Abstract
Vacuolar proton-translocating ATPases (V-ATPases) are highly conserved proton pumps consisting of a peripheral membrane subcomplex called V1, which contains the sites of ATP hydrolysis, attached to an integral membrane subcomplex called Vo, which encompasses the proton pore. V-ATPase regulation by reversible dissociation, characterized by release of assembled V1 sectors into the cytosol and inhibition of both ATPase and proton transport activities, was first identified in tobacco hornworm and yeast. It has since become clear that modulation of VATPase assembly level is also a regulatory mechanism in mammalian cells. In this review, the implications of reversible disassembly for V-ATPase structure are discussed, along with insights into underlying subunit-subunit interactions provided by recent structural work. Although initial experiments focused on glucose deprivation as a trigger for disassembly, it is now clear that V-ATPase assembly can be regulated by other extracellular conditions. Consistent with a complex, integrated response to extracellular signals, a number of different regulatory proteins, including RAVE/rabconnectin, aldolase and other glycolytic enzymes, and protein kinase A have been suggested to control V-ATPase assembly and disassembly. It is likely that multiple signaling pathways dictate the ultimate level of assembly and activity. Tissue-specific V-ATPase inhibition is a potential therapy for osteoporosis and cancer; the possibility of exploiting reversible disassembly in design of novel V-ATPase inhibitors is discussed.
Reversible Disassembly as a Regulatory Mechanism
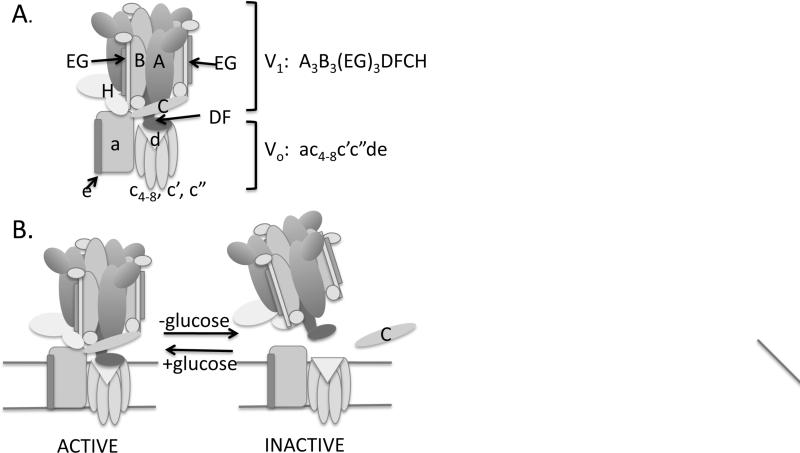
V-ATPases are highly conserved proton pumps that acidify lysosomes, endosomes, Golgi apparatus, and certain secretory granules in all eukaryotes [1, 2]. They can also be recruited to the plasma membrane for proton export in certain cells [2, 3]. V-ATPases are comprised of a peripheral membrane complex (V1), which contains the ATP-binding sites, attached to a membrane-associated complex (Vo), which contains the proton pore. ATP-driven proton pumping requires stable association of these two subcomplexes. A model of the yeast V-ATPase is shown in Fig. 1A; by convention, V1 subunits are denoted by upper-case letters (A-H) and Vo subunits are denoted by lower-case letters (a, c, c’, c”, d, and e). The overall structure of V-ATPases from all eukaryotes is very similar, and there is extensive homology between subunit genes of different species.
Figure 1.
Model of V-ATPase structure and reversible disassembly: A. Model of V-ATPase subunit composition and structure; an estimate of stoichiometry based on Ref. [27] is shown. V1 subunits are designated in upper case and Vo subunits are indicated in lower case.. B. Diagram of reversible disassembly in response to glucose deprivation and readdition. The C subunit is released from both sectors upon disassembly.
A number of years ago, the V1 subcomplex was shown to detach from the Vo complex in response to glucose deprivation, both on internal membranes of yeast cells and at the apical plasma membrane of the tobacco hornworm (Manduca sexta) intestine (Figure 1B) [4, 5]. Disassembly of V1 from Vo inhibits the activity of both sectors, and remarkably, proved to be fully reversible [4, 6, 7]. These initial studies laid the groundwork for reversible disassembly as a major regulatory mechanism for V-ATPases. Because this process has been extensively reviewed [2, 8, 9], I will only summarize the major features, then focus on more recent data including structural implications, potential signaling pathways, and prospects for targeting reversible disassembly as a means of controlling V-ATPase activity therapeutically.
