Abstract
Mixed lineage leukemia (MLL) fusion protein (FP)-induced acute leukemia is highly aggressive and often refractory to therapy. Recent progress in the field has unraveled novel mechanisms and targets to combat this disease. Menin, a nuclear protein, interacts with wild-type (WT) MLL, MLL-FPs and other partners such as the chromatin-associated protein LEDGF and the transcription factor c-myb to promote leukemogenesis. The newly solved co-crystal structure illustrating the menin-MLL interaction, coupled with the role of menin in recruiting both WT MLL and MLL-FPs to target genes, highlights menin as a scaffold protein and a central hub controlling this type of leukemia. The menin/WT MLL/MLL-FP hub may also cooperate with several signaling pathways, including Wnt, GSK3, and bromodomain-containing Brd4-related pathways to sustain MLL-FP-induced leukemogenesis, revealing new therapeutic targets to improve the treatment of MLL-FP leukemias.
Keywords: Menin, MLL, Chromatin, Leukemia, Therapy
Introduction
Chromosomal translocations involving the Mixed Lineage Leukemia (MLL) gene occur with one of multiple partner genes, leading to the formation and expression of MLL fusion proteins (MLL-FPs) and the development of acute leukemias [1]. These acute leukemias can be lymphoid (ALL), myeloid (AML), or biphenotypic in nature [2]. MLL translocations are found in ~10% of all leukemias and the majority of infant leukemia cases, and patients harboring this genetic abnormality have a particularly poor prognosis [3,4].
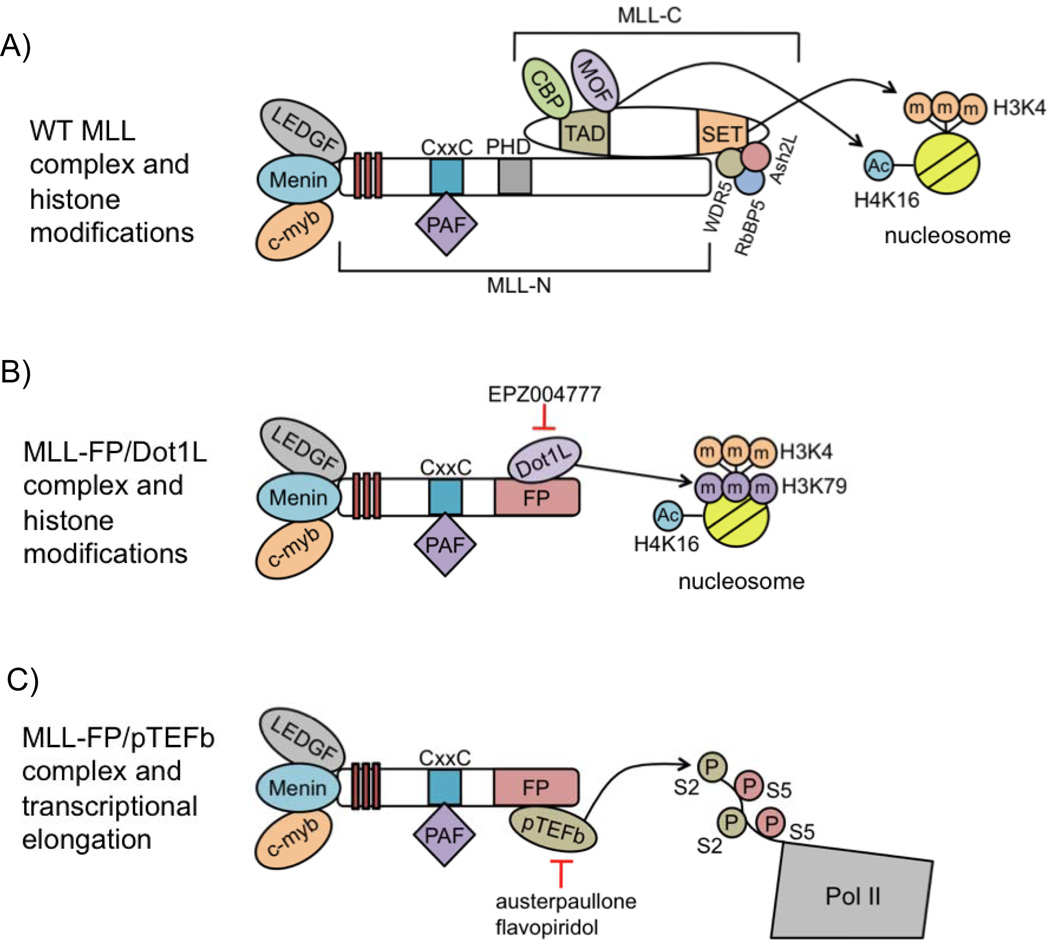
The wild-type (WT) MLL protein is post-translationally cleaved by the protease taspase-1 into N-terminal (MLL-N) and C-terminal (MLL-C) fragments, which then re-associate to form the MLL complex [5]. WT MLL is recruited via MLL-N to target genes through interactions with various proteins, including menin, LEDGF, and C-Myb, as well as its CxxC domain, leading to transcriptional activation of these targets (Figure 1A) [6–9]. Target gene activation is mediated at least in part through histone H3 lysine 4 trimethylation (H3K4m3) catalyzed by the MLL C-terminal SET domain, in concert with the cofactors Wdr5, Ash2L and Rbbp5 (Figure 1A) [10–12]. Direct WT MLL targets include HOX genes, which are activated in a controlled manner during normal hematopoiesis, with high expression in progenitor cells and decreased expression correlating with cell differentiation [13,14].
Figure 1.
MLL-FPs retain a portion of MLL-N, including domains important for its recruitment to MLL target genes, but lack a large C-terminal portion of the protein, including the H3K4-methylating SET domain (Figure 1B, C). Although MLL-C is lacking in MLL-FPs, MLL-FP expression still leads to the upregulation of HOX genes [15,16], which restrain progenitor cells from differentiation and cause leukemia when overexpressed in mouse bone marrow (BM) [17]. These findings appear to suggest that leukemogenesis does not require the C-terminal portion of MLL (Figure 1B, C).
However, one allele of WT MLL remains intact in MLL-FP-expressing leukemia cells. Recent work has demonstrated a critical role for the remaining WT allele of MLL in MLL-AF9-mediated leukemogenesis [8,18]. The biochemical mechanisms by which MLL-FPs affect chromatin modification and transcriptional elongation have been extensively reviewed [19–23]. This review focuses on the roles of, and interplay between, menin, WT MLL and MLL-FPs in gene transcription and leukemic transformation in MLL-FP-mediated leukemia, as well as their potential interplay with other proteins and signaling pathways. Recent findings highlight menin as a central component regulating MLL-FP-mediated leukemia, and a potential target for the pharmacological inhibition of MLL-FP leukemias.
