Abstract
Background and Aim of the Study
Use of genetically-altered small animal models is a powerful strategy for elucidating the mechanisms of heart valve disease. However, while the ability to manipulate genes in rodent models is well established, there remains a significant obstacle in determining the functional mechanical properties of the genetically mutated leaflets. Here, we present a feasibility study using micromechanical analysis via atomic force microscopy to determine the stiffness of mouse heart valve leaflets in the context of age and disease states.
Materials and Methods
A novel atomic force microscopy imaging technique for the quantification of heart valve leaflet stiffness was performed on cryosectioned tissues. Heart valve leaflet samples were obtained from wild type mice (2 month old and 17 month old) and genetically altered mice (10 month old Notch1 heterozygous and 20 month old ApoE homozygous). Histology was performed on adjacent sections to determine the ECM characteristics of scanned areas.
Results
17 month old wild type, 10 month old Notch1, and 20 month old ApoE aortic valve leaflets were significantly stiffer than leaflets from 2 month old wild type mice. Notch1 leaflets were significantly stiffer than all other leaflets examined, indicating that the Notch1 heterozygous mutation may alter leaflet stiffness, both earlier and to a greater degree than the homozygous ApoE mutation; however, these conclusions are preliminary due to small sample size used in this proof of concept study.
Conclusions
We believe that this technique provides a powerful end-point analysis for determining the mechanical properties of heart valve leaflets from genetically-altered mice. Further, this analysis technique is complementary to standard histological processing and does not require excess tissue for mechanical testing. In this proof-of-concept study, we show that AFM can be a powerful tool for heart valve researchers who develop genetically-altered animals for their studies.
Keywords: AFM, Aortic Valve, ApoE, Notch1
Introduction
Acquired heart valve disease research has been limited due to lack of appropriate animal models; however, in recent years, the ApoE and Notch1 genetically-altered animal models have been found to acquire aortic valve (AV) sclerosis, calcification, and stenosis with age similar to humans (1-5). Specifically, AV sclerosis in ApoE homozygous mice is characterized by increased AV flow velocity after 43 weeks of age compared to age-matched wild type (WT) animals (2) with areas of ectopic calcification, cells positive for smooth muscle α-actin and osteoblast related proteins, and frequent apoptosis, all common markers of degenerative AV disease in humans (5). Notch1 heterozygous mutations have been shown to cause bicuspid AVs and subsequently increase risk for calcific valve disease in humans (1, 3). Similar to ApoE animals, 10 month old Notch1 leaflets have been found to have significant calcification, over 5-fold greater when compared to age-matched WT littermates. While ApoE and Notch1 mice have an apparent change in morphology and hemodynamics, the resulting functional biomechanical changes have not been quantified. The primary reason for this is that biomechanical assessment of mouse AV leaflets is very difficult due to the small size of the leaflet tissue. Recently, micropipette aspiration was utilized to quantify the mechanical properties of mouse AV leaflets (6) and while this technique was successful, it is laborious and requires entire leaflets. Therefore, we have developed the following atomic force microscopy (AFM) strategy to quantify the mechanical properties of mouse AV leaflets and demonstrate this proof-of-principle technique using WT, ApoE, and Notch1 mice. The AFM technique described is also advantageous in that leaflet tissue mechanical properties of genetically-altered animals can be performed in conjunction with standard histological cryosectioning techniques.
AFM was developed nearly thirty years ago as an extension of scanning tunneling microscopy where forces as small as 10−18 N were used to achieve angstrom level resolution in the study of sample surfaces (7). Although it was developed primarily for studying topography on hard surfaces such as ordered graphite and silicate compounds, biopolymers soon became an interest of examination with AFM (8). These few initial studies demonstrated the promise of AFM as a way to obtain measurements of mechanical and adhesive properties of tissues, cells, and even biomolecules that had previously been impossible. However, it has taken many years to optimize AFM to the point where mechanical measurements can be determined due to limitations and complications of extremely soft and thin samples (9). In AFM, a nanoscale tip is attached to a cantilever with a reflective coating on the reverse side. The movement of the tip, and sample deformation, is measured by the deflection of a laser off of the cantilever captured by a sensor. However, in extremely soft samples the tip often punctures the sample, resulting in quantification of the substrate beneath it. Therefore, tip and scan parameter selection is paramount in assuring accurate measurements of biological samples.