Early studies of V-ATPase disassembly and reassembly in yeast and M. sexta indicated a number of common features of this process. V1 subunits were released as a complex from the membrane, with the exception of V1 subunit C which was largely detached from both the V1 and Vo subcomplexes following glucose deprivation [4, 6, 10]. Released V1 sectors do not hydrolyze MgATP, the physiological substrate of assembled V-ATPases, suggesting that released V1 sectors may be inhibited to protect cytosolic ATP stores from ATP hydrolysis uncoupled from proton transport [6, 7]. Subsequent data suggested that the V1 H subunit is responsible for this inhibition [7, 11, 12]. In yeast, both disassembly and reassembly are complete within 5 minutes of glucose deprivation and readdition, respectively, and neither requires new protein synthesis [4], suggesting that preexisting V-ATPase complexes are disassembled and reassembled. In addition, intermediate levels of V-ATPase assembly are observed in yeast cells grown in poor carbon sources, indicating that disassembly was not an “all-or-none” phenomenon, but rather a means of tuning V-ATPase assembly and activity to carbon source availability. The overall picture that emerged from both yeast and M. sexta studies was that the V1- Vo interaction could be a major target of V-ATPase regulation, and that V-ATPase activity could be rapidly adjusted in response to nutrient availability.
The parallels between reversible dissociation in the M. sexta systems suggested that reversible disassembly was a highly conserved regulatory mechanism, but evidence for this mechanism in mammalian cells was slower to emerge. Regulated V-ATPase assembly in response to maturation signals was first reported in dendritic cells [13]. In this setting, lysosomal acidification was partially suppressed in immature dendritic cells as a mechanism for inhibiting antigen processing and allowing storage in lysosomes; reduced lysosomal acidification was accompanied by reduced assembly of V1 and Vo sectors. As the cells were triggered to mature and induce lysosomal proteolysis of antigen, V-ATPases were assembled and the lysosome was acidified. More recently, secretagogue stimulation of alveolar epithelial type II cells was shown to trigger disassembly of V1 from Vo as a means of inhibiting V-ATPase activity [14]. Interestingly, these two cases both demonstrate control of V-ATPase activity at the level of enzyme assembly working in opposite directions, but have no obvious link to glucose. The pathways transmitting the assembly signals are not well understood in either case. Some of the first evidence of glucose-dependent control of V-ATPase assembly in mammalian cells was obtained in cultured kidney cells [15, 16]. Reversible, glucose-dependent changes in V-ATPase assembly and activity were demonstrated in LLC-PK1 cells, a porcine proximal tubule cell model. In these cells, glucose deprivation resulted in both loss of organelle acidification and release of V1 subunits into the cytosol; readdition of glucose restored both organelle acidification, localization of V1 to internal membranes, and V-ATPase activity. In a human proximal tubule cell model, HK-2 cells, glucose stimulated recruitment of V1 to the apical plasma membrane, but also altered trafficking of the intact complex, suggesting that V1 might reach the plasma membrane by vesicular transport rather than by recruitment from a cytosolic pool [16]. Interestingly, both glucose-induced trafficking and assembly in these kidney cells were dependent on activity of phosphatidyl inositol-3-kinase (PI3 kinase)[15, 16]. In addition, glycolytic enzymes, particularly aldolase, were found to associate with the V-ATPase in kidney cells, suggesting a link between glycolysis and VATPase assembly/activity (described below) [17-19].
Taken together, these experiments clearly establish that the extent of V1- Vo assembly can be an important regulatory target of in mammalian cells, although many questions remain to be answered. In addition, these data suggest that glucose may be only one of several signals able to regulate V1- Vo assembly. This conclusion is also supported by recent evidence in the yeast system suggesting that other conditions such as altered extracellular pH can also dictate V-ATPase assembly level [20, 21], and by previous evidence in plants that other signals such as salt stress can modulate V-ATPase assembly [22]. Thus, while glucose induced changes in VATPase assembly are still the major paradigm for reversible disassembly, it is now clear that multiple pathways and signals can impact assembly and activity of V-ATPases.
Structural implications of reversible disassembly
V-ATPases are complex, multisubunit enzymes, and the existence of reversible disassembly as a regulatory mechanism has important implications for the enzyme's structure. The catalytic and regulatory ATP-binding (V1 subunits A and B) and the proteolipid subunits (Vo subunits c, c’, and c”) of V-ATPases have significant homology to subunits of the F-type ATP synthases of mitochondria, chloroplasts, and bacteria as well as to archaeal ATPases (called both A-ATPases and archaeal V-ATPases) [23, 24]. These subunits are central components of the rotational catalytic machinery of both enzymes, suggesting a common evolutionary history that diverged to make eukaryotic V-ATPases dedicated proton pumps and F-ATPases predominantly ATP synthases. In both enzyme classes, ATP-binding and hydrolysis functions reside in peripheral membrane subunits that are attached to the proton pore subunits by multiple “stalk” subunits. These stalk subunits are generally highly conserved among eukaryotic V-ATPases, but show much less homology to the F-ATPases. In addition, while the overall structure of the F-ATPases includes one central (rotor) stalk and one peripheral (stator) stalk, V-ATPases have one central stalk and multiple peripheral stalks [25-30]. The eukaryotic V-ATPases, in particular, appear to have three peripheral stalks (comprised of V1 subunits E and G) that interact with the Vo sector in part through two “ bridging” subunits (V1 subunits C and H)[27-32]. In F-ATPases, the single peripheral stalk is sufficient to give stable assembly of the ATP synthase and to resist the torque that arises from rotational catalysis [33-35]. In contrast, the essential “ stator” functions of any rotary motor, stable assembly and resistance to torque, appear to be divided among three weaker peripheral connecting stalks in V-ATPases [27-30, 36]. This distribution of the stator function may allow rapid release of V1 under disassembly conditions.