MLL Fusion Proteins Promote HOX Gene Expression and Leukemogenesis
The driving force behind MLL-FP leukemia is the fusion protein itself, as its expression causes HOX gene upregulation and leukemic transformation [16,24,25]. Leukemogenesis is achieved even though a large C-terminal portion of WT MLL, which is normally required for the activation of HOX gene transcription, is lost due to the chromosomal translocation. Some less frequently occurring MLL-FPs have cytosolic fusion partners, such as AF6, GAS7, EEN, and Septin proteins, and MLL-GAS7 does not require Hoxa9 for BM transformation [20,26]. However, the most common MLL-FPs have nuclear fusion partners, and these MLL-FPs upregulate HOX genes to promote leukemogenesis [20]. It was initially puzzling how these MLL-FPs activate HOX genes to an even greater extent than WT MLL. Several important studies have since led to a greater understanding of the mechanism of MLL-FP-mediated HOX gene upregulation and leukemogenesis.
Dot1L interacts with MLL-FPs, and is required for MLL-FP-Mediated Transformation
Many MLL-FPs recruit the histone H3 lysine 79 (H3K79) methyltransferase Dot1L to target genes, promoting leukemogenesis. MLL translocation partners, including AF9, ENL, and AF10, exist in a complex with Dot1L, and MLL-FPs containing these fusion partners retain the ability to interact with Dot1L, leading to increased H3K79 methylation at target genes [27,28]. Although it is not yet known how H3K79 methylation regulates transcription, it is frequently found at active genes [29].
Enhanced transcription of MLL targets is associated with MLL-FP recruitment of Dot1L to these genes. Hox gene upregulation and leukemic transformation is contingent upon the interaction between MLL-AF10 and Dot1L [30,31]. Additionally, direct fusion of Dot1L to MLL-N transforms mouse BM, while a Dot1L catalytic mutant fused to MLL-N fails to do so [30]. These findings suggest that Dot1L catalytic activity is necessary for MLL-FP-mediated leukemogenesis. Also, MLL-FP-mediated bone marrow transformation is suppressed by deletion or catalytic inhibition of Dot1L [32–36]. These observations support a model whereby MLL-FPs recruit Dot1L to target genes, leading to enhanced H3K79 methylation, upregulation of these genes, and leukemogenesis (Figure 1B).
However, it is unclear how H3K79m2 might promote HOX gene expression, and an MLL-Dot1L fusion that retains its catalytic activity but cannot recruit pTEFb (see below) to target genes is unable to transform mouse BM [37], suggesting that recruitment of Dot1L catalytic activity alone is insufficient for leukemic transformation. In addition, MLL-AF4, which interacts with pTEFb but not Dot1L, causes leukemia [37], suggesting that direct Dot1L recruitment is not absolutely required for leukemogenesis mediated by at least some MLL-FPs. Even so, H3K79 methylation is still enriched at MLL targets in MLL-AF4 cells [38], and Dot1L catalytic inhibition is effective in reducing Hox gene expression and viability in these cells [36]. Future work may provide a clearer understanding as to how Dot1L regulates transcription and its precise role in MLL-FP leukemic transformation.
PTEFb interacts with MLL-FPs, and promotes HOX gene transcription and leukemogenesis
MLL-FP recruitment of the pTEFb complex to target genes promotes transcriptional elongation, providing another mechanism for transformation by MLL-FPs. Some of the same MLL fusion partners that interact with Dot1L, such as AF9 and ENL, as well as other fusion partners, including AF4, are part of a distinct complex containing pTEFb, a kinase consisting of the Cyclin T and CDK9 proteins, that promotes transcriptional elongation [27,39,40]. Transcriptional elongation is regulated at many developmentally relevant genes that have an initiated or “poised” RNA polymerase II (Pol II) resting at their promoters [41]. Initiated Pol II is phosphorylated by pTEFb at serine 2 of its C-terminal domain (CTD), causing Pol II to be released from the promoter, and allowing transcriptional elongation [42].
The expression of MLL-FPs, including MLL-AF9, -ENL, and –AF4, leads to pTEFb recruitment to MLL target genes and enhanced transcriptional elongation (Figure 1C) [37,43]. MLL-FP leukemia cell lines are more sensitive to the CDK9 inhibitors flavopiridol and alsterpaullone than non-MLL-FP cell lines, suggesting that dysregulated transcriptional elongation at MLL targets is at least one mechanism for leukemic transformation by MLL-FPs (Figure 1C) [43]. The finding that MLL-FPs recruit Dot1L and/or pTEFb to target genes, leading to their upregulation, has provided insight into the mechanism of MLL-FP-mediated leukemogenesis.
Wild-Type MLL is Required for MLL-FP-Mediated HOX Gene Upregulation and Leukemogenesis
In addition to the MLL-FP, expression of non-translocated WT MLL is required to maintain the transformed state of MLL-FP leukemia cells, likely through C-terminal domains that are lacking in the fusion protein. While MLL-FP cells have one translocated allele, which results in the expression of MLL-FPs, the other allele of MLL remains intact and expresses full-length WT MLL. WT MLL is not only expressed, but binds target genes in MLL-FP cell lines [18,44]. MLL-FPs lack a large C-terminal portion of WT MLL that is normally necessary for target gene activation. However, MLL-C function is still carried out in MLL-FP cells by expression of WT MLL from the non-translocated allele.
Non-translocated WT MLL is critical for maintaining the transformed state of MLL-FP leukemias, and the survival of MLL-AF9 mice is prolonged by WT MLL knockout [18]. WT MLL depletion in human MLL-AF9 cells not only leads to a decrease in HOX gene transcription and WT MLL-mediated H3K4m3, but also reduces MLL-AF9-induced H3K79m2, indicating that MLL-FP function requires WT MLL [18]. WT MLL is necessary for MLL-AF9 recruitment to Hox genes in MEFs, but there is no physical interaction between MLL-AF9 and WT MLL [8]. Therefore, WT MLL function may indirectly lead to MLL-FP recruitment through chromatin modifications mediated by C-terminal domains that are lacking in MLL-FPs, such as the trans-activation domain (TAD) and SET domain (Figure 1A).
The potential role of WT MLL-mediated H3K4m3 in MLL-FP leukemias
MLL-C SET domain-catalyzed H3K4m3 is associated with transcriptional activation, and may be important for the activation of WT MLL/MLL-FP target genes through the recruitment of certain proteins that specifically bind this chromatin modification. Mice expressing MLL lacking the SET domain are viable, but exhibit skeletal defects and decreased Hox expression, suggesting that the SET domain and H3K4m3 are required for optimal expression of MLL target genes during development [12].