In this report, we present a novel AFM technique for the mechanical analysis of mouse AV leaflets. Further, to ensure that our mechanical quantification of AV leaflet sections is accurate, we validate our AFM tip and scanning parameters on homogenous and linear isotropic polymer standards over the range of leaflet stiffness values. The polymer standards were examined both with AFM and bulk mechanical testing methods to establish the validity of the quantitative method described in the feasibility study presented below. For a more thorough discussion of AFM techniques in the analysis of biological samples, the authors recommend the following review (10).
Methods
Mice Leaflets
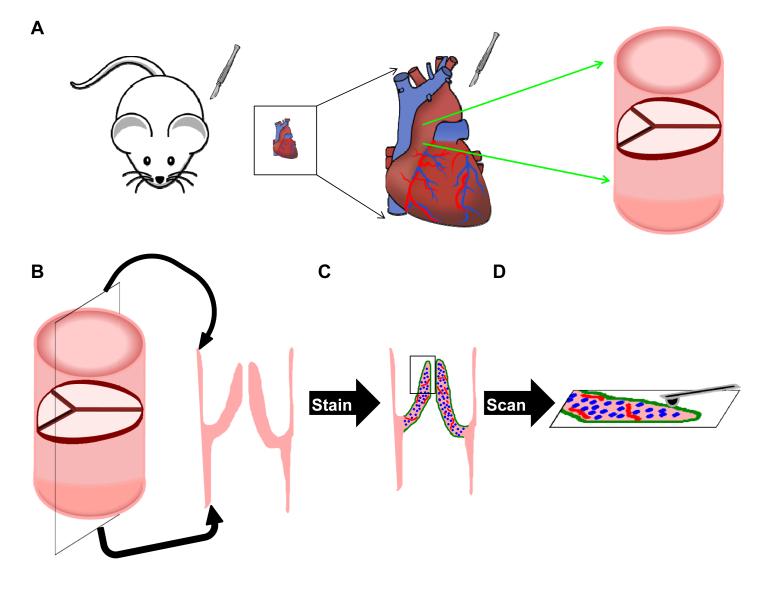
2 and 17 month old WT C57blk6 mice and 20 month old ApoE homozygous mice on C57blk6 were obtained from Jackson Laboratory (Bar Harbor, ME, USA). A Notch1 heterozygous male retired breeder on a mixed CD-1 background was a generous gift from Dr. Stacey Huppert (Vanderbilt University). All animals were maintained on a standard diet under approved IACUC protocols. Mice were euthanized by CO2 asphyxiation and hearts were excised into cold PBS. AVs were removed in cold PBS, flash frozen in OCT, and sectioned at 10 μm. Sections were stored at −20°C until analyzed, and were then removed and stained for 30 min with 1:300 FITC CD31 (558738 BD Pharmingen, San Diego CA, USA) and Cy3 alpha smooth muscle actin (C6198 Sigma, St. Louis MO, USA) in Hank’s buffered salt solution containing 0.2% FBS and 0.1 μg/ml DAPI nuclear stain. Slides were then washed 3 times in PBS, rinsed in diH2O, and air dried for 10 min prior to AFM scanning (Fig. 1). Adjacent tissue sections from the same animals used in the AFM study were also stained using Movat’s Pentachrome (K042 Poly Scientific R&D Corporation, Bay Shore NY, USA). All histological images were taken using a Nikon Eclipse E800 microscope (Nikon Inc., Melville NY, USA) with a Spot RT3 camera (Spot Imaging Solutions, Sterling Heights, MI, NY).