Understanding the structural basis of reversible disassembly is the focus of current research in a number of labs, but recent pseudo-atomic models, mostly arising from electron microscopy of the full V-ATPase and subcomplexes, have focused attention on a limited set of protein-protein interactions that must be altered in reversible dissociation [28-31]. The C subunit, which is released from both the V1 and Vo sectors by reversible disassembly is particularly interesting in this regard. Both previous crosslinking studies and recent biochemical evidence with purified subunits indicate that this subunit interacts with two of the three EG peripheral stalks as well as the Vo subunit a [30, 37, 38]. One of the two EG stalks has a much higher affinity for subunit C than the other, and the expressed subunits form a 1:1:1 complex [30, 38]. Subunit C has an extended structure that forms part of a “ collar” at the V1- Vo interface, and at one end of subunit C, a quaternary complex consisting of an EG stalk, subunit C, and Vo subunit a may form [30, 31]. From this information, it would appear that for V1 disassembly to occur, the interactions of V1 subunits E, G, and C with Vo subunit a must be disrupted and that subunit C must also be released from its tight interaction from V1 subunits E and G. It is still not clear how this occurs, but transient phosphorylation of subunit C during reassembly of the disassembled enzyme has been reported in insects [10] (discussed below). If this phosphorylation proves to be general, it is possible that the phosphorylation state of subunit C determines its interaction with V1 and/or Vo subunits, and thus could serve as a primary determinant of V-ATPase assembly state.
In addition to critical interactions with subunit C, there is evidence that the H subunit, also located at the V1- Vo interface, alters its interactions with V1 and Vo subunits during reversible disassembly [11, 12]. The H subunit is located in proximity to the Vo a subunit in current models of the assembled V-ATPase and these two subunits interact in vitro [11, 29]. The H subunit appears to lose its interaction with Vo when the V1 sector disassembles [11]. The ability of the H subunit to silence ATPase activity in free V1 sectors likely arises from interactions with the central stalk of V1 that occur in free V1, but not in V1 Vo [11, 12].
It is also interesting that inhibition of V-ATPase activity can inhibit disassembly of the enzyme, suggesting that catalysis can make the enzyme more susceptible to disassembly [39, 40]. Disassembly of the yeast V-ATPase is inhibited by concanamycin A, which binds to the Vo sector of the enzyme, or by mutations in the Vo sector that do not permit proton transport [39]. In addition, a different class of mammalian-specific V-ATPase inhibitors, the salicylihalimides, causes increased V1 assembly on the membrane in mammalian cells [41]. In each of these cases, a change in conformation in the Vo sector, as a result of inhibitor binding or mutation, appears to tighten the binding of V1 to Vo. The results with the salicylihalimides also suggest that there might be ongoing disassembly and reassembly of V-ATPases in certain mammalian cells, so that the full capacity for V1 assembly with Vo is only recognized when V-ATPase activity is inhibited.
Regulatory proteins and signals for reversible disassembly
Reversible disassembly requires that extracellular conditions be communicated to V-ATPases that are often on internal organelles. This suggests involvement of one or more signaling pathways and the possibility of assembly factors to assist with reassembly. Although there are multiple assembly factors implicated in V-ATPase biosynthesis, the RAVE (regulator of H+-ATPase of vacuolar and endosomal membranes) complex was the first assembly complex to be implicated in reassembly of dissociated V1 and Vo [42]. In yeast, the RAVE complex consists of the ubiquitous adaptor protein Skp1p, along with two other subunits Rav1p and Rav2p [42]. SKP1 is an essential gene and Skp1 protein plays an important role in other complexes [42], but rav1Δ and rav2Δ mutants have a milder version of the Vma- phenotype characteristic of loss of V-ATPase function in yeast [42, 43]. RAV2 homologs are found only in fungi, but RAV1 homologs are found in virtually all eukaryotes. Rav1p is the central component of the RAVE complex, interacting not only with Skp1p and Rav2p, but also with V1 complexes via the E and/or G subunits, and independently with subunit C [43, 44]. RAVE- V1 complexes can be isolated from cytosolic fractions, and it has been suggested that Skp1 mediates transient interactions with membranes to help support assembly with Vo [45]. The interaction between RAVE and V1 is not intrinsically glucose-sensitive, suggesting that this complex may not be the glucose sensor [43]. The RAVE complex was first characterized in yeast, but more recently, Rav1 homologs in Drosophila and mice (called rabconnectins) were shown to be important for organelle acidification, suggesting that this complex has a conserved role in regulating V-ATPase activity and assembly [46, 47].