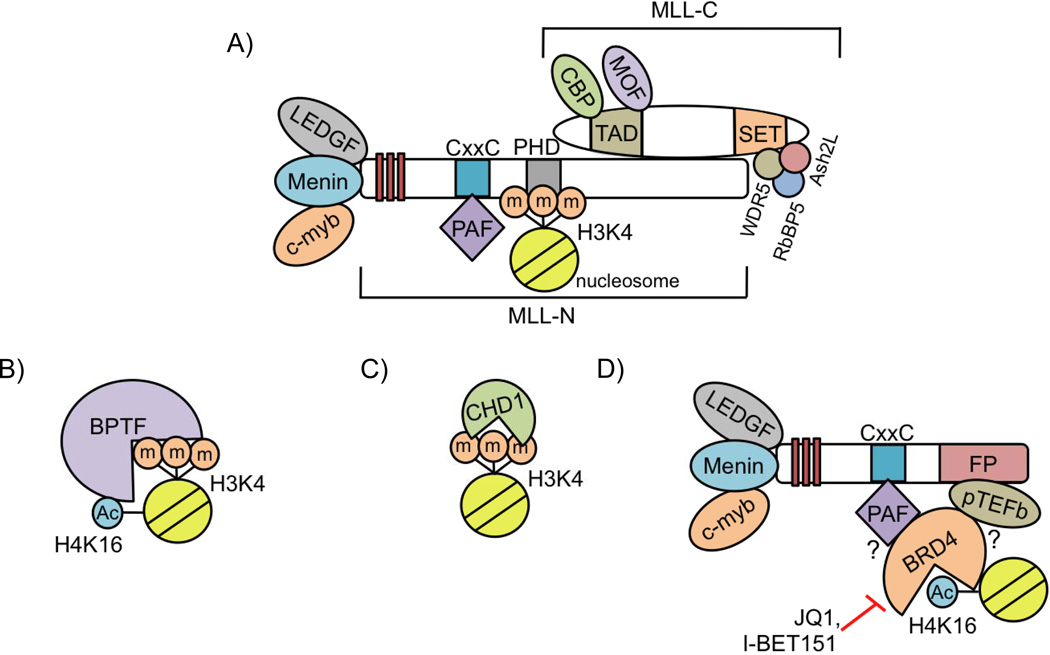
H3K4m3 is specifically recognized by various “reader” proteins. One of these readers is WT MLL itself, which recognizes H3K4m3 via its third PHD (PHD3) domain at target promoters (Figure 2A) [45,46]. The H3K4m3 mark is recognized by PHD3 through a hydrophobic cavity"reading” the gene as active, and retaining the active chromatin state [45]. It is likely that the recognition of H3K4m3 by PHD3 allows the propagation of active histone modifications catalyzed by WT MLL and its interacting partners, maintaining the chromatin in a conformation conducive to MLL-FP binding. MLL-FPs invariably lack the WT MLL PHD domains. Interestingly, overexpression of MLL-ENL containing the PHD3 domain fails to transform mouse BM [47]. However, it is not yet understood why MLL-FPs must contain MLL-N lacking the PHD domains.
Figure 2.
Another reader of H3K4m3 is BPTF, a subunit of the NuRF chromatin-remodeling complex. BPTF specifically binds nucleosomes that contain both H3K4m3 and acetylated histone H4 at lysine 16 (H4K16Ac) via adjacent PHD- and bromo-domains (Figure 2B) [48–50]. BPTF has also been found to bind Hox loci [49]. Hox gene expression is altered by BPTF knockdown in Xenopus during development, but it is not yet known whether BPTF regulates HOX gene expression in MLL-FP leukemia cells [48].
In MLL-AF9 leukemia cells, the chromodomain-containing protein CHD1 also recognizes H3K4m3, and localizes to Hox genes, but has not yet been implicated in MLL-FP-mediated leukemogenesis (Figure 2C) [9,51]. Future studies will determine whether WT MLL complex-catalyzed H3K4m3 and the reading of this mark by CHD1 and BPTF are involved in restructuring chromatin to allow MLL-FP binding and HOX gene upregulation (Figure 2B,C).
The potential role of the WT MLL TAD and histone acetylation in MLL-FP leukemias
In addition to the SET domain, the WT MLL TAD, which promotes histone acetylation, a feature of actively transcribed genes, may have a role in the activation of MLL target genes in MLL-FP cells. WT MLL interacts with the histone acetyltransferase (HAT) enzymes MOF and CREB binding protein (CBP) (Figure 1A) [52,53]. CBP directly interacts with the WT MLL TAD, and CBP is required for MLL transactivation activity in normal cells [52]. An MLL-CBP fusion aberrantly recruits CBP to MLL targets, causing myeloproliferative disease, supporting a potential role for CBP in MLL-FP pathogenesis [54]. The WT MLL TAD also interacts with MOF, an H4K16 acetyltransferase [53].
The dual bromodomain-containing protein Brd4 recognizes acetylated histones [55]. However, it is not known whether Brd4 recognizes histone acetylation mediated by WT MLL at target genes. Nonetheless, Brd4 may have a role in MLL-FP recruitment to target genes. Brd4 recruits PAFc and pTEFb to gene loci [56,57], and PAFc/pTEFb interact with MLL-FPs [37,58], providing a link between MLL-FPs and chromatin (Figure 2D). In support of this model, Brd4 inhibition causes a decrease in PAFc, pTEFb, and PolII CTD phospho-S2 at the promoter of Bcl-2, a direct MLL-FP target gene [57].
Also, BPTF recognizes H3K4m3 and H4K16Ac simultaneously, suggesting that the TAD may act in concert with the SET domain to maintain chromatin in a conformation conducive to MLL-FP binding [48,49]. Future studies will determine whether WT MLL SET domain-catalyzed H3K4m3 and/or WT MLL TAD-mediated histone acetylation are involved in MLL-FP-mediated leukemogenesis.
WT MLL and MLL-FPs Share N-terminal Domains Critical For Their Recruitment to Target Genes
Although the C-terminal portions of WT MLL and MLL-FPs possess quite distinct activities, the N-terminal regions of these proteins, which are responsible for recruitment to target genes, are mostly shared and function in a similar manner with only subtle differences (Figure 1).