Figure 1.
A) Schematic showing how mouse AV leaflets are harvested and sectioned. (B) Heart valve sections are positioned such that the cross-section of each leaflet is available for scanning, eliminating the concern of only observing surface phenomena with the AFM. (C) Slides are stained without fixation to ensure minimal disruption of the ECM. (D) Slides are subjected to scanning via PF-QNM mode of an AFM.
PDMS Standards and Bulk Modulus Measurements
Poly(dimethyl siloxane) (PDMS, Sylgard 184, Dow Corning, Midland MI,USA) standards were prepared by mixing the base and curing agent in 10:1, 15:1, and 30:1 ratios before curing overnight under vacuum at 60°C. Bulk modulus measurements were made under uniaxial tension with standard masses (0.05-0.5g for 10:1 and 15:1 gels, 0.01-0.2g for 30:1 gels) used to deform the PDMS strips in a free-hanging setup. Bulk modulus was calculated as the slope of the line generated by graphing stress versus strain (n=4).
Atomic Force Microscopy
A Catalyst Bioscope AFM (Bruker AXS, Madison WI, USA) was used for all measurements described here. The system was operated in Peak Force Quantitative Nanomechanical Mapping (PF-QNM) mode, which is a modified tapping-mode modality in a non-fluid environment. The sample modulus (E) is calculated by the software using the following equation after setting Poisson’s ratio (ν) = 0.5:
where Ftip is the force on the tip, R is the radius of the tip, d is the distance between the tip and sample, and Fadh is the adhesive force between tip and sample. Borosilicate glass tips (Novascan Technologies, Inc., Ames IA, USA) with a nominal diameter of 5 μm and nominal spring constant of 0.03 N/m were used. Prior to sample measurements, tip radii were calibrated using a PDMS calibration standard sample (E = 2.5 MPa) provided by Bruker. Individual probe spring constants were calculated using the thermal tune method built into the AFM software; these values typically varied between ±25% from the nominal spring constant due to manufacturing variations. This parameter is crucial for accurate calculation of Ftip and Fadh used in the above equation.
AFM Scanning Parameters
AV leaflet sections were scanned at 20 × 20 μm to 30 × 30 μm areas, 0.1 Hz scan rate, and peak force set-points between 0.1-1 nN, with peak force set-points varied to ensure proper calibration on the standard 2.5 MPa PDMS sample. Scan areas were varied to ensure proper alignment of features between AFM images and fluorescent microscopy images; changing this parameter had no effect on stiffness values measured by the AFM. For AV leaflet sections, at least two scans were taken on adjacent areas of each section of leaflet, and multiple sections and animals were used where possible. For PDMS standards, scan parameters were 0.5 × 1 μm scan areas, 0.25 Hz scan rate, and peak force set-points between 10-100 pN. On each PDMS sample, three adjacent areas were scanned to determine PDMS modulus.
Porcine Leaflet Analysis
To compare our AFM micromechanical analysis technique to other methods, we quantified the stiffness of porcine AV leaflets, which have been extensively mechanically characterized by other techniques (11-15). An AV leaflet from the heart of a mature pig was obtained from a local abattoir and treated in the same manner as the mouse leaflets, except that 3 μL of Hoechst 33342 (Invitrogen, Eugene OR, USA) staining was substituted for DAPI and no CD31 stain was utilized. For AFM micromechanical analysis, a borosilicate particle tip with a nominal diameter of 5 μm and a nominal spring constant of 0.06 N/m was used. The tip was calibrated in the method described previously; scan parameters were the same. Extracellular matrix composition (ECM) of the sample was determined using Movat’s Pentachrome stain.
Statistics
Because the scan regions of the AV leaflets are not homogenous, AFM data is represented as modulus versus percent of scan area. The median of each scan distribution was taken as the representative modulus for that particular scan; variability within specimens was determined by averaging median values (each representing a single scan) for a given sample of AV leaflets. Average median values of all groups were compared with ANOVA and pair-wise multiple comparisons were done using the Holm-Sidak post-hoc testing method.