A number of glycolytic enzymes bind to the V-ATPase, and a model in which these enzymes provide a localized source of ATP to the enzyme has been proposed [17-19, 48, 49]. Such a glycolytically active complex would also be an attractive target for V-ATPase regulation in response to glucose levels, and indeed, interactions of the V-ATPase with aldolase appear to be both glucose-sensitive and important for maintaining V1- Vo assembly [17, 18, 48]. The V-ATPase/aldolase interaction was first described in kidney cells, and subsequently extended to yeast. The yeast V-ATPase is disassembled in aldolase deletion mutants, but loss of function mutations in glycolytic enzymes arrest glucose metabolism in yeast and thus would likely mimic glucose deprivation [17, 18]. However, a point mutation in aldolase that preserves its enzymatic function in vitro but disrupts interactions with the V-ATPase, also reduces assembly of V1 with Vo, indicating a signaling role for aldolase beyond its enzymatic role in glycolysis[48]. Another glycolytic enzymes, phosphofructokinase (PFK), was shown to interact with the Vo a subunit in both human kidney and yeast cells [19, 49]. Interestingly, mutations in the human kidney a4 subunit isoform that cause distal renal tubule acidosis, suggesting loss of V-ATPase function in kidney, also disrupt the PFK interaction and affect V-ATPase expression, assembly, or function when introduced into yeast cells [49]. These results argue for a conserved regulatory connection between glycolytic enzymes and the V-ATPase.
There is also substantial evidence that protein kinase A (PKA) and its upstream activation pathways are involved in regulation of V-ATPase assembly. Compelling evidence for PKA involvement in signaling V-ATPase reassembly has been reported in the blowfly (Calliphora vicina) [50-52], and phosphorylation of V1 subunit C under conditions that promote enzyme assembly has been reported in both C. vicina and M. sexta [10]. Subunit C phosphorylation is not required for association of subunit C with V1 and is not detected after reassembly of V1 Vo complexes, but subunit C does become phosphorylated when PKA is activated by cAMP analogs or hormonal stimulation [10]. In yeast, a genetic screen for mutants that failed to disassemble the V-ATPase under conditions of reduced glucose identified mutations in IRA1 and IRA2, two negative regulators of yeast Ras, suggesting that the Ras/cAMP pathway might regulate V-ATPase assembly [53]. In subsequent experiments, it was shown that a number of conditions that increase the level of PKA activity, ranging from constitutive activation of Ras to deletion of the negative regulatory subunit of PKA itself, also prevent disassembly of the yeast V-ATPase with glucose deprivation [53]. As in the insect system, these results suggest that activation of PKA, an established glucose-sensitive enzyme [54], promotes V-ATPase assembly, although no PKA-dependent phosphorylation of the yeast V1 C subunit was shown. However, questions remain about the exact role of PKA in regulation of V-ATPase assembly. More recently, PKA was proposed to act downstream from V-ATPase assembly in both yeast and pancreatic beta cells, while cytosolic pH changes associated with active glucose metabolism were invoked as the signal promoting V-ATPase assembly [21]. These results are compatible with data showing that disassembly of the V-ATPase is suppressed at high extracellular pH [20], but are not easily reconciled with results suggesting that PKA activity stimulates V-ATPase assembly and activity [10, 53]. Additionally, in both clear cells of the epididymus and intercalated cells of the kidney collecting duct, PKA upregulates V-ATPase activity at the plasma membrane by recruitment of intact enzyme in vesicles from an intracellular compartment, rather than by reassembly of V1 with Vo; this mobilization of V-ATPase to the plasma membrane involves PKA-dependent phosphorylation on the V1 A subunit [55-57]. Thus, while there is certainly substantial evidence for PKA as an important V-ATPase regulator, the full spectrum and mechanisms of its regulatory roles are still areas of active research.
Finally, it should be mentioned that there are a number of other signaling pathways that have been implicated in regulation of V-ATPase assembly. As described above, PI 3-kinase activity was required for reassembly of V1 sectors with Vo sectors upon glucose readdition in kidney tissue culture models [15, 16]. PI 3-kinase phosphorylates phosphatidyl inositol lipid headgroups, and thus would affect the lipid environment rather than the V-ATPase directly. In yeast cells, mutation of either the PI 3-kinase Vps34p or the phosphatidylinositol 3-phosphate 5-kinase Fab1p reduce vacuolar acidification [58-60], but at least in the vps34Δ mutant, this primarily reflects a trafficking, rather than a V-ATPase assembly, defect [59]. Furthermore, trapping V-ATPases with the same isoform composition in different organelles alters their response to glucose deprivation [61], suggesting that some aspect of the reversible disassembly process is sensitive to the membrane environment.