Menin captures the N-terminus of MLL through a deep pocket
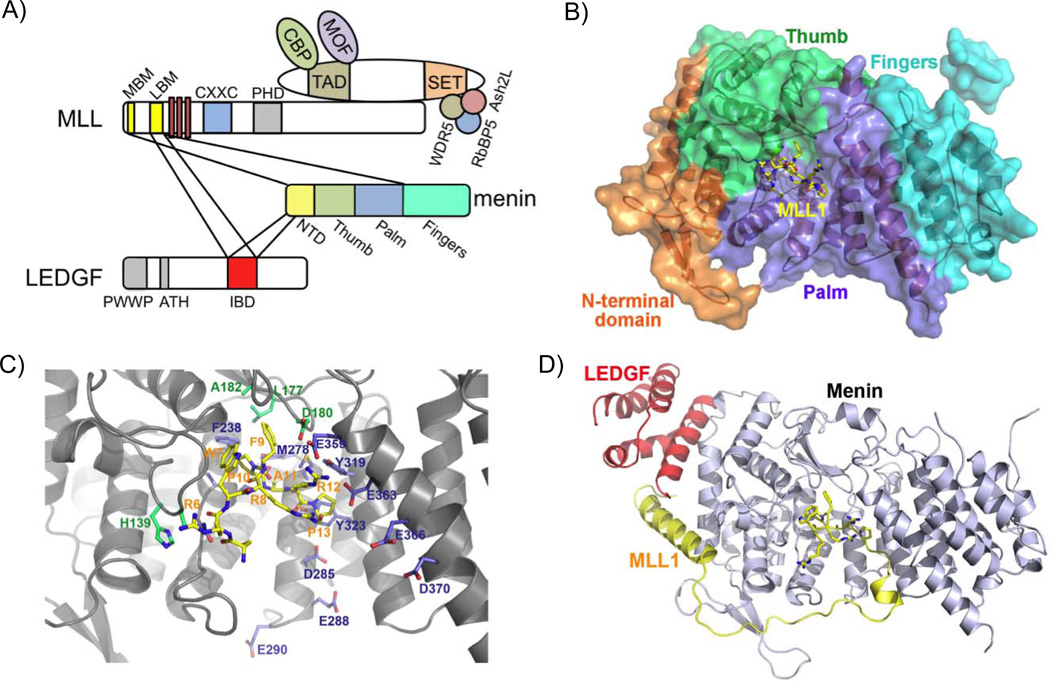
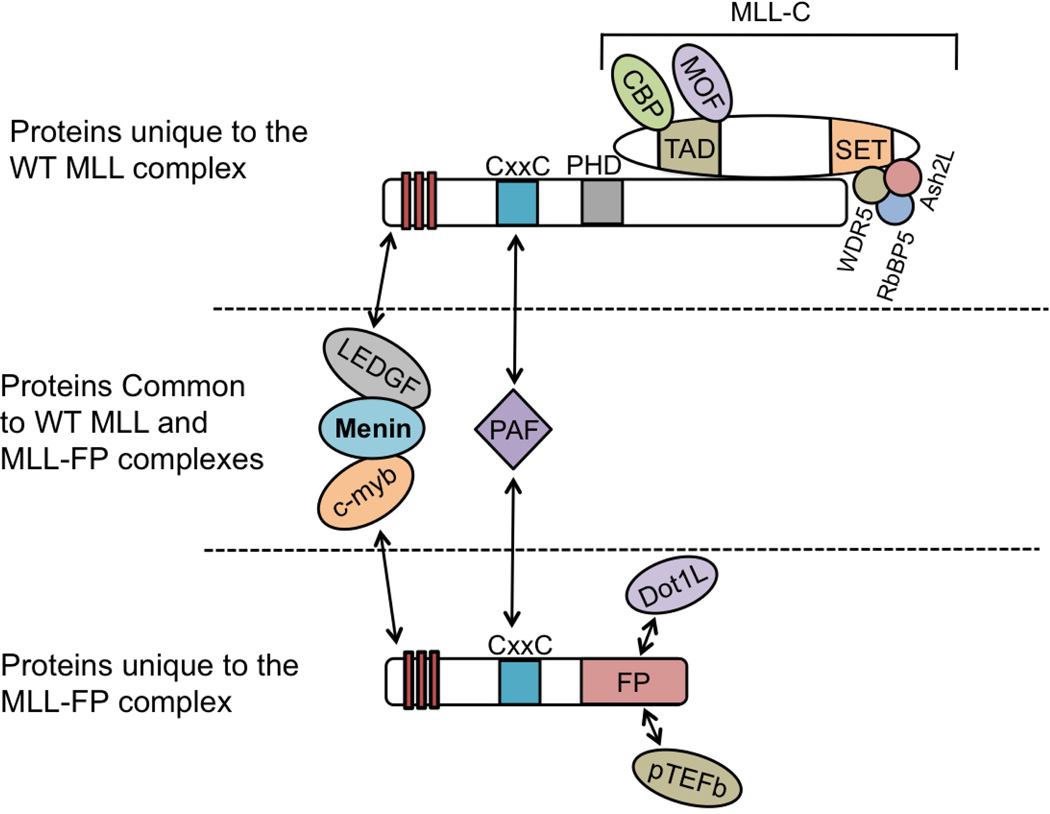
The interaction between MLL-N and menin is pivotal to the physiological as well as the pathological roles of MLL [9,59–61]. MLL-dependent transcription and MLL-FP-mediated leukemic transformation also require the LEDGF protein, which interacts with MLL-N and menin [6]. Recently, crystallographic studies of the structures of menin in its free form and in complex with MLL or an MLL-LEDGF heterodimer explain how menin acts as a central “hub” through its role in recruiting both WT MLL and MLL-FPs to target genes [62,63].
Menin interacts with a bipartite motif at the extreme N-terminus of MLL consisting only of residues 6–25 (MLL1MBM: menin-binding motif), a region that is found in both WT MLL and MLL-FPs. The crystal structures of menin alone and in complex with the MLLMBM peptide reveal that menin adopts a rectangular-shaped conformation, resembling a curved left hand, with a deep pocket formed by its thumb and palm domains (Figure 3A, B). The thumb domain of menin is structurally similar to the transglutaminase-like fold of the transglutaminase superfamily, and the palm domain contains three TPR motifs that usually serve as protein-protein interaction modules. The MLLMBM peptide adopts a highly coiled conformation and plugs into the deep pocket of menin. The interaction affinity and specificity between menin and MLLMBM is mainly determined by extensive hydrophobic contacts at the bottom of the deep pocket of menin (Figure 3C). Disruption of the interaction between menin and MLL-N diminishes WT MLL-mediated H3K4m3 and MLL-AF9 enrichment at the Hoxa9 promoter and decreases Hoxa9 expression in transformed mouse BM, highlighting the importance of the menin-MLL-N interaction for MLL-FP-mediated leukemogenesis (Figure 4) [60].
Figure 3.
Figure 4.
Menin serves as a scaffold protein to assemble a menin-MLL-LEDGF ternary complex
Menin/WT MLL and menin/MLL-FP complexes are guided to MLL target genes through additional interactions with the chromatin-associated LEDGF protein. [6,62]. Notably, LEDGF binds to menin/MLL-N with high affinity, whereas neither menin nor MLL-N alone stably associate with LEDGF. The crystal structure of the menin/MLL-N/LEDGF complex also indicates that the integrase binding domain of LEDGF (LEDGFIBD) binds to a V-shaped groove formed by both menin and MLL-N (Figure 3D). Menin acting as a scaffold protein to mediate the MLL-N/LEDGF interaction underscores the central role of menin in MLL-FP leukemias (Figure 4).
C-Myb interacts with menin and is critical for MLL-FP-mediated transformation
In MLL-FP leukemia cells, menin also interacts with the C-Myb transcription factor, which is essential for WT MLL, and presumably, MLL-FP recruitment to target genes (Figure 1) [7]. C-Myb is required for optimal HOX gene expression in human MLL-FP leukemia cell lines, and MLL-ENL-mediated transformation is suppressed by expression of a C-Myb fragment that binds DNA but lacks the menin-interaction domain [7]. The role of the menin/C-Myb interaction in MLL-FP leukemias again highlights menin as a central player in this disease (Figure 4).