Results
PDMS Standard Bulk Modulus and AFM Modulus Measurements
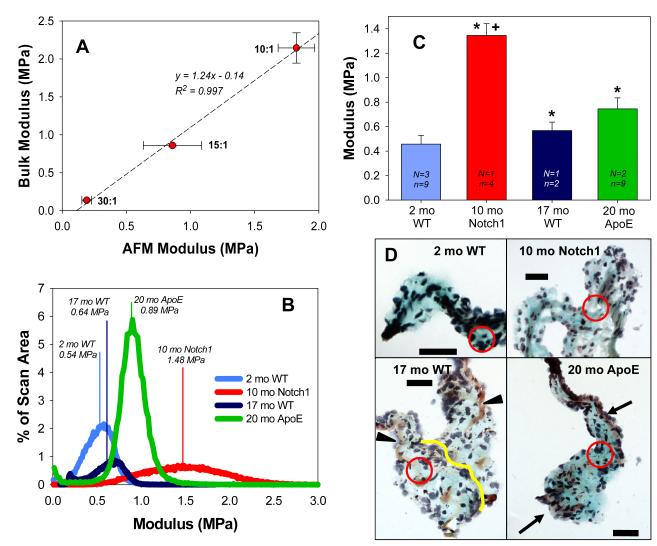
For PDMS, AFM modulus was highly correlated (R2 = 0.997) with bulk modulus (Fig. 2A), indicating that the PF-QNM scanning and calibration methods described above are appropriate for analyzing soft tissues such as AV leaflets, which have modulus values < 2 MPa. The use of PDMS as a standard and as discussed in this paper is not specific to the type of samples to be analyzed by AFM; it is used to verify that the AFM system accurately reports nanomechanical properties that are highly correlated with bulk modulus mechanical properties. Therefore, this demonstrates that once a tip is accurately calibrated, the software is capable of analyzing a wide variety of topographies, provided that the mechanical properties are similar to the calibration standard and force-displacement curves exhibit the same trend.
Figure 2.
A) Bulk modulus and AFM median modulus of PDMS gels show close agreement Data shown as mean ± SE; n = 3(AFM) or 4(bulk). (B) Modulus distributions of AV leaflet AFM scans from WT, Notch1, and ApoE animals. Drawn vertical lines represent median modulus values, which are aggregated and plotted in (C). N = number of animals and n = number of scanned regions on sections of tissue used for analysis. AV leaflets from 10 month old Notch1, 17 month old WT, and 20 month old ApoE animals were significantly stiffer (* = p<0.05) versus 2 month old WT animals. The AV leaflets from the 10 month old Notch1 animal were significantly stiffer than all other groups (+ = p<0.01). (D) Histological images of section adjacent to those subjected to AFM analysis. Arrowheads indicate regions of collagen (yellow); arrows indicate regions of elastin (purple/black) present in older animal sections. Line in 17mo WT added to show demarcation between the two leaflets. Black = nuclei; Purple = elastin; Blue = ground substance; Yellow = collagen; Red = Fibrous regions. Scale bar = 50 μm.