Reversible disassembly as a target for drug development
Although there are a number of situations where inhibition of V-ATPase activity could have therapeutic benefits, including osteoporosis [62],[63], cancer [64-66], and even viral infection [67, 68], there are serious barriers to development of therapeutic V-ATPase inhibitors [69]. The most serious barrier may be the problem of selectivity, because inhibition of V-ATPases outside the target tissue can cause serious side effects [70-72]. Deletion of constitutive V-ATPase subunits is lethal in mammals, suggesting that V-ATPases are essential for viability in some locales [73]. Therefore, useful therapeutics targeting the V-ATPase will need selectivity (the ability to distinguish target V-ATPases from those in other tissues), and this problem is being addressed in a number of ways, including targeting of specific subunit isoforms/interactions and exploitation of enzymatic differences of V-ATPases in different environments [71, 72, 74]. Variations in sensitivity of V-ATPases in different tissues and organelles to reversible disassembly could provide an extra layer of specificity for drug development. In addition, the ability of reversible disassembly to “ turn down” V-ATPase activity rather than completely eliminating it could also be a useful characteristic to exploit in inhibitor design. Finally, the regulators involved in reversible disassembly also provide additional targets for therapies aimed at modulating V-ATPase activity.
Drug development strategies that target the V-ATPase directly or indirectly through reversible disassembly will benefit from a better understanding of VATPase structure and subunit interaction. As described above, the critical subunit-subunit interactions that support V-ATPase assembly and activity are emerging from structural studies, but additional high resolution structural information is required for drug design. Recently, Kartner et al. screened for chemical inhibitors of an osteoclast-specific V1- Vo subunit interaction and found a novel inhibitor of bone resorption, suggesting that they had identified an inhibitor that targeted V-ATPases in the osteoclast plasma membrane [72]. Preliminary evidence in yeast suggests that different subunit isoforms may vary in their sensitivity to reversible disassembly; specifically, complexes containing the vacuole-specific subunit a isoform , Vph1p, are more responsive to glucose deprivation than complexes containing the Golgi/endosome-specific isoform, Stv1p [75]. If the four human subunit a isoforms also vary in their sensitivity to reversible disassembly, then targeting this process could provide tissue-specific V-ATPase inhibition. As subunit-subunit interactions critical for V-ATPase reassembly become clear, the potential for isoform-specific regulation will likely become clearer. Approaches similar to that used by Kartner et al. [72] could then be used to design or screen for inhibitors that prevent reassembly and trap the V-ATPase in its disassembled and naturally inhibited state. As described above, current V-ATPase inhibitors tend to trap the enzyme in an assembled state [39, 41], but given the varied levels of assembly of VATPases in different tissues and cell types [13, 14], it could be an advantage to have classes of inhibitors able to target either assembled or disassembled V-ATPases.
As more information becomes available about signaling pathways that underly reversible disassembly, regulators involved in this process will also emerge as candidates for drug design or screening. At present, the RAVE complex appears to be the most attractive target for drug development, since it has a rather specific role in V-ATPase assembly, while other potential regulators such as PKA and glycolytic enzymes certainly have multiple functions. In yeast cells, loss of RAVE function gives a less severe phenotype than complete loss of V-ATPase function [42, 43]. This may suggest that some populations of V-ATPases do not require RAVE for assembly and function. It has not yet been determined whether the RAVE complex shows any subunit isoform- or organelle-specificity, but the lack of reversible disassembly in Golgi V-ATPases might suggest a reduced requirement for RAVE function for these pumps [61]. This in turn suggests that synthetic inhibitors of RAVE could target a subpopulation of V-ATPases, possibly with less severe side-effects than a general V-ATPase inhibitor. In Drosophila, loss of function mutations in the RAV1 homolog Rabconnectin 3 (Rbcn-3) resulted in similar developmental phenotypes as loss of function mutations in V-ATPase subunits, including inhibition of Notch signaling [47]. Similar effects of Rbcn-3 silencing were observed mammalian cells[46]. At first glance, these results suggest that advantages of RAVE inhibition in yeast will not extend to mammalian cells, although the full consequences of RAVE loss on multiple tissues have not been assessed. However, these rabconnectin studies have further stimulated interest in V-ATPase or RAVE inhibitors as cancer therapeutics, since a number of malignancies require Notch signaling. Endosomal acidification via V-ATPases appears to be a novel and essential requirement of the Notch pathway [46, 76].
In summary, V-ATPases are versatile, ubiquitous, and highly regulated pumps that impact many aspects of mammalian physiology. Their full physiological impact is still being revealed, as suggested by the recent, surprising evidence implicating them in the Notch pathway and independently, in Wnt signaling, another essential pathway in development and cancer [76, 77]. As a conserved and ubiquitous mechanism of V-ATPase regulation, reversible disassembly provides an attractive target for pharmacological intervention in V-ATPase function. To realize this potential, future experiments will need to target the molecular basis of reversible disassembly, both in terms of V-ATPase structure and of signaling
Acknowledgements
This work was supported by NIH grant R01 GM50322.
