | >400 | 1.0 × 105 | 7 | 11 | 6.6 × 105 |
| Imipenem | 153 | 5.1 | 3.0 × 107 | 20 | 94 | 2.1 × 105 | 34 | 9 | 3.8 × 106 | 46 | 39 | 1.2 × 106 |
| Meropenem* | 19 | 2.2 | 8.6 × 106 | 12 | 49 | 2.5 × 105 | 5 | 2 | 2.5 × 106 | 5 | 10 | 5.0 × 105 |
| Ertapenem* | 8 | 0.5 | 1.6 × 107 | – | – | – | – | – | – | 16 | 21 | 7.6 × 105 |
*, The Km was obtained with an inhibition assay using cefoxitin at 200 μM as the reporter substrate; †, pseudo-first-order kinetics in the range of the tested concentrations were observed with these substrates.
In comparison to representatives of the most widespread MBL types (e.g., IMP-1, VIM-2, and NDM-1), the FIM-1 enzyme appeared to be the most efficient carbapenemase, with an unusually high turnover rate for imipenem (kcat, 150 s−1), which translated into relatively high MICs when the enzyme was expressed in the E. coli laboratory strain (Table 2). Another peculiar feature of FIM-1 was represented by the high apparent affinity for ampicillin (Km, ∼8 μM). On the other hand, the oxyimino-cephalosporins ceftazidime and cefepime were poorly recognized by FIM-1 (Km values of >200 μM), possibly depending on the positively charged substituent found in the R2 position. This situation is, to some extent, similar to that of VIM-2 and NDM-1, being very different from that observed with IMP-1, underlining the structural and functional heterogeneity found in subclass B1 acquired MBLs.
Concluding remarks.
FIM-1 is a new acquired MBL detected in a P. aeruginosa clinical isolate from Italy, and its identification underscores the increasing diversity of acquired MBLs that can be encountered in the clinical setting. The patient's history suggested an autochthonous origin for the blaFIM-1 gene, while the similarity of its product with resident proteins of myxobacteria and alphaproteobacteria suggested that progenitors of FIM-1 could be found among members of those taxa.
Thus far we have no information about additional infections caused by P. aeruginosa or other Gram-negative pathogens producing the FIM-1 MBL, suggesting a low spreading potential. However, the association of blaFIM-1 with a P. aeruginosa strain belonging to the ST-235 epidemic lineage could entail some risk for dissemination, and it will be of interest to screen for this new MBL gene in collections of P. aeruginosa and other Gram-negatives from this and other hospitals.
Similar to other acquired MBLs, FIM-1 exhibited an extended substrate specificity, including most β-lactams. However, the enzyme also showed some peculiar functional features (e.g., the marked preference for penicillins and carbapenems and the remarkably high carbapenemase activity) that could make it an interesting model for further investigation of structure-function relationships of MBLs.
ACKNOWLEDGMENTS
This study was partially supported by the EU 7th Framework Programme projects TROCAR (HEALTH-F3-2008-223031) and TEMPOtest-QC (HEALTH-F3-2009-241742).
Footnotes
Published ahead of print 31 October 2012
REFERENCES
- 1. Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect. Dis. 11:381–393 [DOI] [PubMed] [Google Scholar]
- 2. Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. 1991. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35:147–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini GM. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giani T, Marchese A, Coppo E, Kroumova V, Rossolini GM. 2012. VIM-1-producing Pseudomonas mosselii isolates in Italy, predating known VIM-producing index strains. Antimicrob. Agents Chemother. 56:2216–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wachino J, Yoshida H, Yamane K, Suzuki S, Matsui M, Yamagishi T, Tsutsui A, Konda T, Shibayama K, Arakawa Y. 2011. SMB-1, a novel subclass B3 metallo-β-lactamase, associated with ISCR1 and a class 1 integron, from a carbapenem-resistant Serratia marcescens clinical isolate. Antimicrob. Agents Chemother. 55:5143–5149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. El Salabi A, Borra PS, Toleman MA, Samuelsen Ø, Walsh TR. 2012. Genetic and biochemical characterization of a novel metallo-β-lactamase, TMB-1, from an Achromobacter xylosoxidans strain isolated in Tripoli, Libya. Antimicrob. Agents Chemother. 56:2241–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cornaglia G, Akova M, Amicosante G, Cantón R, Cauda R, Docquier JD, Edelstein M, Frère JM, Fuzi M, Galleni M, Giamarellou H, Gniadkowski M, Koncan R, Libisch B, Luzzaro F, Miriagou V, Navarro F, Nordmann P, Pagani L, Peixe L, Poirel L, Souli M, Tacconelli E, Vatopoulos A, Rossolini GM, ESCMID Study Group for Antimicrobial Resistance Surveillance 2007. Metallo-β-lactamases as emerging resistance determinants in Gram-negative pathogens: open issues. Int. J. Antimicrob. Agents 29:380–388 [DOI] [PubMed] [Google Scholar]
- 8. Bebrone C. 2007. Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 74:1686–1701 [DOI] [PubMed] [Google Scholar]
- 9. Partridge SR. 2011. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol. Rev. 35:820–855 [DOI] [PubMed] [Google Scholar]
- 10. Clinical and Laboratory Standards Institute 2012. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically; approved standard, 9th ed CLSI document M07-A9. CLSI, Wayne, PA [Google Scholar]