Mouse AV Leaflet Modulus
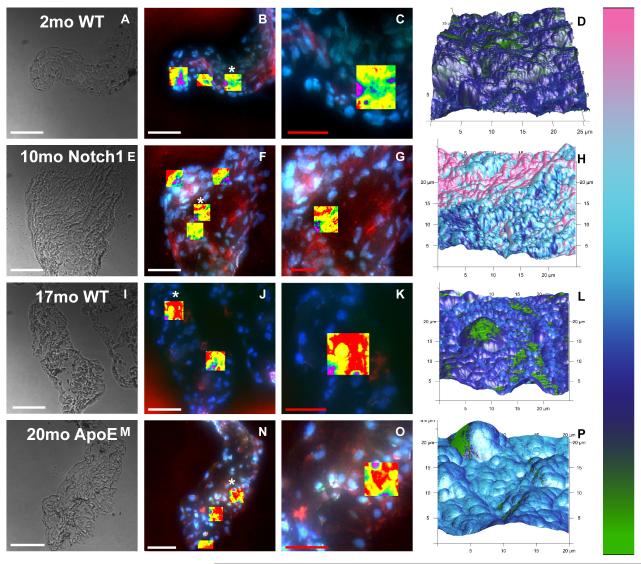
Mouse AV leaflets have a heterogeneous ECM with various proteins of different stiffness. Thus, it is not surprising that the modulus of the leaflet tissues have wide distributions (Fig. 2B). As the shape and skew of the distributions vary between scans, median modulus values were chosen to compare differences between groups (Fig. 2C). All leaflets were significantly stiffer compared to the 2 month old WT leaflets (Fig. 2C; * = p<0.05). The 10 month old Notch1 leaflet modulus was significantly greater than all other groups (Fig. 2C; + = p<0.01). This particular AFM system has a unique feature that allows for 3-channel fluorescence image guidance and the ability to create topographical maps with color-coated modulus values overlaid to compare or correlate regional morphologies and stiffness values (Fig. 3).
Figure 3.
Bright field and three channel fluorescent images (red = αSMA, green = CD31, and blue = nuclei) of leaflet sections analyzed by AFM. Rows correspond to sample group, in order of increasing age: 2 mo WT, 10 mo Notch1, 17 mo WT, and 20 mo ApoE. Bright field images at 40X: A,F,K,P. Fluorescent images showing all areas scanned by AFM, highlighted by overlay of height sensor data: B,F,J,N. Close-ups of areas marked by * in B,F,J,N, showing a scan region in more detail: C,G,K,O. Stiffness values overlaid on three-dimensional topographical maps corresponding to regions shown in third column: D,H,L,P. White scale bar = 50 μm; red scale bar =25 μm.
Mouse AV Leaflet Histology
As stated above, this AFM technique is particularly powerful as it allows for mechanical quantification of leaflets without requiring or destroying the entire leaflet. Specifically, this technique dovetails with standard histological processing and we therefore used Movat’s Pentachrome to analyze adjacent sections to those subjected to AFM scanning. The most common feature in all leaflets was an abundance of ground substance near the tip of the leaflet and surrounding the scan regions (Fig. 2D, circled regions approximate scan region on adjacent section). The older animals show more structural ECM components, such as areas of collagen or elastin (indicated by arrows). This can be explained by the changes in valve structure that occurs over the lifespan of the mouse (16). Note that there is not a well-defined tri-layered matrix as seen in large mammals and human leaflets; this has been previously observed by Hinton et al. (17).
Porcine AV Leaflet Modulus and Histology
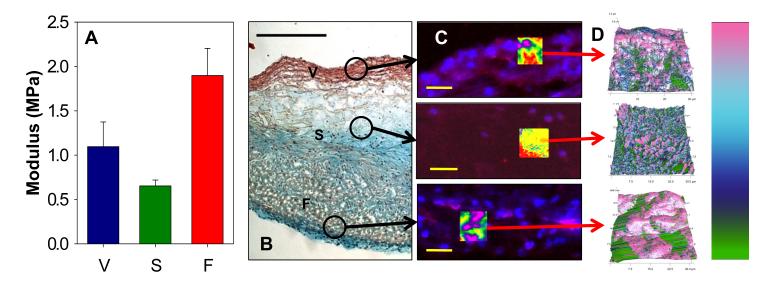
Results show that there are slight differences in stiffness across the thickness of the leaflet, corresponding to the spongiosa region being less stiff than both the fibrosa and ventricularis of the leaflet (Fig. 4A) although not significantly. Our histological slides illustrate the definitive tri-laminar structure of the porcine leaflets, which is similar to human valve leaflets (Fig. 4B). The stiffness values obtained in this study are in the same range as data collected from the mouse valves, reinforcing the value of using this AFM technique to perform quantitative biomechanical analyses.