References
- 1.Kane PM. Microbiol Mol Biol Rev. 2006;70:177–91. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forgac M. Nat Rev Mol Cell Biol. 2007;8:917–29. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 3.Nelson N, Harvey WR. Physiol Rev. 1999;79:361–85. doi: 10.1152/physrev.1999.79.2.361. [DOI] [PubMed] [Google Scholar]
- 4.Kane PM. J Biol Chem. 1995;270:17025–17032. [PubMed] [Google Scholar]
- 5.Sumner JP, Dow JA, Earley FG, Klein U, Jager D, Wieczorek H. J Biol Chem. 1995;270:5649–5653. doi: 10.1074/jbc.270.10.5649. [DOI] [PubMed] [Google Scholar]
- 6.Graf R, Harvey WR, Wieczorek H. J Biol Chem. 1996;271:20908–20913. doi: 10.1074/jbc.271.34.20908. [DOI] [PubMed] [Google Scholar]
- 7.Parra KJ, Keenan KL, Kane PM. J Biol Chem. 2000;275:21761–21767. doi: 10.1074/jbc.M002305200. [DOI] [PubMed] [Google Scholar]
- 8.Merzendorfer H, Graf R, Huss M, Harvey WR, Wieczorek H. J Exp Biol. 1997;200:225–35. doi: 10.1242/jeb.200.2.225. [DOI] [PubMed] [Google Scholar]
- 9.Kane PM, Smardon AM. J Bioenerg Biomembr. 2003;35:313–21. doi: 10.1023/a:1025724814656. [DOI] [PubMed] [Google Scholar]
- 10.Voss M, Vitavska O, Walz B, Wieczorek H, Baumann O. J Biol Chem. 2007;282:33735–42. doi: 10.1074/jbc.M703368200. [DOI] [PubMed] [Google Scholar]
- 11.Diab H, Ohira M, Liu M, Cobb E, Kane PM. J Biol Chem. 2009 doi: 10.1074/jbc.M900475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jefferies KC, Forgac M. J Biol Chem. 2008;283:4512–9. doi: 10.1074/jbc.M707144200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. Science. 2003;299:1400–3. doi: 10.1126/science.1080106. [DOI] [PubMed] [Google Scholar]
- 14.Chintagari NR, Mishra A, Su L, Wang Y, Ayalew S, Hartson SD, Liu L. PLoS One. 2010;5:e9228. doi: 10.1371/journal.pone.0009228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura S. Am J Physiol Cell Physiol. 2004;287:C97–105. doi: 10.1152/ajpcell.00469.2003. [DOI] [PubMed] [Google Scholar]
- 16.Sautin YY, Lu M, Gaugler A, Zhang L, Gluck SL. Mol Cell Biol. 2005;25:575–89. doi: 10.1128/MCB.25.2.575-589.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu M, Holliday LS, Zhang L, Dunn WA, Jr., Gluck SL. J Biol Chem. 2001;276:30407–13. doi: 10.1074/jbc.M008768200. [DOI] [PubMed] [Google Scholar]
- 18.Lu M, Sautin YY, Holliday LS, Gluck SL. J. Biol. Chem. 2003:M303871200. doi: 10.1074/jbc.M303871200. [DOI] [PubMed] [Google Scholar]
- 19.Su Y, Zhou A, Al-Lamki RS, Karet FE. J Biol Chem. 2003;278:20013–8. doi: 10.1074/jbc.M210077200. [DOI] [PubMed] [Google Scholar]
- 20.Diakov TT, Kane PM. J Biol Chem. 2010;285:23771–8. doi: 10.1074/jbc.M110.110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dechant R, Binda M, Lee SS, Pelet S, Winderickx J, Peter M. Embo J. 2010;29:2515–26. doi: 10.1038/emboj.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva P, Geros H. Plant Signal Behav. 2009;4:718–26. doi: 10.4161/psb.4.8.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson N, Taiz L. Trends Biochem Sci. 1989;14:113–116. doi: 10.1016/0968-0004(89)90134-5. [DOI] [PubMed] [Google Scholar]
- 24.Gruber G, Marshansky V. Bioessays. 2008;30:1096–109. doi: 10.1002/bies.20827. [DOI] [PubMed] [Google Scholar]
- 25.Bernal RA, Stock D. Structure. 2004;12:1789–98. doi: 10.1016/j.str.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 26.Boekema EJ, van Breemen JF, Brisson A, Ubbink-Kok T, Konings WN, Lolkema JS. Nature. 1999;401:37–38. doi: 10.1038/43369. [DOI] [PubMed] [Google Scholar]
- 27.Kitagawa N, Mazon H, Heck AJ, Wilkens S. J Biol Chem. 2008;283:3329–37. doi: 10.1074/jbc.M707924200. [DOI] [PubMed] [Google Scholar]