The CxxC Domain is critical for WT MLL and MLL-FP recruitment to target genes
Another common feature of WT MLL and MLL-FPs is the CxxC domain, which interacts with the PAFc complex and is required for WT MLL and MLL-FP binding to Hox genes (Figure 1) [8,58]. The CxxC domain also interacts with unmethylated CpG DNA sites [64–66]. Interestingly, disruption of the CxxC-DNA interaction with a point mutation (K1176A) abrogates MLL-AF9, but not WT MLL binding to the Hoxa9 promoter [64]. This discrepancy could be due to the fact that MLL-FPs lack PHD3, which may stabilize WT MLL at target genes, further suggesting that an “open” chromatin conformation is critical for MLL-FP binding and stabilization at target genes.
However, the necessity of the CxxC-DNA interaction for MLL-AF9 recruitment is still unclear, as another point mutation abrogating CxxC DNA binding (C1188D) has no effect on MLL-AF9 binding in MEFs, but is still required for MLL-AF9-mediated leukemogenesis, potentially through affecting methylation of CpG islands at the Hoxa9 promoter [66]. As WT MLL and MLL-FP recruitment are controlled by the same interacting proteins, which are largely coordinated by menin, menin may be particularly important for regulating two distinct functions controlling HOX gene upregulation and leukemogenesis (Figure 4).
Direct Transcriptional Targets of Menin/WT MLL/MLL-FPs
MLL-FP recruitment to HOX genes and other recently identified target genes causes enhanced transcriptional elongation and subsequent overexpression of these genes. These direct MLL-FP targets play various roles in promoting leukemic disease, highlighting the importance of investigating genes that are upregulated by MLL-FPs. Understanding MLL-FP downstream effectors has yielded further insight into the mechanism by which expression of these fusion proteins causes leukemia.
HOX genes are critical downstream effectors of MLL-FP-mediated leukemogenesis
In MLL-FP leukemias, HOX genes have been implicated in promoting cell survival and blocking differentiation to promote leukemogenesis. In mammalian cells, there are 39 Hox genes residing in 4 distinct clusters on 4 respective chromosomes [67]. In particular, HOXA9 and its dimerization partner MEIS1 are the most well characterized direct targets of the MLL-FP pathway. MLL-AF9 leukemic disease can be mimicked by overexpression HOXA9 and MEIS1 [16,17]. Also, Hoxa9/Meis1 overexpression is able to rescue BM colony formation defects caused by menin or WT MLL depletion, demonstrating that Hox genes are critical downstream mediators of MLL-FP-mediated leukemia [9,18]. In addition, shRNA-mediated depletion of HOXA9 in human MLL-AF9 cells causes growth arrest, increased expression of differentiation-associated genes, and apoptosis, indicating various roles for HOX genes in leukemogenesis [68]. As HOX genes themselves are transcription factors, it is likely that MLL-FP-mediated HOX gene upregulation leads to increased transcription of multiple HOX target genes involved in leukemogenesis. Further investigation into the direct downstream targets of HOXA9/MEIS1 will yield more insight into the yet uncharacterized mechanisms by which these genes function to regulate critical processes in MLL-FP leukemia cells.
MLL-FPs directly activate additional genes that have a role in leukemogenesis
Recently, global approaches have led to the discovery of previously unrecognized direct MLL-FP target genes that promote MLL-FP-mediated transformation. A comparison of MLL-FP-expressing cells to cells containing only WT MLL has demonstrated that MLL-FPs regulate a subset (~15%) of WT MLL target genes [44]. Two of these genes, Six1 and Eya1, are able to transform mouse BM, demonstrating a role for these proteins in leukemogenesis [44]. Additionally, Evi1, a gene that was discovered as a common retroviral integration site in murine leukemias, was found to be a direct MLL-AF9 target in mouse hematopoietic stem cells (HSCs) [69]. Future experiments may lead to the discovery of more direct MLL-FP targets involved in leukemogenesis.
Signaling Pathways Regulating MLL-FP Leukemias
In addition to direct downstream targets of MLL-FPs, which are critical for leukemic transformation, signaling pathways have been found that cooperate with MLL-FP function to regulate leukemogenesis. These pathways include GSK-3 signaling, which modulates Hoxa9/Meis1 function, Wnt signaling, and the Akt/Foxo pathway. Understanding pathways that function in concert with, or in parallel to MLL-FPs, has led to a broader understanding of the cellular context in which MLL-FPs function.
GSK-3 Regulates HOX function downstream of MLL-FPs
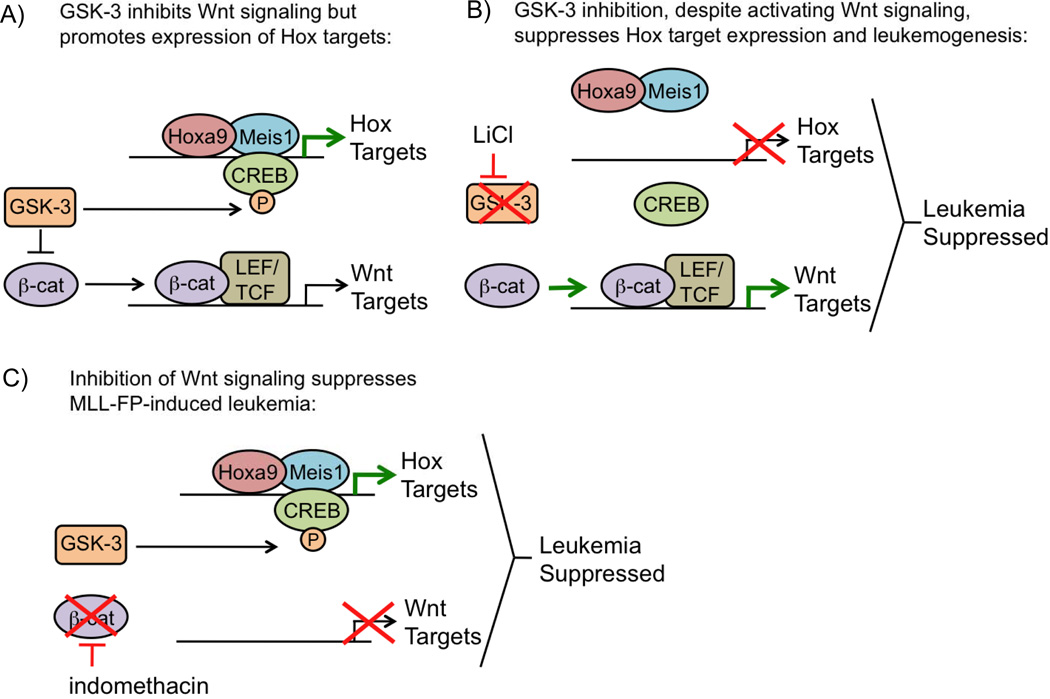
The GSK-3 serine/threonine kinase promotes MLL-FP-mediated leukemogenesis by increasing the recruitment of HOXA9 and its cofactor MEIS1 to target genes. GSK3 has a role in multiple cellular pathways, such as Wnt signaling. Wnt signaling is repressed by GSK-3, which induces β-catenin phosphorylation and degradation [70,71]. Interestingly, independent of its role in Wnt signaling, GSK-3 has an opposing pro-oncogenic role in MLL-FP leukemia, as its inhibition leads to G1 arrest [72]. Further investigation into how the cell cycle is regulated by GSK-3 demonstrated its ability to directly phosphorylate CREB, leading to the formation of a complex including CREB, MEIS1, HOXA9, and CBP to activate HOX/MEIS1 downstream targets (Figure 5A) [73]. These results identify GSK-3-mediated HOX protein recruitment as a critical pathway for MLL-FP-mediated leukemogenesis.