Figure 4.
Micromechanical and histological analysis of a porcine AV leaflet (representative images shown) showing ECM influence on measured stiffness values. V = ventricularis, S = spongiosa, F = fibrosa. (A) Average median stiffness values of the three layers of the porcine AV leaflet, with the spongiosa the most compliant and the fibrosa the stiffest layer. (B) Histological image of porcine AV leaflet; circled areas indicated approximate regions scanned by AFM. Scale bar = 500μm. (C) Fluorescent images (red = αSMA, blue = Hoescht) of AV leaflet layer regions scanned by AFM. Areas highlighted are overlays of AFM height data, to indicate scan region. Scale bar = 30μm. (D) Call-outs show three-dimensional topographical map with stiffness values overlaid. Modulus scale bar: 0-2 MPa.
Discussion
AV Leaflet Stiffness Increases with Age and ApoE or Notch1 Mutation
As expected, the AFM technique was able to detect slight variations in leaflet stiffness. Specifically, aged mice had stiffer AV leaflets than the young WT animals (Fig. 2C), and the Notch1 animal had the stiffest AV leaflets, with a modulus more than 2-fold higher than the aged WT animal (p<0.01). The ApoE leaflets were slightly stiffer than the “age-matched” WT leaflets, although not significantly. The finding that ApoE leaflets are not significantly stiffer, even in this preliminary study with minimal samples, might indicate that the hemodynamic changes observed in these animals (2) may be caused by minor increases in AV leaflet stiffness. Finally, while the precise role of Notch1 in causing AV disease is unclear, the leaflets from the 10 month old Notch1 animal in this study were significantly stiffer than the much older WT or ApoE leaflets, indicating that the Notch1 mutation may significantly alter AV leaflet ECM stiffness, perhaps at earlier time points than the ApoE mutation. Interestingly, the results of WT tissue analysis are in the range of results from micropipette aspiration measurements of mouse AV cusps, which were roughly 300kPa for a wide age range of animals (6) and thus support the use of AFM as a technique to analyze the biomechanical properties of mouse heart valve leaflets.
Differences in AV Stiffness Is Not Revealed by Histology
Histology results indicated heterogeneous profiles across all of the leaflets imaged (Fig. 2D). Despite this variation, the thickest portions of the leaflets, those scanned by AFM, are composed primarily of ground substance (blue) with little amounts of structural proteins such as collagen (yellow) or elastin (purple/black). Although leaflets from older animals show more of these proteins than young WT animals, the histological images indicate similar areas were scanned in all samples (Fig. 2D). Further work is needed to determine specific causes of stiffness differences between samples, although this initial study shows promising results in the ability to characterize biomechanical properties of tissues.
Regional Variations in Stiffness of Porcine AV Leaflets
Porcine valve leaflets are more similar to human leaflets than those from mice in that they exhibit definitive tri-laminar structure. The spongiosa, rich in glycosaminoglycans, is the softest portion of the valve leaflet. The fibrosa and ventricularis, rich in collagen and elastin respectively, exhibit higher stiffness values than the spongiosa. The differences between the regions of the leaflet tissue highlight the importance of coupling histology with the AFM technique for measuring leaflet stiffness. Without the guidance of histology, the differences between measured stiffness values could not be attributed to physiological composition of the tissue. By using Movat’s Pentachrome to determine the boundaries between fibrosa, spongiosa, and ventricularis, the selection of appropriate regions for AFM analysis can be accomplished. Additionally, the values measured in this study are comparable to those determined via other methods such as micropipette aspiration and three-point bending (6, 14).