- 28.Muench SP, Huss M, Song CF, Phillips C, Wieczorek H, Trinick J, Harrison MA. J Mol Biol. 2009;386:989–99. doi: 10.1016/j.jmb.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Zheng Y, Mazon H, Milgrom E, Kitagawa N, Kish-Trier E, Heck AJ, Kane PM, Wilkens S. J Biol Chem. 2008;283:35983–95. doi: 10.1074/jbc.M805345200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diepholz M, Venzke D, Prinz S, Batisse C, Florchinger B, Rossle M, Svergun DI, Bottcher B, Fethiere J. Structure. 2008;16:1789–98. doi: 10.1016/j.str.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 31.Hildenbrand ZL, Molugu SK, Stock D, Bernal RA. PLoS One. 2010;5:e12588. doi: 10.1371/journal.pone.0012588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diepholz M, Borsch M, Bottcher B. Biochem Soc Trans. 2008;36:1027–31. doi: 10.1042/BST0361027. [DOI] [PubMed] [Google Scholar]
- 33.Dickson VK, Silvester JA, Fearnley IM, Leslie AG, Walker JE. Embo J. 2006;25:2911–8. doi: 10.1038/sj.emboj.7601177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubinstein JL, Walker JE, Henderson R. Embo J. 2003;22:6182–92. doi: 10.1093/emboj/cdg608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkens S, Capaldi RA. Biochim Biophys Acta. 1998;1365:93–97. doi: 10.1016/s0005-2728(98)00048-6. [DOI] [PubMed] [Google Scholar]
- 36.Ohira M, Smardon AM, Charsky CM, Liu J, Tarsio M, Kane PM. J Biol Chem. 2006 doi: 10.1074/jbc.M601441200. [DOI] [PubMed] [Google Scholar]
- 37.Inoue T, Forgac M. J Biol Chem. 2005;280:27896–903. doi: 10.1074/jbc.M504890200. [DOI] [PubMed] [Google Scholar]
- 38.Oot RA, Wilkens S. J Biol Chem. 2010;285:24654–64. doi: 10.1074/jbc.M110.136960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parra KJ, Kane PM. Mol Cell Biol. 1998;18:7064–7074. doi: 10.1128/mcb.18.12.7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao E, Forgac M. J Biol Chem. 2004;279:48663–70. doi: 10.1074/jbc.M408278200. [DOI] [PubMed] [Google Scholar]
- 41.Xie XS, Padron D, Liao X, Wang J, Roth MG, De Brabander JK. J Biol Chem. 2004;279:19755–63. doi: 10.1074/jbc.M313796200. [DOI] [PubMed] [Google Scholar]
- 42.Seol JH, Shevchenko A, Deshaies RJ. Nat Cell Biol. 2001;3:384–91. doi: 10.1038/35070067. [DOI] [PubMed] [Google Scholar]
- 43.Smardon AM, Tarsio M, Kane PM. J Biol Chem. 2002;13:13. doi: 10.1074/jbc.M200682200. [DOI] [PubMed] [Google Scholar]
- 44.Smardon AM, Kane PM. J Biol Chem. 2007;282:26185–94. doi: 10.1074/jbc.M703627200. [DOI] [PubMed] [Google Scholar]
- 45.Brace EJ, Parkinson LP, Fuller RS. Eukaryot Cell. 2006;5:2104–13. doi: 10.1128/EC.00347-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sethi N, Yan Y, Quek D, Schupbach T, Kang Y. J Biol Chem. 2010;285:34757–64. doi: 10.1074/jbc.M110.158634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan Y, Denef N, Schupbach T. Dev Cell. 2009;17:387–402. doi: 10.1016/j.devcel.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu M, Ammar D, Ives H, Albrecht F, Gluck SL. J Biol Chem. 2007;282:24495–503. doi: 10.1074/jbc.M702598200. [DOI] [PubMed] [Google Scholar]
- 49.Su Y, Blake-Palmer KG, Sorrell S, Javid B, Bowers K, Zhou A, Chang SH, Qamar S, Karet FE. Am J Physiol Renal Physiol. 2008;295:F950–8. doi: 10.1152/ajprenal.90258.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dames P, Zimmermann B, Schmidt R, Rein J, Voss M, Schewe B, Walz B, Baumann O. Proc Natl Acad Sci U S A. 2006;103:3926–31. doi: 10.1073/pnas.0600011103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rein J, Voss M, Blenau W, Walz B, Baumann O. Am J Physiol Cell Physiol. 2008;294:C56–65. doi: 10.1152/ajpcell.00041.2007. [DOI] [PubMed] [Google Scholar]
- 52.Voss M, Schmidt R, Walz B, Baumann O. Cell Tissue Res. 2009;335:657–62. doi: 10.1007/s00441-008-0673-x. [DOI] [PubMed] [Google Scholar]