Figure 5.
Wnt Signaling is required for MLL-FP-mediated leukemogenesis
The Wnt pathway is also crucial for MLL-FP-mediated leukemogenesis, as β-catenin knockout impairs the ability of MLL-AF9-expressing cells to cause leukemia (Figure 5C) [74,75]. Although Wnt signaling is essential for MLL-FP leukemia, inhibition of GSK-3, which activates Wnt signaling, leads to MLL-FP cell death through inhibiting HOXA9/MEIS1 binding to target genes (Figure 5B) [72,73]. These results suggest that in the absence of HOXA9/MEIS1 function, Wnt signaling is unable to sustain MLL-FP-mediated leukemogenesis.
MLL-FPs may independently regulate Wnt signaling through their interaction with Dot1L. The native Dot1L complex, in addition to MLL fusion partners, also contains Wnt pathway components TRRAP, Skp1, and β-catenin in 293T cells [28]. This finding suggests that MLL-FPs may regulate Wnt signaling at least in part through their interaction with Dot1L in MLL-FP leukemia cells, possibly through stabilizing or aiding in the recruitment of the Dot1L complex to Wnt targets. Although it is unclear whether Dot1L regulates Wnt signaling in MLL-FP cells, Wnt signaling is upregulated by MLL-FPs, and is essential for MLL-FP leukemia.
Low Akt signaling, resulting in Foxo activation, is required for MLL-FP leukemia stem cells
Akt activity is low in MLL-AF9 leukemia stem cells (LSCs), and overexpression of a constitutively active form of Akt leads to MLL-AF9 cell differentiation [76]. In MLL-FP leukemias, low Akt signaling leads to nuclear translocation and activation of Foxo proteins. Depletion of Foxo1/3/4 in vivo increases the survival rate of mice transplanted with MLL-AF9 transduced primary BM cells, demonstrating a pro-oncogenic role for Foxo genes in MLL-AF9 leukemia, in part through activation of the JNK/Jun pathway [76]. In addition to GSK-3, Wnt, and Akt/Foxo, it is likely that yet unidentified signaling pathways are involved in MLL-FP-mediated leukemogenesis.
Potential Therapeutic Targets of the Menin-WT MLL-MLL-FP pathway
Substantial progress has been made in understanding the mechanism of MLL-FP-mediated leukemogenesis and critical signaling pathways in these cells. Pharmacological inhibition of these targets may be effective in treating patients with this disease.
Inhibition of Wnt signaling as a potential therapy for MLL-FP leukemias
Inhibition of Wnt signaling by β-catenin knockout depletes MLL-FP LSCs and increases the survival of MLL-FP leukemic mice, highlighting the Wnt pathway as a potential target for treating this disease [74,75]. Importantly, Wnt signaling does not affect adult HSCs in mice [77], decreasing the likelihood of toxicity due to Wnt inhibition. The Wnt pathway inhibitor indomethacin, a commonly used anti-inflammatory drug, inhibits Cox1/Cox2 and causes β -catenin degradation in MLL-FP cells [75,78]. Treatment of MLL-AF9 leukemic mice with indomethacin leads to a modest yet significant increase in survival time, suggesting that MLL-FP leukemia can be suppressed by pharmacological inhibition of the Wnt pathway (Figure 5C) [75].
Since Wnt pathway components are found in a complex with Dot1L [28], it is possible that Dot1L inhibition could decrease not only HOX gene expression, but also Wnt signaling. It should be noted, however, that inhibition of Wnt signaling is less effective in later stages of the disease [75], suggesting that a Wnt inhibitor may only be useful in combination with other therapies to effectively treat this disease.
Inhibition of GSK-3 as a potential therapy for MLL-FP leukemias
GSK-3, through phosphorylation of CREB, leads to MEIS1 recruitment to target genes [73]. Since HOXA9/MEIS1 are critical mediators of MLL-FP-mediated leukemogenesis, inhibition of GSK-3 could be a viable treatment option for this type of leukemia. In support of this theory, GSK-3 inhibition with lithium chloride, a common treatment for bipolar disorder, prolongs the survival of MLL-AF4 leukemic mice (Figure 5B) [73].
Although GSK-3 inhibition is initially effective, mice eventually become resistant to therapy and succumb to disease [73]. One possible mechanism for resistance is that in addition to inhibiting HOXA9/MEIS1 targets with GSK-3 inhibition, inhibiting GSK-3 also activates the Wnt pathway, which is essential for MLL-FP leukemia (Figure 5B). MLL-FP cells are re-sensitized to GSK-3 inhibition by Wnt pathway disruption through depletion of β -catenin, suggesting that a combination of GSK-3 and Wnt inhibition may increase treatment efficacy [74].
Directly targeting MLL-FP function to treat MLL-FP leukemias
As MLL-FPs are the driving force behind leukemogenesis, it seems logical to directly target fusion protein function through inhibition of Dot1L or pTEFb. Along these lines, mice xenografted with MLL-AF4 leukemia cells have a modest but significant increase in survival when treated with EPZ004777, a Dot1L catalytic inhibitor, underscoring the potential for targeting Dot1L to treat MLL-FP leukemia (Figure 1B) [36].
Also, pTEFb inhibition with the CDK9 inhibitors flavopiridol or alsterpaullone shows some selectivity toward MLL-FP cell lines (Figure 1C) [43], and clinical trials involving flavopiridol and other CDK9 inhibitors are ongoing for the treatment of chronic lymphocytic leukemia and acute leukemias, presenting a possible avenue for MLL-FP leukemia therapy in the future [79,80].
The use of BET bromodomain inhibitors to treat MLL-FP-induced leukemi a
The potential for treating MLL-FP leukemia patients by blocking Brd4 binding to acetylated histone through chemical inhibition is promising. Two specific inhibitors, which bind the first bromodomain of Brd4 with strong affinity, JQ1 and I-BET151 have been developed (Figure 2D) [57,81,82]. These drugs have shown efficacy in cell culture and animal models, leading to differentiation, cell cycle arrest, and apoptosis of MLL-FP leukemia cells [57,81]. Although the life span of leukemic mice is extended by treatment with Brd4 inhibitors, the animals do succumb to disease, suggesting that the inhibitor needs to be improved or used in combination with other therapies to treat MLL-FP leukemias.