Limitations in Micromechanical Analysis of AV Leaflets
The micromechanical analysis of mouse AV leaflets with AFM presents limitations worth discussing. One limitation is identifying appropriate and comparable scan regions between leaflet samples, ensuring that differences seen between samples are significant and not an artifact of variation between scan locations. This is a result of the complex, heterogeneous nature of the heart valve leaflets. The free-edge of mouse heart valve leaflets is composed of a poorly defined bilaminar structure of fibrosa and spongiosa (16), providing comparable regions for analysis in this study. It is important to note that in larger mammals such as pigs and sheep, the tri-laminar structure of AV leaflets is more similar to human AV leaflets. In the case of the porcine AV leaflet sample, differences in ECM composition showed differences in stiffness values. Multiple scans per section, as well as multiple sections per animal are recommended to ensure correct characterization of leaflet mechanical properties. Additionally, histological analysis of adjacent tissue sections is critical to determining relevant ECM characteristics of scanned regions.
Finally, sample treatment prior to AFM scanning is an important factor to consider. After cryopreservation, heart valve leaflet tissues retain their ECM composition, although the elastin and collagen fibers may be deformed to some extent (18). As the tissue is frozen, the formation of ice crystals can disrupt the complex ECM network within the tissue (18-20). If such processes disrupt a large portion of the valve leaflet section, leaflet stiffness could be under-estimated by the AFM. Conversely, chemical fixation permeates cell membranes and crosslinks ECM proteins, increasing tissue stiffness. Other sectioning techniques, specifically paraffin embedding, utilize tissue dehydration and temperatures above 50°C, which would lead to protein denaturation and thus altered mechanical characteristics. Therefore, we selected the method of sample collection and processing that we believe has the minimal possible effect on relevant biomechanical properties of valve leaflets. Finally, as our technique corresponds to recent data of mice leaflets analyzed with micropipette aspiration, we feel confident that this processing strategy causes minimal change in mechanical properties.
Concluding Remarks
We believe that strategy shown here for determining the stiffness of mouse AV leaflets with AFM provides a novel tool for researchers who utilize genetically-altered animal models, and as the preparation described above employs standard cryosectioning techniques, this analysis can be added to research projects without a significant loss of tissues typically required for biomechanical analyses. Moreover, because these AVs were obtained from animals that exhibit diseased valve states similar to humans (1-3, 5), the observed differences in stiffness values between sample types describes, for the first time, the biomechanical properties of these tissues as they relate to pathological states. Although this study is small in scope, it shows quantification of valve leaflet stiffness via AFM is feasible as a novel technique for the biomechanical analysis of heart valve tissues as it relates to heart valve disease.
Acknowledgements
We thank Dr. Stacey Huppert for providing the Notch1 animal used in this study. We also thank Stephanie C. Preston for assistance in quantifying PDMS bulk modulus. Finally, we wish to thank Dan DeLaughter, Joseph Love, and Dr. Joey Barnett for their assistance with the histology. This work was supported by the following grants from the NIH: HL094707, RL1HL0952551, and U01HL100398.