- 11. Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 42:5644–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pitout JD, Gregson DB, Poirel L, McClure JA, Le P, Church DL. 2005. Detection of Pseudomonas aeruginosa producing metallo-β-lactamases in a large centralized laboratory. J. Clin. Microbiol. 43:3129–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 14. Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70:119–123 [DOI] [PubMed] [Google Scholar]
- 15. Johnson JL. 1994. Similarity analysis of DNAs, p 655–682 In Gerhardt P, Murray RGE, Wood WA, Krieg NR. (ed), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC [Google Scholar]
- 16. Rossolini GM, Condemi MA, Pantanella F, Docquier JD, Amicosante G, Thaller MC. 2001. Metallo-β-lactamase producers in environmental microbiota: new molecular class B enzyme in Janthinobacterium lividum. Antimicrob. Agents Chemother. 45:837–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38(webserver issue):W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu SL, Hessel A, Sanderson KE. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for rRNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:6874–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. García A, Navarro F, Miró E, Villa L, Mirelis B, Coll P, Carattoli A. 2007. Acquisition and diffusion of blaCTX-M-9 gene by R478-IncHI2 derivative plasmids. FEMS Microbiol. Lett. 271:71–77 [DOI] [PubMed] [Google Scholar]
- 20. Docquier JD, Riccio ML, Mugnaioli C, Luzzaro F, Endimiani A, Toniolo A, Amicosante G, Rossolini GM. 2003. IMP-12, a new plasmid-encoded metallo-β-lactamase from a Pseudomonas putida clinical isolate. Antimicrob. Agents Chemother. 47:1522–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 22. Docquier JD, Lamotte-Brasseur J, Galleni M, Amicosante G, Frère JM, Rossolini GM. 2003. On functional and structural heterogeneity of VIM-type metallo-β-lactamases. J. Antimicrob. Chemother. 51:257–266 [DOI] [PubMed] [Google Scholar]
- 23. Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35:736–755 [DOI] [PubMed] [Google Scholar]
- 24. Sardelic S, Bedenic B, Colinon-Dupuich C, Orhanovic S, Bosnjak Z, Plecko V, Cournoyer B, Rossolini GM. 2012. Infrequent finding of metallo-β-lactamase VIM-2 in carbapenem-resistant Pseudomonas aeruginosa strains from Croatia. Antimicrob. Agents Chemother. 56:2746–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ranellou K, Kadlec K, Poulou A, Voulgari E, Vrioni G, Schwarz S, Tsakris A. 2012. Detection of Pseudomonas aeruginosa isolates of the international clonal complex 11 carrying the blaPER-1 extended-spectrum β-lactamase gene in Greece. J. Antimicrob. Chemother. 67:357–361 [DOI] [PubMed] [Google Scholar]
- 26. Yoo JS, Yang JW, Kim HM, Byeon J, Kim HS, Yoo JI, Chung GT, Lee YS. 2012. Dissemination of genetically related IMP-6-producing multidrug-resistant Pseudomonas aeruginosa ST235 in South Korea. Int. J. Antimicrob. Agents 39:300–304 [DOI] [PubMed] [Google Scholar]
- 27. Laraki N, Franceschini N, Rossolini GM, Santucci P, Meunier C, de Pauw E, Amicosante G, Frère JM, Galleni M. 1999. Biochemical characterization of the Pseudomonas aeruginosa 101/1477 metallo-β-lactamase IMP-1 produced by Escherichia coli. Antimicrob. Agents Chemother. 43:902–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Franceschini N, Caravelli B, Docquier JD, Galleni M, Frère JM, Amicosante G, Rossolini GM. 2000. Purification and biochemical characterization of the VIM-1 metallo-β-lactamase. Antimicrob. Agents Chemother. 44:3003–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murphy TA, Simm AM, Toleman MA, Jones RN, Walsh TR. 2003. Biochemical characterization of the acquired metallo-β-lactamase SPM-1 from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 47:582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Galleni M, Lamotte-Brasseur J, Rossolini GM, Spencer J, Dideberg O, Frère JM, Metallo-β-Lactamases Working Group 2001. Standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 45:660–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Naas T, Namdari F, Bogaerts P, Huang TD, Glupczynski Y, Nordmann P. 2008. Genetic structure associated with blaOXA-18, encoding a clavulanic acid-inhibited extended-spectrum oxacillinase. Antimicrob. Agents Chemother. 52:3898–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poirel L, Magalhaes M, Lopes M, Nordmann P. 2004. Molecular analysis of metallo-β-lactamase gene blaSPM-1-surrounding sequences from disseminated Pseudomonas aeruginosa isolates in Recife, Brazil. Antimicrob. Agents Chemother. 48:1406–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yong D, Toleman MA, Bell J, Ritchie B, Pratt R, Ryley H, Walsh TR. 2012. Genetic and biochemical characterization an acquired subgroup B3 metallo-β-lactamase gene, blaAIM-1, and its unique genetic context in Pseudomonas aeruginosa from Australia. Antimicrob. Agents Chemother. doi:10.1128/AAC.05654-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Philippon LN, Naas T, Bouthors AT, Barakett V, Nordmann P. 1997. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2188–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]