The potential for targeting WT MLL to treat MLL-FP leukemias
WT MLL is required for MLL-FP-mediated leukemogenesis [18]. Thus, blocking WT MLL function could hold therapeutic value for patients with this disease. The development of specific inhibitors of the WT MLL SET domain may inhibit WT MLL function in MLL-FP-mediated leukemias. Additionally, the TAD or PHD3 could potentially be targeted. However, the exact mechanism by which WT MLL promotes MLL-FP-mediated leukemogenesis is unknown. Also, while pharmacological inhibition rarely completely inhibits the function of a protein, complete ablation of WT MLL can cause defects in adult hematopoiesis, suggesting that inhibition of WT MLL function could lead to modest or even severe suppression of normal BM [83–85].
Targeting menin, a central hub regulating WT MLL and MLL-FPs to treat MLL-FP leukemias
Menin directly interacts with WT MLL and MLL-FPs, and is necessary for directing these two critical components for leukemogenesis to HOX genes, establishing menin as a hub in this disease and potential therapeutic target (Figure 4) [9,18,60–63]. Further, menin directly interacts with LEDGF and C-Myb [6,7], strengthening the view that menin acts as a central “hub” for these various complexes suitable for therapeutic targeting (Figure 4). Therefore, inhibiting menin may more effectively treat MLL-FP leukemias. In addition, inhibition of menin may be less toxic, as menin depletion causes a functional defect in LT-HSCs, but does not affect steady-state hematopoiesis [14].
Crystal structures of menin complexes have revealed promising drug targets for the therapy of MLL-FP-mediated leukemia through menin inhibition (Figure 3). The hydrophobic menin pocket that binds MLLMBM is specifically shaped for the phenyl ring of phenylalanine. Substitution of the phenyl ring with an imidazole ring of histidine or a hydroxyphenyl ring of tyrosine abolishes menin-MLL interaction [62]. Thus, inhibition of the menin-MLL interaction could be achieved with small aromatic compounds.
In addition, two phenol rings of MLLLBM bind the hydrophobic pocket of the LEDGFIBD (Figure 3D). This interaction is another potential target for blocking the formation of the menin-MLL-LEDGF complex. A potent inhibitor of the menin/MLL-N interaction has recently been developed. This thienopyromidine class compound, termed MI-2, inhibits the interaction between MLL-FPs and menin, and blocks MLL-FP induced transformation [60], providing a proof of principle for targeting menin in MLL-FP leukemias.
Conclusions and outlook
Recently, significant progress has been made in understanding the mechanisms by which the menin/WT MLL/MLL-FP hub upregulates a subset of MLL target genes and drives leukemogenesis. In addition to MLL target gene upregulation, MLL-FPs work in concert with various signaling pathways to promote leukemia. Menin acts as a central hub through its role in recruiting WT MLL and MLL-FPs to target genes. Menin also links C-Myb/LEDGF to the MLL N-terminus, further highlighting menin’s central role in this disease. A more detailed understanding of the molecular mechanism of menin/WT MLL/MLL-FP-mediated upregulation of target genes, and potential interactions with crucial signaling pathways may provide additional therapeutic targets for this disease.
Increasing knowledge about the mechanism of MLL-FP-mediated leukemogenesis has led to the possibility of treating this disease with targeted therapeutics. MLL-FP function can be suppressed by Inhibition of Dot1L catalytic activity [36], and MLL-FP cells are also sensitive to the BET bromodomain inhibitors JQ1 and I-BET151 [57,81]. JQ1 and I-BET151 are the first in a new class of small molecules inhibiting readers of epigenetic marks. The development of additional drugs in this class could be used to inhibit other reader proteins that may be important for MLL-FP-mediated leukemogenesis (Figure 2). Inhibition of the menin/MLL-N interaction with the small molecule MI-2 is effective in blocking MLL-FP-induced transformation [60], providing proof of principle for targeting this interaction to treat patients with this disease. Inhibiting the menin/MLL/MLL-FP hub in combination with targeted therapies based on pathways interacting with the menin hub or conventional chemotherapeutic agents will likely lead to a more favorable prognosis for MLL-FP leukemia patients in the future.
Abbreviations
- BM
bone marrow
- HSC
hematopoietic stem cell
- LSC
leukemia stem cell
- MLL-FP
MLL fusion protein.
References
- 1.Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, et al. New insights to the MLL recombinome of acute leukemias. Leukemia. 2009;23:1490–1499. doi: 10.1038/leu.2009.33. [DOI] [PubMed] [Google Scholar]
- 2.Daser A, Rabbitts TH. Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev. 2004;18:965–974. doi: 10.1101/gad.1195504. [DOI] [PubMed] [Google Scholar]
- 3.Holleman A, Cheok MH, den Boer ML, Yang W, Veerman AJ, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351:533–542. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 4.Liu H, Cheng EH, Hsieh JJ. MLL fusions: pathways to leukemia. Cancer Biol Ther. 2009;8:1204–1211. doi: 10.4161/cbt.8.13.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsieh JJ, Ernst P, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol Cell Biol. 2003;23:186–194. doi: 10.1128/MCB.23.1.186-194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin S, Zhao H, Yi Y, Nakata Y, Kalota A, et al. c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis. J Clin Invest. 2010;120:593–606. doi: 10.1172/JCI38030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milne TA, Kim J, Wang GG, Stadler SC, Basrur V, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell. 2010;38:853–863. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen YX, Yan J, Keeshan K, Tubbs AT, Wang H, et al. The tumor suppressor menin regulates hematopoiesis and myeloid transformation by influencing Hox gene expression. Proc Natl Acad Sci U S A. 2006;103:1018–1023. doi: 10.1073/pnas.0510347103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 12.Terranova R, Agherbi H, Boned A, Meresse S, Djabali M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci U S A. 2006;103:6629–6634. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pineault N, Helgason CD, Lawrence HJ, Humphries RK. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp Hematol. 2002;30:49–57. doi: 10.1016/s0301-472x(01)00757-3. [DOI] [PubMed] [Google Scholar]
- 14.Maillard I, Chen YX, Friedman A, Yang Y, Tubbs AT, et al. Menin regulates the function of hematopoietic stem cells and lymphoid progenitors. Blood. 2009;113:1661–1669. doi: 10.1182/blood-2009-01-135012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeisig BB, Milne T, Garcia-Cuellar MP, Schreiner S, Martin ME, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, et al. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiel AT, Blessington P, Zou T, Feather D, Wu X, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17:148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muntean AG, Hess JL. The Pathogenesis of Mixed-Lineage Leukemia. Annu Rev Pathol. 2011 doi: 10.1146/annurev-pathol-011811-132434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 21.Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marschalek R. Mixed lineage leukemia: roles in human malignancies and potential therapy. FEBS J. 2010;277:1822–1831. doi: 10.1111/j.1742-4658.2010.07608.x. [DOI] [PubMed] [Google Scholar]
- 24.Wei J, Wunderlich M, Fox C, Alvarez S, Cigudosa JC, et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell. 2008;13:483–495. doi: 10.1016/j.ccr.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.So CW, Karsunky H, Wong P, Weissman IL, Cleary ML. Leukemic transformation of hematopoietic progenitors by MLL-GAS7 in the absence of Hoxa7 or Hoxa9. Blood. 2004;103:3192–3199. doi: 10.1182/blood-2003-10-3722. [DOI] [PubMed] [Google Scholar]