- 53.Bond S, Forgac M. J Biol Chem. 2008;283:36513–21. doi: 10.1074/jbc.M805232200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dechant R, Peter M. Curr Opin Cell Biol. 2008;20:678–87. doi: 10.1016/j.ceb.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 55.Alzamora R, Thali RF, Gong F, Smolak C, Li H, Baty CJ, Bertrand CA, Auchli Y, Brunisholz RA, Neumann D, Hallows KR, Pastor-Soler NM. J Biol Chem. 2010;285:24676–85. doi: 10.1074/jbc.M110.106278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gong F, Alzamora R, Smolak C, Li H, Naveed S, Neumann D, Hallows KR, Pastor-Soler NM. Am J Physiol Renal Physiol. 2010 doi: 10.1152/ajprenal.00645.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pastor-Soler NM, Hallows KR, Smolak C, Gong F, Brown D, Breton S. Am J Physiol Cell Physiol. 2008;294:C488–94. doi: 10.1152/ajpcell.00537.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sambade M, Alba M, Smardon AM, West RW, Kane PM. Genetics. 2005;170:1539–51. doi: 10.1534/genetics.105.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li SC, Kane PM. Biochim Biophys Acta. 2009;1793:650–63. doi: 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qi J, Forgac M. J Biol Chem. 2007;282:24743–51. doi: 10.1074/jbc.M700663200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu J, Cheng T, Feng HT, Pavlos NJ, Zheng MH. Histol Histopathol. 2007;22:443–54. doi: 10.14670/HH-22.443. [DOI] [PubMed] [Google Scholar]
- 63.Farina C, Gagliardi S. Curr Pharm Des. 2002;8:2033–48. doi: 10.2174/1381612023393369. [DOI] [PubMed] [Google Scholar]
- 64.Sennoune SR, Luo D, Martinez-Zaguilan R. Cell Biochem Biophys. 2004;40:185–206. doi: 10.1385/CBB:40:2:185. [DOI] [PubMed] [Google Scholar]
- 65.Perez-Sayans M, Somoza-Martin JM, Barros-Angueira F, Rey JM, Garcia-Garcia A. Cancer Treat Rev. 2009;35:707–13. doi: 10.1016/j.ctrv.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 66.Sasazawa Y, Futamura Y, Tashiro E, Imoto M. Cancer Sci. 2009;100:1460–7. doi: 10.1111/j.1349-7006.2009.01194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marjuki H, Gornitzky A, Marathe BM, Ilyushina NA, Aldridge JR, Desai G, Webby RJ, Webster RG. Cell Microbiol. 2011;13:587–601. doi: 10.1111/j.1462-5822.2010.01556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muller KH, Kainov DE, El Bakkouri K, Saelens X, De Brabander JK, Kittel C, Samm E, Muller CP. Br J Pharmacol. 2011 doi: 10.1111/j.1476-5381.2011.01346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huss M, Wieczorek H. J Exp Biol. 2009;212:341–6. doi: 10.1242/jeb.024067. [DOI] [PubMed] [Google Scholar]
- 70.Niikura K, Takano M, Sawada M. Br J Pharmacol. 2004;142:558–66. doi: 10.1038/sj.bjp.0705812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nyman JK, Vaananen HK. Calcif Tissue Int. 2010;87:273–83. doi: 10.1007/s00223-010-9395-7. [DOI] [PubMed] [Google Scholar]
- 72.Kartner N, Yao Y, Li K, Crasto GJ, Datti A, Manolson MF. J Biol Chem. 2010;285:37476–90. doi: 10.1074/jbc.M110.123281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun-Wada G, Murata Y, Yamamoto A, Kanazawa H, Wada Y, Futai M. Dev Biol. 2000;228:315–25. doi: 10.1006/dbio.2000.9963. [DOI] [PubMed] [Google Scholar]
- 74.Hinton A, Sennoune SR, Bond S, Fang M, Reuveni M, Sahagian GG, Jay D, Martinez-Zaguilan R, Forgac M. J Biol Chem. 2009;284:16400–8. doi: 10.1074/jbc.M901201200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kawasaki-Nishi S, Nishi T, Forgac M. J Biol Chem. 2001;276:17941–8. doi: 10.1074/jbc.M010790200. [DOI] [PubMed] [Google Scholar]
- 76.Niehrs C, Boutros M. Sci Signal. 2010;3:pe26. doi: 10.1126/scisignal.3134pe26. [DOI] [PubMed] [Google Scholar]
- 77.Cruciat CM, Ohkawara B, Acebron SP, Karaulanov E, Reinhard C, Ingelfinger D, Boutros M, Niehrs C. Science. 2010;327:459–63. doi: 10.1126/science.1179802. [DOI] [PubMed] [Google Scholar]