References
- 1.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–4. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka K, Sata M, Fukuda D, Suematsu Y, Motomura N, Takamoto S, et al. Age-associated aortic stenosis in apolipoprotein E-deficient mice. J Am Coll Cardiol. 2005;46:134–41. doi: 10.1016/j.jacc.2005.03.058. [DOI] [PubMed] [Google Scholar]
- 3.Nigam V, Srivastava D. Notch1 represses osteogenic pathways in aortic valve cells. J Mol Cell Cardiol. 2009;47:828–34. doi: 10.1016/j.yjmcc.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 5.O’Brien KD, Reichenbach DD, Marcovina SM, Kuusisto J, Alpers CE, Otto CM. Apolipoproteins B, (a), and E accumulate in the morphologically early lesion of ‘degenerative’ valvular aortic stenosis. Arterioscler Thromb Vasc Biol. 1996;16:523–32. doi: 10.1161/01.atv.16.4.523. [DOI] [PubMed] [Google Scholar]
- 6.Krishnamurthy VK, Guilak F, Narmoneva DA, Hinton RB. Regional structure-function relationships in mouse aortic valve tissue. J Biomech. 2011;44:77–83. doi: 10.1016/j.jbiomech.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binnig G, Quate CF, Gerber C. Atomic Force Microscope. Phys Rev Lett. 1986;56:930–3. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 8.Rugar D, Hansma P. Atomic Force Microscopy. Phys Today. 1990;43:23–30. [Google Scholar]
- 9.Domke J, Parak WJ, George M, Gaub HE, Radmacher M. Mapping the mechanical pulse of single cardiomyocytes with the atomic force microscope. Eur Biophys J. 1999;28:179–86. doi: 10.1007/s002490050198. [DOI] [PubMed] [Google Scholar]
- 10.Jalili N, Laxminarayana K. A review of atomic force microscopy imaging systems: application to molecular metrology and biological sciences. Mechatronics. 2004;14:907–45. [Google Scholar]
- 11.Duncan AC, Boughner D, Vesely I. Dynamic glutaraldehyde fixation of a porcine aortic valve xenograft. I. Effect of fixation conditions on the final tissue viscoelastic properties. Biomaterials. 1996;17:1849–56. doi: 10.1016/0142-9612(96)00006-3. [DOI] [PubMed] [Google Scholar]
- 12.Vesely I, Lozon A. Natural preload of aortic valve leaflet components during glutaraldehyde fixation: effects on tissue mechanics. J Biomech. 1993;26:121–31. doi: 10.1016/0021-9290(93)90043-e. [DOI] [PubMed] [Google Scholar]
- 13.Liao J, Joyce EM, Sacks MS. Effects of decellularization on the mechanical and structural properties of the porcine aortic valve leaflet. Biomaterials. 2008;29:1065–74. doi: 10.1016/j.biomaterials.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merryman WD, Huang HY, Schoen FJ, Sacks MS. The effects of cellular contraction on aortic valve leaflet flexural stiffness. J Biomech. 2006;39:88–96. doi: 10.1016/j.jbiomech.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Rousseau EP, Sauren AA, van Hout MC, van Steenhoven AA. Elastic and viscoelastic material behaviour of fresh and glutaraldehyde-treated porcine aortic valve tissue. J Biomech. 1983;16:339–48. doi: 10.1016/0021-9290(83)90017-9. [DOI] [PubMed] [Google Scholar]
- 16.Hinton RB, Jr., Alfieri CM, Witt SA, Glascock BJ, Khoury PR, Benson DW, et al. Mouse heart valve structure and function: echocardiographic and morphometric analyses from the fetus through the aged adult. Am J Physiol Heart Circ Physiol. 2008;294:H2480–8. doi: 10.1152/ajpheart.91431.2007. [DOI] [PubMed] [Google Scholar]
- 17.Hinton RB, Jr., Lincoln J, Deutsch GH, Osinska H, Manning PB, Benson DW, et al. Extracellular matrix remodeling and organization in developing and diseased aortic valves. Circ Res. 2006;98:1431–8. doi: 10.1161/01.RES.0000224114.65109.4e. [DOI] [PubMed] [Google Scholar]
- 18.Schenke-Layland K, Madershahian N, Riemann I, Starcher B, Halbhuber KJ, Konig K, et al. Impact of cryopreservation on extracellular matrix structures of heart valve leaflets. Ann Thorac Surg. 2006;81:918–26. doi: 10.1016/j.athoracsur.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 19.Brockbank KG, Song YC. Morphological analyses of ice-free and frozen cryopreserved heart valve explants. J Heart Valve Dis. 2004;13:297–301. [PubMed] [Google Scholar]
- 20.Elder E, Chen Z, Ensley A, Nerem R, Brockbank K, Song Y. Enhanced tissue strength in cryopreserved, collagen-based blood vessel constructs. Transplant Proc. 2005;37:4625–9. doi: 10.1016/j.transproceed.2005.10.033. [DOI] [PubMed] [Google Scholar]