- 27.Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- 28.Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, et al. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24:574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 31.DiMartino JF, Ayton PM, Chen EH, Naftzger CC, Young BD, et al. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. Blood. 2002;99:3780–3785. doi: 10.1182/blood.v99.10.3780. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood. 2011;117:6912–6922. doi: 10.1182/blood-2011-02-334359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang MJ, Wu H, Achille NJ, Reisenauer MR, Chou CW, et al. Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes. Cancer Res. 2010;70:10234–10242. doi: 10.1158/0008-5472.CAN-10-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117:4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, et al. Selective Killing of Mixed Lineage Leukemia Cells by a Potent Small-Molecule DOT1L Inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao SM, Zhang J, Jeffery DA, Koleske AJ, Thompson CM, et al. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 40.Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 41.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 43.Mueller D, Garcia-Cuellar MP, Bach C, Buhl S, Maethner E, et al. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7:e1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang QF, Wu G, Mi S, He F, Wu J, et al. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood. 2011;117:6895–6905. doi: 10.1182/blood-2010-12-324699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang PY, Hom RA, Musselman CA, Zhu L, Kuo A, et al. Binding of the MLL PHD3 finger to histone H3K4me3 is required for MLL-dependent gene transcription. J Mol Biol. 2010;400:137–144. doi: 10.1016/j.jmb.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Song J, Milne TA, Wang GG, Li H, et al. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell. 2010;141:1183–1194. doi: 10.1016/j.cell.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J, Santillan DA, Koonce M, Wei W, Luo R, et al. Loss of MLL PHD finger 3 is necessary for MLL-ENL-induced hematopoietic stem cell immortalization. Cancer Res. 2008;68:6199–6207. doi: 10.1158/0008-5472.CAN-07-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 49.Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, et al. Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol. 2001;21:2249–2258. doi: 10.1128/MCB.21.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 54.Wang J, Iwasaki H, Krivtsov A, Febbo PG, Thorner AR, et al. Conditional MLL-CBP targets GMP and models therapy-related myeloproliferative disease. EMBO J. 2005;24:368–381. doi: 10.1038/sj.emboj.7600521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Z, Yik JH, Chen R, He N, Jang MK, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 57.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17:609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caslini C, Yang Z, El-Osta M, Milne TA, Slany RK, et al. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res. 2007;67:7275–7283. doi: 10.1158/0008-5472.CAN-06-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grembecka J, He S, Shi A, Purohit T, Muntean AG, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol. 2012;8:277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang J, Gurung B, Wan B, Matkar S, Veniaminova NA, et al. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature. 2012;482:542–546. doi: 10.1038/nature10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murai MJ, Chruszcz M, Reddy G, Grembecka J, Cierpicki T. Crystal structure of menin reveals binding site for mixed lineage leukemia (MLL) protein. J Biol Chem. 2011;286:31742–31748. doi: 10.1074/jbc.M111.258186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol Cell Biol. 2004;24:10470–10478. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Allen MD, Grummitt CG, Hilcenko C, Min SY, Tonkin LM, et al. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. EMBO J. 2006;25:4503–4512. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, et al. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol. 2010;17:62–68. doi: 10.1038/nsmb.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He H, Hua X, Yan J. Epigenetic regulations in hematopoietic Hox code. Oncogene. 2011;30:379–388. doi: 10.1038/onc.2010.484. [DOI] [PubMed] [Google Scholar]
- 68.Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arai S, Yoshimi A, Shimabe M, Ichikawa M, Nakagawa M, et al. Evi-1 is a transcriptional target of mixed-lineage leukemia oncoproteins in hematopoietic stem cells. Blood. 2011;117:6304–6314. doi: 10.1182/blood-2009-07-234310. [DOI] [PubMed] [Google Scholar]
- 70.Peifer M, Sweeton D, Casey M, Wieschaus E. wingless signal and Zeste-white 3 kinase trigger opposing changes in the intracellular distribution of Armadillo. Development. 1994;120:369–380. doi: 10.1242/dev.120.2.369. [DOI] [PubMed] [Google Scholar]
- 71.Orford K, Crockett C, Jensen JP, Weissman AM, Byers SW. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J Biol Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 72.Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TC, et al. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455:1205–1209. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Z, Iwasaki M, Ficara F, Lin C, Matheny C, et al. GSK-3 promotes conditional association of CREB and its coactivators with MEIS1 to facilitate HOX-mediated transcription and oncogenesis. Cancer Cell. 2010;17:597–608. doi: 10.1016/j.ccr.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yeung J, Esposito MT, Gandillet A, Zeisig BB, Griessinger E, et al. beta-Catenin mediates the establishment and drug resistance of MLL leukemic stem cells. Cancer Cell. 2010;18:606–618. doi: 10.1016/j.ccr.2010.10.032. [DOI] [PubMed] [Google Scholar]
- 75.Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sykes SM, Lane SW, Bullinger L, Kalaitzidis D, Yusuf R, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011;146:697–708. doi: 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jeannet G, Scheller M, Scarpellino L, Duboux S, Gardiol N, et al. Long-term, multilineage hematopoiesis occurs in the combined absence of beta-catenin and gamma-catenin. Blood. 2008;111:142–149. doi: 10.1182/blood-2007-07-102558. [DOI] [PubMed] [Google Scholar]
- 78.Goessling W, North TE, Loewer S, Lord AM, Lee S, et al. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136:1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karp JE, Smith BD, Resar LS, Greer JM, Blackford A, et al. Phase 1 and pharmacokinetic study of bolus-infusion flavopiridol followed by cytosine arabinoside and mitoxantrone for acute leukemias. Blood. 2011;117:3302–3310. doi: 10.1182/blood-2010-09-310862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Byrd JC, Lin TS, Dalton JT, Wu D, Phelps MA, et al. Flavopiridol administered using a pharmacologically derived schedule is associated with marked clinical efficacy in refractory, genetically high-risk chronic lymphocytic leukemia. Blood. 2007;109:399–404. doi: 10.1182/blood-2006-05-020735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McMahon KA, Hiew SY, Hadjur S, Veiga-Fernandes H, Menzel U, et al. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007;1:338–345. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 84.Jude CD, Climer L, Xu D, Artinger E, Fisher JK, et al. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1:324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gan T, Jude CD, Zaffuto K, Ernst P. Developmentally induced Mll1 loss reveals defects in postnatal haematopoiesis. Leukemia. 2010;24:1732–1741. doi: 10.1038/leu.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]